

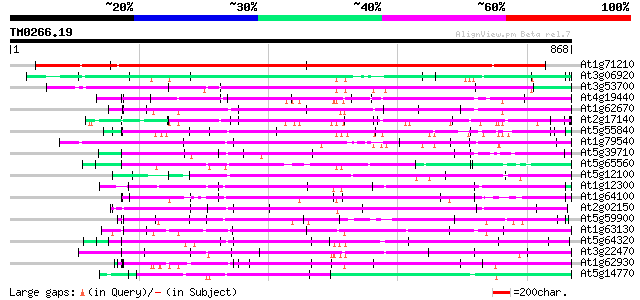
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0266.19
(868 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g71210 unknown protein 613 e-176
At3g06920 hypothetical protein 175 1e-43
At3g53700 putative protein 151 2e-36
At4g19440 unknown protein 149 9e-36
At1g62670 PPR-repeat protein 146 6e-35
At2g17140 hypothetical protein 142 6e-34
At5g55840 putative protein 142 8e-34
At1g79540 unknown protein 141 1e-33
At5g39710 putative protein 140 2e-33
At5g65560 putative protein 140 3e-33
At5g12100 unknown protein (At5g12100) 140 3e-33
At1g12300 hypothetical protein 139 5e-33
At1g64100 hypothetical protein 138 2e-32
At2g02150 hypothetical protein 137 3e-32
At5g59900 putative protein 135 8e-32
At1g63130 unknown protein 134 3e-31
At5g64320 putative protein 133 5e-31
At3g22470 hypothetical protein 132 1e-30
At1g62930 unknown protein 131 2e-30
At5g14770 putative protein 130 4e-30
>At1g71210 unknown protein
Length = 879
Score = 613 bits (1582), Expect = e-176
Identities = 351/807 (43%), Positives = 497/807 (61%), Gaps = 23/807 (2%)
Query: 41 SSFPPPSFAISKTD-VADTINTWFTTRHQSQDPLLI-RIYNIL---SSDDD---FSAALS 92
S+F PS +I+ D + WF R Q LI RI++IL S+D D F LS
Sbjct: 45 STFTKPSSSIAPGDFLVREWKDWFKHRDVKQSHQLIDRIFDILRAPSNDGDDRAFYLHLS 104
Query: 93 ALSLPLSETFVLRVLRHGGDDGDILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLR 152
L L L+E FVL VL H DIL CLKFFDWA RQP F+HTR TF AIF+IL A+L
Sbjct: 105 NLRLRLTEKFVLDVLSH--TRYDILCCLKFFDWAARQPGFHHTRATFHAIFKILRGAKLV 162
Query: 153 PLVFDFLR---DFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGY 209
L+ DFL F SC H R D LVVGYA+AG+ DIAL G MRF+GLDLD FGY
Sbjct: 163 TLMIDFLDRSVGFESCR--HSLRLCDALVVGYAVAGRTDIALQHFGNMRFRGLDLDSFGY 220
Query: 210 HILLNSLAENNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGS 269
H+LLN+L E C+++FDVI +QI +RG+ +T+ I++K CKQG+L+EAE +L L+ +
Sbjct: 221 HVLLNALVEEKCFDSFDVIFDQISVRGFVCAVTHSILVKKFCKQGKLDEAEDYLRALLPN 280
Query: 270 GKELHRSELSFLIGVLCESNRFERAVELVSEFGT--SLPLENAYGVWIRGLVQGGRLDEA 327
S L L+ LC +F+ A +L+ E ++ ++ AY +WIR L++ G L+
Sbjct: 281 DPAGCGSGLGILVDALCSKRKFQEATKLLDEIKLVGTVNMDRAYNIWIRALIKAGFLNNP 340
Query: 328 LEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLC 387
+F ++ EG RYN ++ +LL+EN L VY++L +M + PN TMNA LC
Sbjct: 341 ADFLQKISPLEGCELEVFRYNSMVFQLLKENNLDGVYDILTEMMVRGVSPNKKTMNAALC 400
Query: 388 FFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFP 447
FFCK G VD ALEL+ SRS+ G +P M+Y YLI TLC + ++AY VL+ + G+F
Sbjct: 401 FFCKAGFVDEALELYRSRSEIGFAPTAMSYNYLIHTLCANESVEQAYDVLKGAIDRGHFL 460
Query: 448 DRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMR 507
+TFSTL NALC + K D +L+ A ER +P + +SALC G+VED ++
Sbjct: 461 GGKTFSTLTNALCWKGKPDMARELVIAAAERDLLPKRIAGCKIISALCDVGKVEDALMIN 520
Query: 508 GDLDKV---TARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHM 564
+K T+ + +I G I RGDIAA+L++ M+EKGY RS YR+V+ C+ M
Sbjct: 521 ELFNKSGVDTSFKMFTSLIYGSITLMRGDIAAKLIIRMQEKGYTPTRSLYRNVIQCVCEM 580
Query: 565 DNPRTRFFNLL--EMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASS 622
++ FF L ++ + +N FI+GA A KP LAR V+++M R+GI +S
Sbjct: 581 ESGEKNFFTTLLKFQLSLWEHKVQAYNLFIEGAGFAGKPKLARLVYDMMDRDGITPTVAS 640
Query: 623 QILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLK 682
IL+++SY ++ +I+DAL FF+D+R Q +LY MIVGLCK++K D A+ EM
Sbjct: 641 NILMLQSYLKNEKIADALHFFHDLREQGKTKKRLYQVMIVGLCKANKLDDAMHFLEEMKG 700
Query: 683 VGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYH 742
GL PSIECYEV +QKLC+ ++Y EAV LVN + K+GRR+T+F+GNVLL ++M S VY
Sbjct: 701 EGLQPSIECYEVNIQKLCNEEKYDEAVGLVNEFRKSGRRITAFIGNVLLHNAMKSKGVYE 760
Query: 743 SCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRK 802
+ + R E + + L +IG FSG + + ++ L+E+I KC+P+D+YTYN+L+R
Sbjct: 761 AWTRM-RNIEDKIPEMKSLGELIGLFSGRIDMEVELKRLDEVIEKCYPLDMYTYNMLLRM 819
Query: 803 LTHHDMDKACELFDRMCQRGLEPNRWT 829
+ + + A E+ +R+ +RG PN T
Sbjct: 820 IVMNQAEDAYEMVERIARRGYVPNERT 846
Score = 49.3 bits (116), Expect = 9e-06
Identities = 69/309 (22%), Positives = 121/309 (38%), Gaps = 49/309 (15%)
Query: 156 FDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNS 215
F L F+ + H+ + ++ + G AGKP +A R+ + +D DG +
Sbjct: 588 FTTLLKFQLSLWEHKVQAYNLFIEGAGFAGKPKLA-----RLVYDMMDRDGITPTVA--- 639
Query: 216 LAENNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHR 275
+N+++++ K ++ +A + L GK R
Sbjct: 640 --------------------------SNILMLQSYLKNEKIADALHFFHDLREQGKTKKR 673
Query: 276 SELSFLIGVLCESNRFERAVELVSEF-GTSL-PLENAYGVWIRGLVQGGRLDEALEFFRQ 333
++G LC++N+ + A+ + E G L P Y V I+ L + DEA+ +
Sbjct: 674 LYQVMIVG-LCKANKLDDAMHFLEEMKGEGLQPSIECYEVNIQKLCNEEKYDEAVGLVNE 732
Query: 334 KRDSEGFVPCKVRYNILIGRLLRENRLKD--VYELLMDM-NETCIPPNMVTMNAVLCFFC 390
R S R IG +L N +K VYE M N P M ++ ++ F
Sbjct: 733 FRKSGR------RITAFIGNVLLHNAMKSKGVYEAWTRMRNIEDKIPEMKSLGELIGLFS 786
Query: 391 KLGMVDVALELFNSRSQFGLSPNYMAYKY-LILTLCWDGCPKEAYRVLRSSSGTGYFPDR 449
G +D+ +EL Y Y ++L + ++AY ++ + GY P+
Sbjct: 787 --GRIDMEVELKRLDEVIEKCYPLDMYTYNMLLRMIVMNQAEDAYEMVERIARRGYVPNE 844
Query: 450 RTFSTLANA 458
RT L A
Sbjct: 845 RTDMILERA 853
Score = 41.2 bits (95), Expect = 0.003
Identities = 59/297 (19%), Positives = 111/297 (36%), Gaps = 43/297 (14%)
Query: 576 EMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRR 635
E H CD + + G A + D+A + F M+ G+ ++ +++ + +
Sbjct: 176 ESCRHSLRLCD---ALVVGYAVAGRTDIALQHFGNMRFRGLDLDSFGYHVLLNALVEEKC 232
Query: 636 ISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYE-- 693
F+ I + V ++ ++ CK K D A + +L N C
Sbjct: 233 FDSFDVIFDQISVRGFVCAVTHSILVKKFCKQGKLDEAEDYLRALLP---NDPAGCGSGL 289
Query: 694 -VLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKE 752
+LV LCS +++ EA L++ + G N+ + ++I ++ D ++
Sbjct: 290 GILVDALCSKRKFQEATKLLDEIKLVGTVNMDRAYNIWI-RALIKAGFLNNPADFLQK-- 346
Query: 753 GEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYN-LLMRKLTHHDMDKA 811
I GC ELE ++ YN ++ + L +++D
Sbjct: 347 ------------ISPLEGC--------ELE----------VFRYNSMVFQLLKENNLDGV 376
Query: 812 CELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
++ M RG+ PN+ T F G DEA + GF P + N +
Sbjct: 377 YDILTEMMVRGVSPNKKTMNAALCFFCKAGFVDEALELYRSRSEIGFAPTAMSYNYL 433
>At3g06920 hypothetical protein
Length = 871
Score = 175 bits (443), Expect = 1e-43
Identities = 193/847 (22%), Positives = 341/847 (39%), Gaps = 105/847 (12%)
Query: 26 TLFSNNCSSLSPSSSSSFPPPSFAISKTDVADTINTWFTTRHQSQDPLLIRIYNILSSDD 85
T F N SSLS + + P +F ++ V D I N+L +
Sbjct: 7 TKFCKNLSSLSDNGENHEKPYTFEGNRQTVND-------------------ICNVLETGP 47
Query: 86 DFSAA---LSALSLPLSETFVLRVLRHGGDDGDILSCLKFFDWAGRQPRFYHTRTTFVAI 142
+A LSALS FV+ VLR D + +++F W R+ H ++ ++
Sbjct: 48 WGPSAENTLSALSFKPQPEFVIGVLRRLKD---VNRAIEYFRWYERRTELPHCPESYNSL 104
Query: 143 FRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGL 202
+++ R + L + F +V+G A K ++ MR
Sbjct: 105 LLVMARCRNFDALDQILGEMSVAGFGPSVNTCIEMVLGCVKANKLREGYDVVQMMRKFKF 164
Query: 203 DLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYES--HMTNVIVIKHLCKQGRLEEAE 260
Y L+ + + N + + Q+ GYE H+ + I+ K+GR++ A
Sbjct: 165 RPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELGYEPTVHLFTTL-IRGFAKEGRVDSAL 223
Query: 261 AHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGL 318
+ L+ + S + + I + + + A + E + P E Y I L
Sbjct: 224 SLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGVL 283
Query: 319 VQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPN 378
+ RLDEA+E F + VPC YN +I + + Y LL P+
Sbjct: 284 CKANRLDEAVEMFEHLEKNRR-VPCTYAYNTMIMGYGSAGKFDEAYSLLERQRAKGSIPS 342
Query: 379 MVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLR 438
++ N +L K+G VD AL++F + +PN Y LI LC G A+ +
Sbjct: 343 VIAYNCILTCLRKMGKVDEALKVFEEMKK-DAAPNLSTYNILIDMLCRAGKLDTAFELRD 401
Query: 439 SSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAG 498
S G FP+ RT + + + LC+ K+DE + + + P+ T+ + L + G
Sbjct: 402 SMQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMDYKVCTPDEITFCSLIDGLGKVG 461
Query: 499 RVEDGYLMRG---DLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYR 555
RV+D Y + D D T Y +I F R KE G+++ Y+
Sbjct: 462 RVDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHGR-----------KEDGHKI----YK 506
Query: 556 HVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNG 615
+++ + P + N+++D A +P+ R +FE ++
Sbjct: 507 DMIN-------------------QNCSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKARR 547
Query: 616 IMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIAL 674
+ +A S +++ ++ ++ F ++ Q V+ T+ YN +I G CK K + A
Sbjct: 548 FVPDARSYSILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAY 607
Query: 675 ELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHS 734
+L EM G P++ Y ++ L + R EA L +E+A + NV+++ S
Sbjct: 608 QLLEEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAYML---FEEAKSKRIEL--NVVIYSS 662
Query: 735 MISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIY 794
+I F R+ + LEEL+ K ++Y
Sbjct: 663 LID-----------------------------GFGKVGRIDEAYLILEELMQKGLTPNLY 693
Query: 795 TYNLLMRKLTH-HDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEM 853
T+N L+ L ++++A F M + PN+ TYG++ +G + ++A + EM
Sbjct: 694 TWNSLLDALVKAEEINEALVCFQSMKELKCTPNQVTYGILINGLCKVRKFNKAFVFWQEM 753
Query: 854 LKKGFNP 860
K+G P
Sbjct: 754 QKQGMKP 760
Score = 151 bits (382), Expect = 1e-36
Identities = 162/745 (21%), Positives = 299/745 (39%), Gaps = 91/745 (12%)
Query: 107 LRHGGDDGDILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPLVFDFLRDFRSCS 166
LR G D ++ KF +P F TT + F ++ + + +F +++
Sbjct: 149 LREGYDVVQMMRKFKF------RPAF-SAYTTLIGAFSAVNHSDMMLTLFQQMQEL---G 198
Query: 167 FPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFD 226
+ TL+ G+A G+ D AL LL M+ LD D Y++ ++S
Sbjct: 199 YEPTVHLFTTLIRGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFG--------- 249
Query: 227 VIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLC 286
K G+++ A + + +G + + +IGVLC
Sbjct: 250 -------------------------KVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGVLC 284
Query: 287 ESNRFERAVELVS--EFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCK 344
++NR + AVE+ E +P AY I G G+ DEA ++R ++G +P
Sbjct: 285 KANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMGYGSAGKFDEAYSLLERQR-AKGSIPSV 343
Query: 345 VRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNS 404
+ YN ++ L + ++ + ++ +M + PN+ T N ++ C+ G +D A EL +S
Sbjct: 344 IAYNCILTCLRKMGKVDEALKVFEEMKKDA-APNLSTYNILIDMLCRAGKLDTAFELRDS 402
Query: 405 RSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECK 464
+ GL PN ++ LC EA + PD TF +L + L + +
Sbjct: 403 MQKAGLFPNVRTVNIMVDRLCKSQKLDEACAMFEEMDYKVCTPDEITFCSLIDGLGKVGR 462
Query: 465 IDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGY-------------------- 504
+D+ + + + L+ NS Y+ + GR EDG+
Sbjct: 463 VDDAYKVYEKMLDSDCRTNSIVYTSLIKNFFNHGRKEDGHKIYKDMINQNCSPDLQLLNT 522
Query: 505 -----LMRGDLDKVTARF-------------SYAKMIMGFIKSNRGDIAARLLVEMKEKG 546
G+ +K A F SY+ +I G IK+ + L MKE+G
Sbjct: 523 YMDCMFKAGEPEKGRAMFEEIKARRFVPDARSYSILIHGLIKAGFANETYELFYSMKEQG 582
Query: 547 YELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAR 605
L +Y V+ + L EM T G +P + S IDG ++ D A
Sbjct: 583 CVLDTRAYNIVIDGFCKCGKVNKAYQLLEEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAY 642
Query: 606 EVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGL 664
+FE + I N ++ + + RI +A ++ + + + +N ++ L
Sbjct: 643 MLFEEAKSKRIELNVVIYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNLYTWNSLLDAL 702
Query: 665 CKSDKADIALELCFEMLK-VGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLT 723
K+++ + AL +CF+ +K + P+ Y +L+ LC ++++ +A +K G + +
Sbjct: 703 VKAEEINEAL-VCFQSMKELKCTPNQVTYGILINGLCKVRKFNKAFVFWQEMQKQGMKPS 761
Query: 724 SFLGNVLLFHSMISPEVYHSCVDLRREK-EGEFLDSSMLTLIIGAFSGCLRVSYSIQELE 782
+ ++ + + + R K G DS+ +I S R + E
Sbjct: 762 TISYTTMISGLAKAGNIAEAGALFDRFKANGGVPDSACYNAMIEGLSNGNRAMDAFSLFE 821
Query: 783 ELIAKCFPVDIYTYNLLMRKLTHHD 807
E + P+ T +L+ L +D
Sbjct: 822 ETRRRGLPIHNKTCVVLLDTLHKND 846
Score = 133 bits (335), Expect = 4e-31
Identities = 142/626 (22%), Positives = 246/626 (38%), Gaps = 73/626 (11%)
Query: 281 LIGVLCESNRFERAVELVSEFGTSL--PLENAYGVWIRGLVQGGRLDEALEFFRQKRDSE 338
L+ V+ F+ +++ E + P N + G V+ +L E + + R +
Sbjct: 104 LLLVMARCRNFDALDQILGEMSVAGFGPSVNTCIEMVLGCVKANKLREGYDVVQMMRKFK 163
Query: 339 GFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVA 398
F P Y LIG N + L M E P + ++ F K G VD A
Sbjct: 164 -FRPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELGYEPTVHLFTTLIRGFAKEGRVDSA 222
Query: 399 LELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANA 458
L L + L + + Y I + G A++ G PD T++++
Sbjct: 223 LSLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGV 282
Query: 459 LCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTARFS 518
LC+ ++DE ++ + + R +P TY+
Sbjct: 283 LCKANRLDEAVEMFEHLEKNRRVP--CTYA------------------------------ 310
Query: 519 YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMM 578
Y MIMG+ + + D A LL + KG +Y +L CL M EM
Sbjct: 311 YNTMIMGYGSAGKFDEAYSLLERQRAKGSIPSVIAYNCILTCLRKMGKVDEALKVFEEMK 370
Query: 579 THGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISD 638
P+ +N ID A K D A E+ + MQ+ G+ N + +++ +S+++ +
Sbjct: 371 KDAAPNLSTYNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKSQKLDE 430
Query: 639 ALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQ 697
A F ++ ++V ++ + +I GL K + D A ++ +ML + Y L++
Sbjct: 431 ACAMFEEMDYKVCTPDEITFCSLIDGLGKVGRVDDAYKVYEKMLDSDCRTNSIVYTSLIK 490
Query: 698 KLCSLKR-------YYEAVN--------LVNVYE-------------------KAGRRLT 723
+ R Y + +N L+N Y KA R +
Sbjct: 491 NFFNHGRKEDGHKIYKDMINQNCSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKARRFVP 550
Query: 724 SFLGNVLLFHSMISPEVYHSCVDL--RREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQEL 781
+L H +I + +L +++G LD+ ++I F C +V+ + Q L
Sbjct: 551 DARSYSILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNKAYQLL 610
Query: 782 EELIAKCFPVDIYTYNLLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNH 840
EE+ K F + TY ++ L D +D+A LF+ + +E N Y + GF
Sbjct: 611 EEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRIELNVVIYSSLIDGFGKV 670
Query: 841 GRKDEAKRWVHEMLKKGFNPPENTRN 866
GR DEA + E+++KG P T N
Sbjct: 671 GRIDEAYLILEELMQKGLTPNLYTWN 696
Score = 120 bits (300), Expect = 4e-27
Identities = 139/668 (20%), Positives = 255/668 (37%), Gaps = 80/668 (11%)
Query: 280 FLIGVLCESNRFERAVELVS--EFGTSLP-LENAYGVWIRGLVQGGRLDEALEFFRQKRD 336
F+IGVL RA+E E T LP +Y + + + D AL+ +
Sbjct: 67 FVIGVLRRLKDVNRAIEYFRWYERRTELPHCPESYNSLLLVMARCRNFD-ALDQILGEMS 125
Query: 337 SEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVD 396
GF P ++ ++ N+L++ Y+++ M + P ++ F + D
Sbjct: 126 VAGFGPSVNTCIEMVLGCVKANKLREGYDVVQMMRKFKFRPAFSAYTTLIGAFSAVNHSD 185
Query: 397 VALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLA 456
+ L LF + G P + LI +G A +L + D ++
Sbjct: 186 MMLTLFQQMQELGYEPTVHLFTTLIRGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCI 245
Query: 457 NALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---V 513
++ + K+D W P+ TY+ + LC+A R+++ M L+K V
Sbjct: 246 DSFGKVGKVDMAWKFFHEIEANGLKPDEVTYTSMIGVLCKANRLDEAVEMFEHLEKNRRV 305
Query: 514 TARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFN 573
++Y MIMG+ + + D A LL + KG +Y +L CL M
Sbjct: 306 PCTYAYNTMIMGYGSAGKFDEAYSLLERQRAKGSIPSVIAYNCILTCLRKMGKVDEALKV 365
Query: 574 LLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRS 633
EM P+ +N ID A K D A E+ + MQ+ G+ N + +++ +S
Sbjct: 366 FEEMKKDAAPNLSTYNILIDMLCRAGKLDTAFELRDSMQKAGLFPNVRTVNIMVDRLCKS 425
Query: 634 RRISDALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECY 692
+++ +A F ++ ++V ++ + +I GL K + D A ++ +ML + Y
Sbjct: 426 QKLDEACAMFEEMDYKVCTPDEITFCSLIDGLGKVGRVDDAYKVYEKMLDSDCRTNSIVY 485
Query: 693 EVLVQKLCSLKR-------YYEAVN--------LVNVYE-------------------KA 718
L++ + R Y + +N L+N Y KA
Sbjct: 486 TSLIKNFFNHGRKEDGHKIYKDMINQNCSPDLQLLNTYMDCMFKAGEPEKGRAMFEEIKA 545
Query: 719 GRRLTSFLGNVLLFHSMISPEVYHSCVDL--RREKEGEFLDSSMLTLIIGAFSGCLRVSY 776
R + +L H +I + +L +++G LD+ ++I F C +V+
Sbjct: 546 RRFVPDARSYSILIHGLIKAGFANETYELFYSMKEQGCVLDTRAYNIVIDGFCKCGKVNK 605
Query: 777 SIQELEELIAKCFPVDIYTYNLLMRKLTHHD----------------------------- 807
+ Q LEE+ K F + TY ++ L D
Sbjct: 606 AYQLLEEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRIELNVVIYSSLID 665
Query: 808 -------MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNP 860
+D+A + + + Q+GL PN +T+ + +EA M + P
Sbjct: 666 GFGKVGRIDEAYLILEELMQKGLTPNLYTWNSLLDALVKAEEINEALVCFQSMKELKCTP 725
Query: 861 PENTRNVI 868
+ T ++
Sbjct: 726 NQVTYGIL 733
Score = 89.0 bits (219), Expect = 1e-17
Identities = 96/467 (20%), Positives = 180/467 (37%), Gaps = 55/467 (11%)
Query: 186 KPDIALHLLGRMRFQGLDLDGFGYHILLNSLAE----NNCYNAFDVIANQICMRGYESHM 241
K D A + M ++ D + L++ L + ++ Y ++ + + C
Sbjct: 427 KLDEACAMFEEMDYKVCTPDEITFCSLIDGLGKVGRVDDAYKVYEKMLDSDCR------- 479
Query: 242 TNVIV----IKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVEL 297
TN IV IK+ GR E+ ++ L+ + + ++ E+ +
Sbjct: 480 TNSIVYTSLIKNFFNHGRKEDGHKIYKDMINQNCSPDLQLLNTYMDCMFKAGEPEKGRAM 539
Query: 298 VSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLL 355
E +P +Y + I GL++ G +E E F ++ +G V YNI+I
Sbjct: 540 FEEIKARRFVPDARSYSILIHGLIKAGFANETYELFYSMKE-QGCVLDTRAYNIVIDGFC 598
Query: 356 RENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYM 415
+ ++ Y+LL +M P +VT +V+ K+ +D A LF + N +
Sbjct: 599 KCGKVNKAYQLLEEMKTKGFEPTVVTYGSVIDGLAKIDRLDEAYMLFEEAKSKRIELNVV 658
Query: 416 AYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFA 475
Y LI G EAY +L G P+ T+++L +AL + +I+E
Sbjct: 659 IYSSLIDGFGKVGRIDEAYLILEELMQKGLTPNLYTWNSLLDALVKAEEINEALVCFQSM 718
Query: 476 LERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMIMGFIKSNRG 532
E + PN TY ++ LC+ + ++ ++ K + SY MI G K+
Sbjct: 719 KELKCTPNQVTYGILINGLCKVRKFNKAFVFWQEMQKQGMKPSTISYTTMISGLAKAGNI 778
Query: 533 DIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFI 592
A L K G G P +N+ I
Sbjct: 779 AEAGALFDRFKANG----------------------------------GVPDSACYNAMI 804
Query: 593 DGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDA 639
+G + N+ A +FE +R G+ + + ++++ + ++ + A
Sbjct: 805 EGLSNGNRAMDAFSLFEETRRRGLPIHNKTCVVLLDTLHKNDCLEQA 851
Score = 45.1 bits (105), Expect = 2e-04
Identities = 45/212 (21%), Positives = 84/212 (39%), Gaps = 4/212 (1%)
Query: 660 MIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAG 719
M++G K++K ++ M K P+ Y L+ ++ + L ++ G
Sbjct: 139 MVLGCVKANKLREGYDVVQMMRKFKFRPAFSAYTTLIGAFSAVNHSDMMLTLFQQMQELG 198
Query: 720 RRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSML--TLIIGAFSGCLRVSYS 777
T L L+ S + L E + LD+ ++ + I +F +V +
Sbjct: 199 YEPTVHLFTTLI-RGFAKEGRVDSALSLLDEMKSSSLDADIVLYNVCIDSFGKVGKVDMA 257
Query: 778 IQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHG 836
+ E+ A D TY ++ L + +D+A E+F+ + + P + Y M G
Sbjct: 258 WKFFHEIEANGLKPDEVTYTSMIGVLCKANRLDEAVEMFEHLEKNRRVPCTYAYNTMIMG 317
Query: 837 FSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
+ + G+ DEA + KG P N I
Sbjct: 318 YGSAGKFDEAYSLLERQRAKGSIPSVIAYNCI 349
>At3g53700 putative protein
Length = 754
Score = 151 bits (381), Expect = 2e-36
Identities = 159/710 (22%), Positives = 291/710 (40%), Gaps = 56/710 (7%)
Query: 58 TINTWFTTRHQSQDPLLIRIYNILSSDDDFSAALSALSLPLSETFVLRVLRHGGDDGDIL 117
+I+ T H S L + +S SAALS+ + L ++ LR DD
Sbjct: 13 SISQAVTLTHHSFSLNLTPPSSTISFASPHSAALSSTDVKLLDS-----LRSQPDDS--- 64
Query: 118 SCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTL 177
+ L+ F+ A ++P F + I L + + L D +S L
Sbjct: 65 AALRLFNLASKKPNFSPEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLIL 124
Query: 178 VVGYAIAGKPDIALHLLGRMRFQ-GLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRG 236
+ YA D L ++ M + GL D Y+ +LN L + N ++ ++ + G
Sbjct: 125 IESYAQFELQDEILSVVDWMIDEFGLKPDTHFYNRMLNLLVDGNSLKLVEISHAKMSVWG 184
Query: 237 YESHMTNV-IVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAV 295
+ ++ ++IK LC+ +L A L + G + ++ E + A+
Sbjct: 185 IKPDVSTFNVLIKALCRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGAL 244
Query: 296 ---ELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIG 352
E + EFG S + V + G + GR+++AL F ++ + +GF P + +N L+
Sbjct: 245 RIREQMVEFGCSWS-NVSVNVIVHGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVN 303
Query: 353 RLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSP 412
L + +K E++ M + P++ T N+V+ CKLG V A+E+ + SP
Sbjct: 304 GLCKAGHVKHAIEIMDVMLQEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSP 363
Query: 413 NYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLL 472
N + Y LI TLC + +EA + R + G PD TF++L LC +L
Sbjct: 364 NTVTYNTLISTLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELF 423
Query: 473 DFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKS 529
+ + P+ TY+ + +LC G++++ M ++ + +Y +I GF K+
Sbjct: 424 EEMRSKGCEPDEFTYNMLIDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKA 483
Query: 530 NRGDIAARLLVEMKEKGYELKRSSYRHVLHCL---------------LHMDNPRTRFFNL 574
N+ A + EM+ G +Y ++ L + M+ + +
Sbjct: 484 NKTREAEEIFDEMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTY 543
Query: 575 LEMMTH---------------------GKPHCDIFNSFIDGAMHANKPDLAREVFELMQR 613
++TH +P + + I G A + ++A ++ +Q
Sbjct: 544 NSLLTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQM 603
Query: 614 NGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIV--GLCK-SDKA 670
GI + V++ FR R+ ++A+ F ++ Q + IV GLC
Sbjct: 604 KGINLTPHAYNPVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVSYRIVFRGLCNGGGPI 663
Query: 671 DIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGR 720
A++ E+L+ G P +L + L +L V LVN+ + R
Sbjct: 664 REAVDFLVELLEKGFVPEFSSLYMLAEGLLTLSMEETLVKLVNMVMQKAR 713
Score = 137 bits (344), Expect = 3e-32
Identities = 136/613 (22%), Positives = 247/613 (40%), Gaps = 50/613 (8%)
Query: 246 VIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVS----EF 301
++ L + G ++ + L + S E+ S LI + + + +V EF
Sbjct: 89 ILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYAQFELQDEILSVVDWMIDEF 148
Query: 302 GTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLK 361
G P + Y + LV G L + +E K G P +N+LI L R ++L+
Sbjct: 149 GLK-PDTHFYNRMLNLLVDGNSL-KLVEISHAKMSVWGIKPDVSTFNVLIKALCRAHQLR 206
Query: 362 DVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLI 421
+L DM + P+ T V+ + + G +D AL + +FG S + ++ ++
Sbjct: 207 PAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWSNVSVNVIV 266
Query: 422 LTLCWDGCPKEAYRVLRSSSGT-GYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRF 480
C +G ++A ++ S G+FPD+ TF+TL N LC+ + +++D L+ +
Sbjct: 267 HGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVMLQEGY 326
Query: 481 MPNSSTYSRFVSALCRAGRVEDGY-----LMRGDLDKVTARFS----------------- 518
P+ TY+ +S LC+ G V++ ++ D T ++
Sbjct: 327 DPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLISTLCKENQVEEATE 386
Query: 519 ----------------YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLL 562
+ +I G + +A L EM+ KG E +Y ++ L
Sbjct: 387 LARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFTYNMLIDSLC 446
Query: 563 HMDNPRTRFFNLLEMMTHGKPHCDI-FNSFIDGAMHANKPDLAREVFELMQRNGIMTNAS 621
L +M G I +N+ IDG ANK A E+F+ M+ +G+ N+
Sbjct: 447 SKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKANKTREAEEIFDEMEVHGVSRNSV 506
Query: 622 SQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEM 680
+ ++ +SRR+ DA + + + + K YN ++ C+ A ++ M
Sbjct: 507 TYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTYNSLLTHFCRGGDIKKAADIVQAM 566
Query: 681 LKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVL---LFHSMIS 737
G P I Y L+ LC R A L+ + G LT N + LF +
Sbjct: 567 TSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQMKGINLTPHAYNPVIQGLFRKRKT 626
Query: 738 PEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYN 797
E + ++ + E S + G +G + ++ L EL+ K F + +
Sbjct: 627 TEAINLFREMLEQNEAPPDAVSYRIVFRGLCNGGGPIREAVDFLVELLEKGFVPEFSSLY 686
Query: 798 LLMRKLTHHDMDK 810
+L L M++
Sbjct: 687 MLAEGLLTLSMEE 699
Score = 116 bits (291), Expect = 5e-26
Identities = 124/556 (22%), Positives = 223/556 (39%), Gaps = 19/556 (3%)
Query: 327 ALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVL 386
AL F F P Y ++ RL R D+ ++L DM + T ++
Sbjct: 66 ALRLFNLASKKPNFSPEPALYEEILLRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILI 125
Query: 387 CFFCKLGMVDVALELFNSR-SQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGT-G 444
+ + + D L + + +FGL P+ Y + L L DG + + + G
Sbjct: 126 ESYAQFELQDEILSVVDWMIDEFGLKPDTHFYNRM-LNLLVDGNSLKLVEISHAKMSVWG 184
Query: 445 YFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGY 504
PD TF+ L ALCR ++ +L+ +P+ T++ + G ++
Sbjct: 185 IKPDVSTFNVLIKALCRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGAL 244
Query: 505 LMRGDLDKVTARFSYAKM---IMGFIKSNRGDIAARLLVEMKEK-GYELKRSSYRHVLHC 560
+R + + +S + + GF K R + A + EM + G+ + ++ +++
Sbjct: 245 RIREQMVEFGCSWSNVSVNVIVHGFCKEGRVEDALNFIQEMSNQDGFFPDQYTFNTLVNG 304
Query: 561 LLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTN 619
L + + + M+ G P +NS I G + A EV + M N
Sbjct: 305 LCKAGHVKHAIEIMDVMLQEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPN 364
Query: 620 ASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCF 678
+ ++ + + ++ +A + + ++ +N +I GLC + +A+EL
Sbjct: 365 TVTYNTLISTLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFE 424
Query: 679 EMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLL--FHSMI 736
EM G P Y +L+ LCS + EA+N++ E +G + N L+ F
Sbjct: 425 EMRSKGCEPDEFTYNMLIDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKAN 484
Query: 737 SPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTY 796
D E G +S +I RV + Q ++++I + D YTY
Sbjct: 485 KTREAEEIFD-EMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTY 543
Query: 797 NLLMRKLTHH----DMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHE 852
N L LTH D+ KA ++ M G EP+ TYG + G GR + A + +
Sbjct: 544 NSL---LTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRS 600
Query: 853 MLKKGFNPPENTRNVI 868
+ KG N + N +
Sbjct: 601 IQMKGINLTPHAYNPV 616
>At4g19440 unknown protein
Length = 825
Score = 149 bits (375), Expect = 9e-36
Identities = 140/589 (23%), Positives = 254/589 (42%), Gaps = 16/589 (2%)
Query: 219 NNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSEL 278
+ CY A DV + S T I++ L + ++ +V G
Sbjct: 205 DGCYLALDVFPVLANKGMFPSKTTCNILLTSLVRANEFQKC-CEAFDVVCKGVSPDVYLF 263
Query: 279 SFLIGVLCESNRFERAVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQKRD 336
+ I C+ + E AV+L S+ + N + I GL GR DEA F++K
Sbjct: 264 TTAINAFCKGGKVEEAVKLFSKMEEAGVAPNVVTFNTVIDGLGMCGRYDEAF-MFKEKMV 322
Query: 337 SEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVD 396
G P + Y+IL+ L R R+ D Y +L +M + PPN++ N ++ F + G ++
Sbjct: 323 ERGMEPTLITYSILVKGLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLN 382
Query: 397 VALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLA 456
A+E+ + GLS Y LI C +G A R+L+ G+ ++ +F+++
Sbjct: 383 KAIEIKDLMVSKGLSLTSSTYNTLIKGYCKNGQADNAERLLKEMLSIGFNVNQGSFTSVI 442
Query: 457 NALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDG------YLMRGDL 510
LC D + L R P + +S LC+ G+ +L +G +
Sbjct: 443 CLLCSHLMFDSALRFVGEMLLRNMSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKGFV 502
Query: 511 DKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTR 570
V R S A ++ G ++ + D A R+ E+ +G + R SY ++
Sbjct: 503 --VDTRTSNA-LLHGLCEAGKLDEAFRIQKEILGRGCVMDRVSYNTLISGCCGKKKLDEA 559
Query: 571 FFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKS 629
F L EM+ G KP ++ I G + NK + A + ++ +RNG++ + + +++
Sbjct: 560 FMFLDEMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCKRNGMLPDVYTYSVMIDG 619
Query: 630 YFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPS 688
++ R + FF+++ + V +T +YN +I C+S + +ALEL +M G++P+
Sbjct: 620 CCKAERTEEGQEFFDEMMSKNVQPNTVVYNHLIRAYCRSGRLSMALELREDMKHKGISPN 679
Query: 689 IECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLL-FHSMISPEVYHSCVDL 747
Y L++ + + R EA L G F L+ + + V C+
Sbjct: 680 SATYTSLIKGMSIISRVEEAKLLFEEMRMEGLEPNVFHYTALIDGYGKLGQMVKVECLLR 739
Query: 748 RREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTY 796
+ + T++IG ++ V+ + + L E+ K D TY
Sbjct: 740 EMHSKNVHPNKITYTVMIGGYARDGNVTEASRLLNEMREKGIVPDSITY 788
Score = 143 bits (361), Expect = 4e-34
Identities = 137/552 (24%), Positives = 244/552 (43%), Gaps = 34/552 (6%)
Query: 337 SEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVD 396
++G P K NIL+ L+R N + E D+ + P++ + FCK G V+
Sbjct: 219 NKGMFPSKTTCNILLTSLVRANEFQKCCEAF-DVVCKGVSPDVYLFTTAINAFCKGGKVE 277
Query: 397 VALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLA 456
A++LF+ + G++PN + + +I L G EA+ G P T+S L
Sbjct: 278 EAVKLFSKMEEAGVAPNVVTFNTVIDGLGMCGRYDEAFMFKEKMVERGMEPTLITYSILV 337
Query: 457 NALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGR----VEDGYLMRGDLDK 512
L R +I + + +L ++ F PN Y+ + + AG +E LM
Sbjct: 338 KGLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIKDLMVSKGLS 397
Query: 513 VTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLH---MDNPRT 569
+T+ +Y +I G+ K+ + D A RLL EM G+ + + S+ V+ CLL M +
Sbjct: 398 LTSS-TYNTLIKGYCKNGQADNAERLLKEMLSIGFNVNQGSFTSVI-CLLCSHLMFDSAL 455
Query: 570 RFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKS 629
RF + ++ + P + + I G K A E++ G + + + ++
Sbjct: 456 RFVGEM-LLRNMSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKGFVVDTRTSNALLHG 514
Query: 630 YFRSRRISDALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEMLKVGLNPS 688
+ ++ +A R +I + V ++ YN +I G C K D A EM+K GL P
Sbjct: 515 LCEAGKLDEAFRIQKEILGRGCVMDRVSYNTLISGCCGKKKLDEAFMFLDEMVKRGLKPD 574
Query: 689 IECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLR 748
Y +L+ L ++ + EA+ + ++ G L +V + MI C
Sbjct: 575 NYTYSILICGLFNMNKVEEAIQFWDDCKRNG-----MLPDVYTYSVMID-----GCCKAE 624
Query: 749 REKEG-EFLDSSM----------LTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYN 797
R +EG EF D M +I A+ R+S +++ E++ K + TY
Sbjct: 625 RTEEGQEFFDEMMSKNVQPNTVVYNHLIRAYCRSGRLSMALELREDMKHKGISPNSATYT 684
Query: 798 LLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKK 856
L++ ++ +++A LF+ M GLEPN + Y + G+ G+ + + + EM K
Sbjct: 685 SLIKGMSIISRVEEAKLLFEEMRMEGLEPNVFHYTALIDGYGKLGQMVKVECLLREMHSK 744
Query: 857 GFNPPENTRNVI 868
+P + T V+
Sbjct: 745 NVHPNKITYTVM 756
Score = 99.4 bits (246), Expect = 8e-21
Identities = 104/480 (21%), Positives = 198/480 (40%), Gaps = 70/480 (14%)
Query: 135 TRTTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLL 194
T T+ + + L+ A+ + L++ FP ++ L+ + AG + A+ +
Sbjct: 329 TLITYSILVKGLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIK 388
Query: 195 GRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGY---ESHMTNVIVIKHLC 251
M +GL L Y+ L+ +N + + + ++ G+ + T+VI + LC
Sbjct: 389 DLMVSKGLSLTSSTYNTLIKGYCKNGQADNAERLLKEMLSIGFNVNQGSFTSVICL--LC 446
Query: 252 KQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEF---GTSLPLE 308
+ A + ++ L+ LI LC+ + +A+EL +F G +
Sbjct: 447 SHLMFDSALRFVGEMLLRNMSPGGGLLTTLISGLCKHGKHSKALELWFQFLNKGFVVDTR 506
Query: 309 NAYGVWIRGLVQGGRLDEALEFFRQKRD--SEGFVPCKVRYNILIGRLLRENRLKDVYEL 366
+ + + GL + G+LDEA FR +++ G V +V YN LI + +L + +
Sbjct: 507 TSNAL-LHGLCEAGKLDEA---FRIQKEILGRGCVMDRVSYNTLISGCCGKKKLDEAFMF 562
Query: 367 LMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCW 426
L +M + + P+ T + ++C + V+ A++ ++ + G+ P+ Y +I
Sbjct: 563 LDEMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCKRNGMLPDVYTYSVMI----- 617
Query: 427 DGCPKEAYRVLRSSSGTGYF---------PDRRTFSTLANALCRECKIDEMWDLLDFALE 477
DGC K R+ G +F P+ ++ L A CR ++ +L +
Sbjct: 618 DGCCKAE----RTEEGQEFFDEMMSKNVQPNTVVYNHLIRAYCRSGRLSMALELREDMKH 673
Query: 478 RRFMPNSSTYSRFVSALCRAGRVE-------------------------DGYLMRGDLDK 512
+ PNS+TY+ + + RVE DGY G + K
Sbjct: 674 KGISPNSATYTSLIKGMSIISRVEEAKLLFEEMRMEGLEPNVFHYTALIDGYGKLGQMVK 733
Query: 513 VTA-------------RFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH 559
V + +Y MI G+ + A+RLL EM+EKG +Y+ ++
Sbjct: 734 VECLLREMHSKNVHPNKITYTVMIGGYARDGNVTEASRLLNEMREKGIVPDSITYKEFIY 793
Score = 97.4 bits (241), Expect = 3e-20
Identities = 124/554 (22%), Positives = 223/554 (39%), Gaps = 58/554 (10%)
Query: 327 ALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVL 386
AL+FFR DS F Y +LIG LL N L +L+ + +P VL
Sbjct: 109 ALDFFRLASDSFSFSFSLRSYCLLIGLLLDANLLSAARVVLIRLINGNVP--------VL 160
Query: 387 CFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCW----DGCPKEAYRVLRSSSG 442
+ V +A + + F LI C DGC A V +
Sbjct: 161 PCGLRDSRVAIADAMASLSLCFDEEIRRKMSDLLIEVYCTQFKRDGCYL-ALDVFPVLAN 219
Query: 443 TGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVED 502
G FP + T + L +L R + + + D + P+ ++ ++A C+ G+VE+
Sbjct: 220 KGMFPSKTTCNILLTSLVRANEFQKCCEAFDVVC-KGVSPDVYLFTTAINAFCKGGKVEE 278
Query: 503 GYLMRGDLDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH 559
+ +++ ++ +I G R D A +M E+G E +Y ++
Sbjct: 279 AVKLFSKMEEAGVAPNVVTFNTVIDGLGMCGRYDEAFMFKEKMVERGMEPTLITYSILVK 338
Query: 560 CLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMT 618
L +F L EM G P+ ++N+ ID + A + A E+ +LM G+
Sbjct: 339 GLTRAKRIGDAYFVLKEMTKKGFPPNVIVYNNLIDSFIEAGSLNKAIEIKDLMVSKGLSL 398
Query: 619 NASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCF 678
+S+ YN +I G CK+ +AD A L
Sbjct: 399 TSST----------------------------------YNTLIKGYCKNGQADNAERLLK 424
Query: 679 EMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISP 738
EML +G N + + ++ LCS + A+ V E R ++ G + S +
Sbjct: 425 EMLSIGFNVNQGSFTSVICLLCSHLMFDSALRFVG--EMLLRNMSPGGGLLTTLISGLCK 482
Query: 739 EVYHS-CVDLRRE--KEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYT 795
HS ++L + +G +D+ ++ ++ + + +E++ + +D +
Sbjct: 483 HGKHSKALELWFQFLNKGFVVDTRTSNALLHGLCEAGKLDEAFRIQKEILGRGCVMDRVS 542
Query: 796 YNLLMRKLT-HHDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEML 854
YN L+ +D+A D M +RGL+P+ +TY ++ G N + +EA ++ +
Sbjct: 543 YNTLISGCCGKKKLDEAFMFLDEMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCK 602
Query: 855 KKGFNPPENTRNVI 868
+ G P T +V+
Sbjct: 603 RNGMLPDVYTYSVM 616
Score = 95.1 bits (235), Expect = 1e-19
Identities = 99/361 (27%), Positives = 149/361 (40%), Gaps = 21/361 (5%)
Query: 176 TLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMR 235
TL+ G GK AL L + +G +D + LL+ L E + I +I R
Sbjct: 475 TLISGLCKHGKHSKALELWFQFLNKGFVVDTRTSNALLHGLCEAGKLDEAFRIQKEILGR 534
Query: 236 G-YESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERA 294
G ++ +I C + +L+EA L+ +V G + S LI L N+ E A
Sbjct: 535 GCVMDRVSYNTLISGCCGKKKLDEAFMFLDEMVKRGLKPDNYTYSILICGLFNMNKVEEA 594
Query: 295 VELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIG 352
++ + + LP Y V I G + R +E EFF + S+ P V YN LI
Sbjct: 595 IQFWDDCKRNGMLPDVYTYSVMIDGCCKAERTEEGQEFFDEMM-SKNVQPNTVVYNHLIR 653
Query: 353 RLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSP 412
R RL EL DM I PN T +++ + V+ A LF GL P
Sbjct: 654 AYCRSGRLSMALELREDMKHKGISPNSATYTSLIKGMSIISRVEEAKLLFEEMRMEGLEP 713
Query: 413 NYMAYKYLILTLCWDGCPKEAYRV-----LRSSSGTGYFPDRRTFSTLANALCRECKIDE 467
N Y LI DG K V LR P++ T++ + R+ + E
Sbjct: 714 NVFHYTALI-----DGYGKLGQMVKVECLLREMHSKNVHPNKITYTVMIGGYARDGNVTE 768
Query: 468 MWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTARFSYAKMIMGFI 527
LL+ E+ +P+S TY F+ + G V + + K + +YA +I G+
Sbjct: 769 ASRLLNEMREKGIVPDSITYKEFIYGYLKQGGVLEAF-------KGSDEENYAAIIEGWN 821
Query: 528 K 528
K
Sbjct: 822 K 822
Score = 70.5 bits (171), Expect = 4e-12
Identities = 69/266 (25%), Positives = 110/266 (40%), Gaps = 6/266 (2%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
++TL+ G K D A L M +GL D + Y IL+ L N +
Sbjct: 543 YNTLISGCCGKKKLDEAFMFLDEMVKRGLKPDNYTYSILICGLFNMNKVEEAIQFWDDCK 602
Query: 234 MRGYESHM-TNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
G + T ++I CK R EE + + ++ + + + LI C S R
Sbjct: 603 RNGMLPDVYTYSVMIDGCCKAERTEEGQEFFDEMMSKNVQPNTVVYNHLIRAYCRSGRLS 662
Query: 293 RAVELVSEF---GTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNI 349
A+EL + G S P Y I+G+ R++EA F + R EG P Y
Sbjct: 663 MALELREDMKHKGIS-PNSATYTSLIKGMSIISRVEEAKLLFEEMR-MEGLEPNVFHYTA 720
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFG 409
LI + ++ V LL +M+ + PN +T ++ + + G V A L N + G
Sbjct: 721 LIDGYGKLGQMVKVECLLREMHSKNVHPNKITYTVMIGGYARDGNVTEASRLLNEMREKG 780
Query: 410 LSPNYMAYKYLILTLCWDGCPKEAYR 435
+ P+ + YK I G EA++
Sbjct: 781 IVPDSITYKEFIYGYLKQGGVLEAFK 806
>At1g62670 PPR-repeat protein
Length = 630
Score = 146 bits (368), Expect = 6e-35
Identities = 128/498 (25%), Positives = 218/498 (43%), Gaps = 17/498 (3%)
Query: 212 LLNSLAENNCYNAFDVIANQICMRGY-ESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSG 270
LL+++A+ N ++ + Q+ G +H T I+I C++ +L A A L ++ G
Sbjct: 87 LLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAVLGKMMKLG 146
Query: 271 KELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEAL 328
E + LS L+ C S R AV LV + + P + I GL + EA+
Sbjct: 147 YEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAM 206
Query: 329 EFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCF 388
+ ++G P V Y +++ L + + LL M + + P ++ N ++
Sbjct: 207 ALI-DRMVAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDG 265
Query: 389 FCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPD 448
CK +D AL LF G+ PN + Y LI LC G +A R+L PD
Sbjct: 266 LCKYKHMDDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINPD 325
Query: 449 RRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRG 508
TFS L +A +E K+ E L D ++R P+ TYS ++ C R+++ M
Sbjct: 326 VFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFE 385
Query: 509 DL-------DKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCL 561
+ D VT Y +I GF K R + + EM ++G +Y ++ L
Sbjct: 386 FMVSKHCFPDVVT----YNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGL 441
Query: 562 LHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNA 620
+ EM++ G P+ +N+ +DG K + A VFE +QR+ +
Sbjct: 442 FQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTI 501
Query: 621 SSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFE 679
+ ++++ ++ ++ D F ++ + V YN MI G C+ + A L E
Sbjct: 502 YTYNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKE 561
Query: 680 MLKVGLNPSIECYEVLVQ 697
M + G P+ CY L++
Sbjct: 562 MKEDGTLPNSGCYNTLIR 579
Score = 117 bits (293), Expect = 3e-26
Identities = 116/534 (21%), Positives = 225/534 (41%), Gaps = 33/534 (6%)
Query: 355 LRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNY 414
L E +L D L +M ++ P+++ + +L K+ DV + L G+ N+
Sbjct: 57 LSELKLDDAVALFGEMVKSRPFPSIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNH 116
Query: 415 MAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDF 474
Y LI C A VL GY P+ T S+L N C +I E L+D
Sbjct: 117 YTYSILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQ 176
Query: 475 ALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR------FSYAKMIMGFIK 528
+ PN+ T++ + L + + + +D++ A+ +Y ++ G K
Sbjct: 177 MFVTGYQPNTVTFNTLIHGLFLHNKASEAMAL---IDRMVAKGCQPDLVTYGVVVNGLCK 233
Query: 529 SNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLL---HMDNPRTRFFNLLEMMTHG-KPH 584
D+A LL +M++ E Y ++ L HMD+ F EM T G +P+
Sbjct: 234 RGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHMDDALNLF---KEMETKGIRPN 290
Query: 585 CDIFNSFIDGAMHANK-PDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFF 643
++S I + + D +R + ++++R I + + ++ ++ + ++ +A + +
Sbjct: 291 VVTYSSLISCLCNYGRWSDASRLLSDMIERK-INPDVFTFSALIDAFVKEGKLVEAEKLY 349
Query: 644 ND-IRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSL 702
++ ++ + S Y+ +I G C D+ D A ++ M+ P + Y L++ C
Sbjct: 350 DEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSKHCFPDVVTYNTLIKGFCKY 409
Query: 703 KRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSML- 761
KR E + + + G +GN + ++ +I D+ +E E + +
Sbjct: 410 KRVEEGMEVFREMSQRG-----LVGNTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPP 464
Query: 762 ------TLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDKACEL 814
TL+ G ++ ++ E L IYTYN+++ + ++ +L
Sbjct: 465 NIMTYNTLLDGLCKNG-KLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDL 523
Query: 815 FDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
F + +G++P+ Y M GF G K+EA EM + G P N +
Sbjct: 524 FCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTL 577
Score = 116 bits (290), Expect = 6e-26
Identities = 105/412 (25%), Positives = 180/412 (43%), Gaps = 12/412 (2%)
Query: 175 DTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYN-AFDVIANQIC 233
+TL+ G + K A+ L+ RM +G D Y +++N L + + AF+++
Sbjct: 190 NTLIHGLFLHNKASEAMALIDRMVAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQ 249
Query: 234 MRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFER 293
+ + +I LCK +++A + G + S LI LC R+
Sbjct: 250 GKLEPGVLIYNTIIDGLCKYKHMDDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSD 309
Query: 294 AVELVSEFGTSL--PLENAYGVWIRGLVQGGRLDEALEFFRQ--KRDSEGFVPCKVRYNI 349
A L+S+ P + I V+ G+L EA + + + KR + P V Y+
Sbjct: 310 ASRLLSDMIERKINPDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSID---PSIVTYSS 366
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFG 409
LI +RL + ++ M P++VT N ++ FCK V+ +E+F SQ G
Sbjct: 367 LINGFCMHDRLDEAKQMFEFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRG 426
Query: 410 LSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMW 469
L N + Y LI L G A + + G P+ T++TL + LC+ K+++
Sbjct: 427 LVGNTVTYNILIQGLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAM 486
Query: 470 DLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMIMGF 526
+ ++ + P TY+ + +C+AG+VEDG+ + +L + +Y MI GF
Sbjct: 487 VVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGF 546
Query: 527 IKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMM 578
+ + A L EMKE G Y ++ L D R L++ M
Sbjct: 547 CRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARLR-DGDREASAELIKEM 597
Score = 105 bits (262), Expect = 1e-22
Identities = 103/486 (21%), Positives = 205/486 (41%), Gaps = 8/486 (1%)
Query: 154 LVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILL 213
+V ++ PH + L+ + + +AL +LG+M G + + LL
Sbjct: 99 VVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLL 158
Query: 214 NSLAENNCYNAFDVIANQICMRGYESHMTNVIVIKH-LCKQGRLEEAEAHLNGLVGSGKE 272
N + + + +Q+ + GY+ + + H L + EA A ++ +V G +
Sbjct: 159 NGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAMALIDRMVAKGCQ 218
Query: 273 LHRSELSFLIGVLCESNRFERAVELVS--EFGTSLPLENAYGVWIRGLVQGGRLDEALEF 330
++ LC+ + A L++ E G P Y I GL + +D+AL
Sbjct: 219 PDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHMDDALNL 278
Query: 331 FRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFC 390
F++ +++G P V Y+ LI L R D LL DM E I P++ T +A++ F
Sbjct: 279 FKE-METKGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINPDVFTFSALIDAFV 337
Query: 391 KLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRR 450
K G + A +L++ + + P+ + Y LI C EA ++ FPD
Sbjct: 338 KEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSKHCFPDVV 397
Query: 451 TFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDL 510
T++TL C+ +++E ++ +R + N+ TY+ + L +AG + + ++
Sbjct: 398 TYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGLFQAGDCDMAQEIFKEM 457
Query: 511 --DKVTAR-FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNP 567
D V +Y ++ G K+ + + A + ++ E +Y ++ +
Sbjct: 458 VSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKV 517
Query: 568 RTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILV 626
+ + G KP +N+ I G + A +F+ M+ +G + N+ +
Sbjct: 518 EDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTL 577
Query: 627 MKSYFR 632
+++ R
Sbjct: 578 IRARLR 583
Score = 72.0 bits (175), Expect = 1e-12
Identities = 69/294 (23%), Positives = 125/294 (42%), Gaps = 20/294 (6%)
Query: 177 LVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENN----CYNAFDVIANQI 232
L+ + GK A L M + +D Y L+N ++ F+ + ++
Sbjct: 332 LIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSKH 391
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEA-----EAHLNGLVGSGKELHRSELSFLIGVLCE 287
C ++ T +IK CK R+EE E GLVG+ + LI L +
Sbjct: 392 CFPDVVTYNT---LIKGFCKYKRVEEGMEVFREMSQRGLVGN-----TVTYNILIQGLFQ 443
Query: 288 SNRFERAVELVSEFGTSLPLEN--AYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKV 345
+ + A E+ E + N Y + GL + G+L++A+ F + S+ P
Sbjct: 444 AGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSK-MEPTIY 502
Query: 346 RYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSR 405
YNI+I + + +++D ++L +++ + P++V N ++ FC+ G + A LF
Sbjct: 503 TYNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEM 562
Query: 406 SQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANAL 459
+ G PN Y LI DG + + +++ G+ D T + N L
Sbjct: 563 KEDGTLPNSGCYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGLVTNML 616
>At2g17140 hypothetical protein
Length = 903
Score = 142 bits (359), Expect = 6e-34
Identities = 153/639 (23%), Positives = 276/639 (42%), Gaps = 57/639 (8%)
Query: 247 IKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVL---CESNRFERAV---ELV-S 299
I + + ++ E L+ L+ S + +++LS L+ V+ +SN ++A +LV S
Sbjct: 45 IARILVRAKMHEEIQELHNLILSSS-IQKTKLSSLLSVVSIFAKSNHIDKAFPQFQLVRS 103
Query: 300 EFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENR 359
F + P Y + + ++ R+ E + + + G P +N+LI L +
Sbjct: 104 RFPENKPSVYLYNLLLESCIKERRV-EFVSWLYKDMVLCGIAPQTYTFNLLIRALCDSSC 162
Query: 360 LKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKY 419
+ EL +M E PN T ++ +CK G+ D LEL N+ FG+ PN + Y
Sbjct: 163 VDAARELFDEMPEKGCKPNEFTFGILVRGYCKAGLTDKGLELLNAMESFGVLPNKVIYNT 222
Query: 420 LILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKI-DEMWDLLDFALER 478
++ + C +G ++ +++ G PD TF++ +ALC+E K+ D D L+
Sbjct: 223 IVSSFCREGRNDDSEKMVEKMREEGLVPDIVTFNSRISALCKEGKVLDASRIFSDMELDE 282
Query: 479 RF---MPNSSTYSRFVSALCRAGRVEDGYLMRGDL---DKVTARFSYAKMIMGFIKSNRG 532
PNS TY+ + C+ G +ED + + D + + SY + G ++ +
Sbjct: 283 YLGLPRPNSITYNLMLKGFCKVGLLEDAKTLFESIRENDDLASLQSYNIWLQGLVRHGKF 342
Query: 533 DIAARLLVEMKEKGYELKRSSYRHVLH--CLLHMDNPRTRFFNLLEMMTHGK-PHCDIFN 589
A +L +M +KG SY ++ C L M + L M +G P +
Sbjct: 343 IEAETVLKQMTDKGIGPSIYSYNILMDGLCKLGMLSDAKTIVGL--MKRNGVCPDAVTYG 400
Query: 590 SFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ 649
+ G K D A+ + + M RN + NA + +++ S ++ RIS+A + +
Sbjct: 401 CLLHGYCSVGKVDAAKSLLQEMMRNNCLPNAYTCNILLHSLWKMGRISEAEELLRKMNEK 460
Query: 650 -VVVSTKLYNRMIVGLCKSDKADIALELCFEM-----------------------LKVGL 685
+ T N ++ GLC S + D A+E+ M ++
Sbjct: 461 GYGLDTVTCNIIVDGLCGSGELDKAIEIVKGMRVHGSAALGNLGNSYIGLVDDSLIENNC 520
Query: 686 NPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRL--TSFLGNVLLFHSMISPEVYHS 743
P + Y L+ LC R+ EA NL E G +L S N+ + H ++ S
Sbjct: 521 LPDLITYSTLLNGLCKAGRFAEAKNL--FAEMMGEKLQPDSVAYNIFIHHFCKQGKI-SS 577
Query: 744 CVDLRREKEGEFLDSSMLT---LIIGAFSGCLRVSYSIQEL-EELIAKCFPVDIYTYNLL 799
+ ++ E + S+ T LI+G G + I L +E+ K +I TYN
Sbjct: 578 AFRVLKDMEKKGCHKSLETYNSLILGL--GIKNQIFEIHGLMDEMKEKGISPNICTYNTA 635
Query: 800 MRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGF 837
++ L + ++ A L D M Q+ + PN +++ + F
Sbjct: 636 IQYLCEGEKVEDATNLLDEMMQKNIAPNVFSFKYLIEAF 674
Score = 120 bits (302), Expect = 3e-27
Identities = 129/535 (24%), Positives = 211/535 (39%), Gaps = 56/535 (10%)
Query: 342 PCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALEL 401
P YN+L+ ++E R++ V L DM I P T N ++ C VD A EL
Sbjct: 110 PSVYLYNLLLESCIKERRVEFVSWLYKDMVLCGIAPQTYTFNLLIRALCDSSCVDAAREL 169
Query: 402 FNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCR 461
F+ + G PN + L+ C G + +L + G P++ ++T+ ++ CR
Sbjct: 170 FDEMPEKGCKPNEFTFGILVRGYCKAGLTDKGLELLNAMESFGVLPNKVIYNTIVSSFCR 229
Query: 462 ECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGD--LDKVTA---- 515
E + D+ +++ E +P+ T++ +SALC+ G+V D + D LD+
Sbjct: 230 EGRNDDSEKMVEKMREEGLVPDIVTFNSRISALCKEGKVLDASRIFSDMELDEYLGLPRP 289
Query: 516 -RFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNL 574
+Y M+ GF K + A L ++E SY L L+ L
Sbjct: 290 NSITYNLMLKGFCKVGLLEDAKTLFESIRENDDLASLQSYNIWLQGLVRHGKFIEAETVL 349
Query: 575 LEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRS 633
+M G P +N +DG A+ + LM+RNG+ +A +
Sbjct: 350 KQMTDKGIGPSIYSYNILMDGLCKLGMLSDAKTIVGLMKRNGVCPDAVT----------- 398
Query: 634 RRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYE 693
Y ++ G C K D A L EM++ P+
Sbjct: 399 -----------------------YGCLLHGYCSVGKVDAAKSLLQEMMRNNCLPNAYTCN 435
Query: 694 VLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEG 753
+L+ L + R EA L+ + G L + N+++ S E+ +K
Sbjct: 436 ILLHSLWKMGRISEAEELLRKMNEKGYGLDTVTCNIIVDGLCGSGEL---------DKAI 486
Query: 754 EFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAK-CFPVDIYTYNLLMRKLTHHD-MDKA 811
E + + + A G L SY + LI C P D+ TY+ L+ L +A
Sbjct: 487 EIVKG--MRVHGSAALGNLGNSYIGLVDDSLIENNCLP-DLITYSTLLNGLCKAGRFAEA 543
Query: 812 CELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRN 866
LF M L+P+ Y + H F G+ A R + +M KKG + T N
Sbjct: 544 KNLFAEMMGEKLQPDSVAYNIFIHHFCKQGKISSAFRVLKDMEKKGCHKSLETYN 598
Score = 115 bits (287), Expect = 1e-25
Identities = 145/664 (21%), Positives = 258/664 (38%), Gaps = 66/664 (9%)
Query: 245 IVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS 304
++++ K+ R+E +V G + LI LC+S+ + A EL E
Sbjct: 117 LLLESCIKERRVEFVSWLYKDMVLCGIAPQTYTFNLLIRALCDSSCVDAARELFDEMPEK 176
Query: 305 --LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKD 362
P E +G+ +RG + G D+ LE +S G +P KV YN ++ RE R D
Sbjct: 177 GCKPNEFTFGILVRGYCKAGLTDKGLELLNAM-ESFGVLPNKVIYNTIVSSFCREGRNDD 235
Query: 363 VYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS---QFGLS-PNYMAYK 418
+++ M E + P++VT N+ + CK G V A +F+ GL PN + Y
Sbjct: 236 SEKMVEKMREEGLVPDIVTFNSRISALCKEGKVLDASRIFSDMELDEYLGLPRPNSITYN 295
Query: 419 YLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALER 478
++ C G ++A + S ++++ L R K E +L ++
Sbjct: 296 LMLKGFCKVGLLEDAKTLFESIRENDDLASLQSYNIWLQGLVRHGKFIEAETVLKQMTDK 355
Query: 479 RFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKSNRGDIA 535
P+ +Y+ + LC+ G + D + G + + +Y ++ G+ + D A
Sbjct: 356 GIGPSIYSYNILMDGLCKLGMLSDAKTIVGLMKRNGVCPDAVTYGCLLHGYCSVGKVDAA 415
Query: 536 ARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIF--NSFID 593
LL EM + +LH L M + LL M D N +D
Sbjct: 416 KSLLQEMMRNNCLPNAYTCNILLHSLWKMGRI-SEAEELLRKMNEKGYGLDTVTCNIIVD 474
Query: 594 GAMHANKPDLAREVFELMQRNG--IMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVV 651
G + + D A E+ + M+ +G + N + + + + D+L I + +
Sbjct: 475 GLCGSGELDKAIEIVKGMRVHGSAALGNLGNSYIGL--------VDDSL-----IENNCL 521
Query: 652 VSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNL 711
Y+ ++ GLCK+ + A L EM+ L P Y + + C + A +
Sbjct: 522 PDLITYSTLLNGLCKAGRFAEAKNLFAEMMGEKLQPDSVAYNIFIHHFCKQGKISSAFRV 581
Query: 712 VNVYEKAGRRLTSFLGNVLLFHSMISPEVY--HSCVDLRREK-----------------E 752
+ EK G + N L+ I +++ H +D +EK E
Sbjct: 582 LKDMEKKGCHKSLETYNSLILGLGIKNQIFEIHGLMDEMKEKGISPNICTYNTAIQYLCE 641
Query: 753 GE-------FLDSSM----------LTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYT 795
GE LD M +I AF + + E ++ C +
Sbjct: 642 GEKVEDATNLLDEMMQKNIAPNVFSFKYLIEAFCKVPDFDMAQEVFETAVSICGQKE-GL 700
Query: 796 YNLLMRKL-THHDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEML 854
Y+L+ +L + KA EL + + RG E + Y + + A +H+M+
Sbjct: 701 YSLMFNELLAAGQLLKATELLEAVLDRGFELGTFLYKDLVESLCKKDELEVASGILHKMI 760
Query: 855 KKGF 858
+G+
Sbjct: 761 DRGY 764
Score = 112 bits (281), Expect = 7e-25
Identities = 147/661 (22%), Positives = 256/661 (38%), Gaps = 112/661 (16%)
Query: 118 SCLK-----FFDW-------AGRQPRFYHTRTTFVAIFRIL---SCARLRPLVFDFLRDF 162
SC+K F W G P+ Y TF + R L SC +FD + +
Sbjct: 121 SCIKERRVEFVSWLYKDMVLCGIAPQTY----TFNLLIRALCDSSCVDAARELFDEMPE- 175
Query: 163 RSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCY 222
+ C P+ + LV GY AG D L LL M G+ ++ N++ + C
Sbjct: 176 KGCK-PNEFTF-GILVRGYCKAGLTDKGLELLNAMESFGV----LPNKVIYNTIVSSFCR 229
Query: 223 NAFDVIANQICMRGYESHMTNVIV-----IKHLCKQGRLEEA-----EAHLNGLVGSGKE 272
+ + ++ + E + IV I LCK+G++ +A + L+ +G +
Sbjct: 230 EGRNDDSEKMVEKMREEGLVPDIVTFNSRISALCKEGKVLDASRIFSDMELDEYLGLPRP 289
Query: 273 LHRSELSFLIGVLCESNRFERAVELVSEFGTSLPLEN--AYGVWIRGLVQGGRLDEALEF 330
+ + ++ C+ E A L + L + +Y +W++GLV+ G+ EA
Sbjct: 290 -NSITYNLMLKGFCKVGLLEDAKTLFESIRENDDLASLQSYNIWLQGLVRHGKFIEAETV 348
Query: 331 FRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFC 390
+Q D +G P YNIL+ L + L D ++ M + P+ VT +L +C
Sbjct: 349 LKQMTD-KGIGPSIYSYNILMDGLCKLGMLSDAKTIVGLMKRNGVCPDAVTYGCLLHGYC 407
Query: 391 KLGMVDVALELFNSRSQFGLSPNYMAYKYLILTL-------------------------- 424
+G VD A L + PN L+ +L
Sbjct: 408 SVGKVDAAKSLLQEMMRNNCLPNAYTCNILLHSLWKMGRISEAEELLRKMNEKGYGLDTV 467
Query: 425 ---------CWDGCPKEAYRVLRS----------SSGTGYF-------------PDRRTF 452
C G +A +++ + G Y PD T+
Sbjct: 468 TCNIIVDGLCGSGELDKAIEIVKGMRVHGSAALGNLGNSYIGLVDDSLIENNCLPDLITY 527
Query: 453 STLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK 512
STL N LC+ + E +L + + P+S Y+ F+ C+ G++ + + D++K
Sbjct: 528 STLLNGLCKAGRFAEAKNLFAEMMGEKLQPDSVAYNIFIHHFCKQGKISSAFRVLKDMEK 587
Query: 513 VTARFS---YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRT 569
S Y +I+G N+ L+ EMKEKG +Y + L +
Sbjct: 588 KGCHKSLETYNSLILGLGIKNQIFEIHGLMDEMKEKGISPNICTYNTAIQYLCEGEKVED 647
Query: 570 RFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELM-----QRNGIMTNASSQ 623
L EMM P+ F I+ D+A+EVFE Q+ G+ + ++
Sbjct: 648 ATNLLDEMMQKNIAPNVFSFKYLIEAFCKVPDFDMAQEVFETAVSICGQKEGLYSLMFNE 707
Query: 624 ILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKV 683
+L ++ + +A+ + + T LY ++ LCK D+ ++A + +M+
Sbjct: 708 LLAAGQLLKATELLEAV-----LDRGFELGTFLYKDLVESLCKKDELEVASGILHKMIDR 762
Query: 684 G 684
G
Sbjct: 763 G 763
Score = 84.3 bits (207), Expect = 3e-16
Identities = 90/434 (20%), Positives = 164/434 (37%), Gaps = 51/434 (11%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
++ L+ G G A ++G M+ G+ D Y LL+ +A + ++
Sbjct: 364 YNILMDGLCKLGMLSDAKTIVGLMKRNGVCPDAVTYGCLLHGYCSVGKVDAAKSLLQEMM 423
Query: 234 MRG-YESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
+ T I++ L K GR+ EAE L + G L + ++ LC S +
Sbjct: 424 RNNCLPNAYTCNILLHSLWKMGRISEAEELLRKMNEKGYGLDTVTCNIIVDGLCGSGELD 483
Query: 293 RAVELVSEFGTSLPLENAYGVWIRGLVQGGRLDEA-LEFFRQKRDSEGFVPCKVRYNILI 351
+A+E+V G+ + G G L + + +P + Y+ L+
Sbjct: 484 KAIEIVK------------GMRVHGSAALGNLGNSYIGLVDDSLIENNCLPDLITYSTLL 531
Query: 352 GRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLS 411
L + R + L +M + P+ V N + FCK G + A + + G
Sbjct: 532 NGLCKAGRFAEAKNLFAEMMGEKLQPDSVAYNIFIHHFCKQGKISSAFRVLKDMEKKGCH 591
Query: 412 PNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDL 471
+ Y LIL L E + ++ G P+ T++T LC K+++ +L
Sbjct: 592 KSLETYNSLILGLGIKNQIFEIHGLMDEMKEKGISPNICTYNTAIQYLCEGEKVEDATNL 651
Query: 472 LDFALERRFMPNSSTYSRFVSALCR-----------------AGRVEDGY-------LMR 507
LD +++ PN ++ + A C+ G+ E Y L
Sbjct: 652 LDEMMQKNIAPNVFSFKYLIEAFCKVPDFDMAQEVFETAVSICGQKEGLYSLMFNELLAA 711
Query: 508 GDLDKVT-------------ARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSY 554
G L K T F Y ++ K + ++A+ +L +M ++GY ++
Sbjct: 712 GQLLKATELLEAVLDRGFELGTFLYKDLVESLCKKDELEVASGILHKMIDRGYGFDPAAL 771
Query: 555 RHVLHCLLHMDNPR 568
V+ L M N +
Sbjct: 772 MPVIDGLGKMGNKK 785
Score = 83.2 bits (204), Expect = 6e-16
Identities = 112/551 (20%), Positives = 214/551 (38%), Gaps = 72/551 (13%)
Query: 354 LLRENRLKDVYELL-MDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNS-RSQFGLS 411
L+R +++ EL + ++ + + ++ +V+ F K +D A F RS+F +
Sbjct: 49 LVRAKMHEEIQELHNLILSSSIQKTKLSSLLSVVSIFAKSNHIDKAFPQFQLVRSRFPEN 108
Query: 412 -PNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWD 470
P+ Y L+ + + + + + G P TF+ L ALC +D +
Sbjct: 109 KPSVYLYNLLLESCIKERRVEFVSWLYKDMVLCGIAPQTYTFNLLIRALCDSSCVDAARE 168
Query: 471 LLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFI 527
L D E+ PN T+ V C+AG + G + ++ + + Y ++ F
Sbjct: 169 LFDEMPEKGCKPNEFTFGILVRGYCKAGLTDKGLELLNAMESFGVLPNKVIYNTIVSSFC 228
Query: 528 KSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI 587
+ R D + +++ +M+E+G P
Sbjct: 229 REGRNDDSEKMVEKMREEGLV----------------------------------PDIVT 254
Query: 588 FNSFIDGAMHANKPDLAREVFELMQRNGIM----TNASSQILVMKSYFRSRRISDALRFF 643
FNS I K A +F M+ + + N+ + L++K + + + DA F
Sbjct: 255 FNSRISALCKEGKVLDASRIFSDMELDEYLGLPRPNSITYNLMLKGFCKVGLLEDAKTLF 314
Query: 644 NDIR-HQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSL 702
IR + + S + YN + GL + K A + +M G+ PSI Y +L+ LC L
Sbjct: 315 ESIRENDDLASLQSYNIWLQGLVRHGKFIEAETVLKQMTDKGIGPSIYSYNILMDGLCKL 374
Query: 703 KRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRRE--KEGEFLDSSM 760
+A +V + ++ G + LL H S + L +E + ++
Sbjct: 375 GMLSDAKTIVGLMKRNGVCPDAVTYGCLL-HGYCSVGKVDAAKSLLQEMMRNNCLPNAYT 433
Query: 761 LTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHH-DMDKACELF---- 815
+++ + R+S + + L ++ K + +D T N+++ L ++DKA E+
Sbjct: 434 CNILLHSLWKMGRISEAEELLRKMNEKGYGLDTVTCNIIVDGLCGSGELDKAIEIVKGMR 493
Query: 816 -------------------DRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKK 856
D + + P+ TY + +G GR EAK EM+ +
Sbjct: 494 VHGSAALGNLGNSYIGLVDDSLIENNCLPDLITYSTLLNGLCKAGRFAEAKNLFAEMMGE 553
Query: 857 GFNPPENTRNV 867
P N+
Sbjct: 554 KLQPDSVAYNI 564
Score = 74.7 bits (182), Expect = 2e-13
Identities = 87/366 (23%), Positives = 155/366 (41%), Gaps = 20/366 (5%)
Query: 518 SYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLL-- 575
S ++ F KSN D A ++ + E K S Y + L + R F + L
Sbjct: 77 SLLSVVSIFAKSNHIDKAFPQFQLVRSRFPENKPSVYLYNLLLESCIKERRVEFVSWLYK 136
Query: 576 EMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSR 634
+M+ G P FN I ++ D ARE+F+ M G N + ++++ Y ++
Sbjct: 137 DMVLCGIAPQTYTFNLLIRALCDSSCVDAARELFDEMPEKGCKPNEFTFGILVRGYCKAG 196
Query: 635 RISDALRFFNDIRHQVVVSTK-LYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYE 693
L N + V+ K +YN ++ C+ + D + ++ +M + GL P I +
Sbjct: 197 LTDKGLELLNAMESFGVLPNKVIYNTIVSSFCREGRNDDSEKMVEKMREEGLVPDIVTFN 256
Query: 694 VLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLG----NVLLFHSMISPEVYHSCVDLRR 749
+ LC + +A + + E L +LG N + ++ M+ ++ +
Sbjct: 257 SRISALCKEGKVLDASRIFSDME-----LDEYLGLPRPNSITYNLMLKGFCKVGLLEDAK 311
Query: 750 ---EKEGEFLDSSMLTLIIGAFSGCLRVSYSIQE---LEELIAKCFPVDIYTYNLLMRKL 803
E E D + L G +R I+ L+++ K IY+YN+LM L
Sbjct: 312 TLFESIRENDDLASLQSYNIWLQGLVRHGKFIEAETVLKQMTDKGIGPSIYSYNILMDGL 371
Query: 804 THHDM-DKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPE 862
M A + M + G+ P+ TYG + HG+ + G+ D AK + EM++ P
Sbjct: 372 CKLGMLSDAKTIVGLMKRNGVCPDAVTYGCLLHGYCSVGKVDAAKSLLQEMMRNNCLPNA 431
Query: 863 NTRNVI 868
T N++
Sbjct: 432 YTCNIL 437
Score = 60.8 bits (146), Expect = 3e-09
Identities = 63/308 (20%), Positives = 127/308 (40%), Gaps = 32/308 (10%)
Query: 563 HMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASS 622
H+D +F + KP ++N ++ + + + +++ M GI +
Sbjct: 90 HIDKAFPQFQLVRSRFPENKPSVYLYNLLLESCIKERRVEFVSWLYKDMVLCGIAPQTYT 149
Query: 623 QILVMKSYFRSRRISDALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEML 681
L++++ S + A F+++ + + + ++ G CK+ D LEL M
Sbjct: 150 FNLLIRALCDSSCVDAARELFDEMPEKGCKPNEFTFGILVRGYCKAGLTDKGLELLNAME 209
Query: 682 KVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVY 741
G+ P+ Y +V C R ++ +V + G + +++ F+S IS
Sbjct: 210 SFGVLPNKVIYNTIVSSFCREGRNDDSEKMVEKMREEG-----LVPDIVTFNSRISALC- 263
Query: 742 HSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMR 801
KEG+ LD+S + +S EL+E + P I TYNL+++
Sbjct: 264 ---------KEGKVLDASRI--------------FSDMELDEYLGLPRPNSI-TYNLMLK 299
Query: 802 KLTHHDM-DKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNP 860
+ + A LF+ + + + +Y + G HG+ EA+ + +M KG P
Sbjct: 300 GFCKVGLLEDAKTLFESIRENDDLASLQSYNIWLQGLVRHGKFIEAETVLKQMTDKGIGP 359
Query: 861 PENTRNVI 868
+ N++
Sbjct: 360 SIYSYNIL 367
>At5g55840 putative protein
Length = 1274
Score = 142 bits (358), Expect = 8e-34
Identities = 159/700 (22%), Positives = 293/700 (41%), Gaps = 51/700 (7%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
++T++ Y G+ A+ LL M+ +G+D D Y++L++ L +N ++ +
Sbjct: 231 YNTVLHWYCKKGRFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMR 290
Query: 234 MRG-YESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
R + + +T +I +G++ A LN ++ G + + LI F+
Sbjct: 291 KRMIHPNEVTYNTLINGFSNEGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFK 350
Query: 293 RAVEL--VSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFF-RQKRDSEGFVPCKVRYNI 349
A+++ + E P E +YGV + GL + D A F+ R KR+ G ++ Y
Sbjct: 351 EALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKRN--GVCVGRITYTG 408
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFG 409
+I L + L + LL +M++ I P++VT +A++ FCK+G A E+ + G
Sbjct: 409 MIDGLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVG 468
Query: 410 LSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMW 469
LSPN + Y LI C GC KEA R+ + G+ D TF+ L +LC+ K+ E
Sbjct: 469 LSPNGIIYSTLIYNCCRMGCLKEAIRIYEAMILEGHTRDHFTFNVLVTSLCKAGKVAEAE 528
Query: 470 DLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKV---TARFSYAKMIMGF 526
+ + +PN+ ++ ++ +G + + ++ KV F+Y ++ G
Sbjct: 529 EFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFDEMTKVGHHPTFFTYGSLLKGL 588
Query: 527 IKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHC 585
K A + L + + Y +L + N EM+ P
Sbjct: 589 CKGGHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDS 648
Query: 586 DIFNSFIDGAMHANKPDLA-REVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRF-- 642
+ S I G K +A E R ++ N + F++ + + F
Sbjct: 649 YTYTSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFRE 708
Query: 643 -FNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCS 701
+++ H + T N MI G + K + +L EM P++ Y +
Sbjct: 709 QMDNLGHTPDIVTT--NAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNI------- 759
Query: 702 LKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSML 761
L++ Y K TSF LL+ S+I + + G +S+ML
Sbjct: 760 ---------LLHGYSKRKDVSTSF----LLYRSIILNGILPDKLTCHSLVLG-ICESNML 805
Query: 762 TLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRK-LTHHDMDKACELFDRMCQ 820
+ ++ L+ I + VD YT+N+L+ K + +++ A +L M
Sbjct: 806 EI-------------GLKILKAFICRGVEVDRYTFNMLISKCCANGEINWAFDLVKVMTS 852
Query: 821 RGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNP 860
G+ ++ T M + + R E++ +HEM K+G +P
Sbjct: 853 LGISLDKDTCDAMVSVLNRNHRFQESRMVLHEMSKQGISP 892
Score = 128 bits (322), Expect = 1e-29
Identities = 137/636 (21%), Positives = 242/636 (37%), Gaps = 48/636 (7%)
Query: 279 SFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRD 336
+ LI VLC FE++ L+ + S P Y + + GR A+E +
Sbjct: 197 NILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWYCKKGRFKAAIELLDHMK- 255
Query: 337 SEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVD 396
S+G YN+LI L R NR+ Y LL DM + I PN VT N ++ F G V
Sbjct: 256 SKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTYNTLINGFSNEGKVL 315
Query: 397 VALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLA 456
+A +L N FGLSPN++ + LI +G KEA ++ G P ++ L
Sbjct: 316 IASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFYMMEAKGLTPSEVSYGVLL 375
Query: 457 NALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR 516
+ LC+ + D TY+ + LC+ G +++ ++ ++ K
Sbjct: 376 DGLCKNAEFDLARGFYMRMKRNGVCVGRITYTGMIDGLCKNGFLDEAVVLLNEMSKDGID 435
Query: 517 ---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH--CLLHMDNPRTRF 571
+Y+ +I GF K R A ++ + G Y +++ C + R
Sbjct: 436 PDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSPNGIIYSTLIYNCCRMGCLKEAIRI 495
Query: 572 FNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYF 631
+ + + H + H FN + A K A E M +GI+ N S ++ Y
Sbjct: 496 YEAMILEGHTRDHF-TFNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLINGYG 554
Query: 632 RSRRISDALRFFNDIR----------------------------------HQV--VVSTK 655
S A F+++ H V V T
Sbjct: 555 NSGEGLKAFSVFDEMTKVGHHPTFFTYGSLLKGLCKGGHLREAEKFLKSLHAVPAAVDTV 614
Query: 656 LYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVY 715
+YN ++ +CKS A+ L EM++ + P Y L+ LC + A+
Sbjct: 615 MYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDSYTYTSLISGLCRKGKTVIAILFAKEA 674
Query: 716 EKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKE--GEFLDSSMLTLIIGAFSGCLR 773
E G L + + M + + + R + + G D +I +S +
Sbjct: 675 EARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFREQMDNLGHTPDIVTTNAMIDGYSRMGK 734
Query: 774 VSYSIQELEELIAKCFPVDIYTYNLLMRKLT-HHDMDKACELFDRMCQRGLEPNRWTYGL 832
+ + L E+ + ++ TYN+L+ + D+ + L+ + G+ P++ T
Sbjct: 735 IEKTNDLLPEMGNQNGGPNLTTYNILLHGYSKRKDVSTSFLLYRSIILNGILPDKLTCHS 794
Query: 833 MAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
+ G + + + + +G T N++
Sbjct: 795 LVLGICESNMLEIGLKILKAFICRGVEVDRYTFNML 830
Score = 118 bits (296), Expect = 1e-26
Identities = 165/760 (21%), Positives = 295/760 (38%), Gaps = 84/760 (11%)
Query: 159 LRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNS-LA 217
LRD R ++TL+ G++ GK IA LL M GL + ++ L++ ++
Sbjct: 286 LRDMRKRMIHPNEVTYNTLINGFSNEGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHIS 345
Query: 218 ENNCYNA------------------FDVIANQIC-------MRGYESHM----------T 242
E N A + V+ + +C RG+ M T
Sbjct: 346 EGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKRNGVCVGRIT 405
Query: 243 NVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEF- 301
+I LCK G L+EA LN + G + S LI C+ RF+ A E+V
Sbjct: 406 YTGMIDGLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIY 465
Query: 302 --GTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENR 359
G S P Y I + G L EA+ + + EG +N+L+ L + +
Sbjct: 466 RVGLS-PNGIIYSTLIYNCCRMGCLKEAIRIY-EAMILEGHTRDHFTFNVLVTSLCKAGK 523
Query: 360 LKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKY 419
+ + E + M I PN V+ + ++ + G A +F+ ++ G P + Y
Sbjct: 524 VAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFDEMTKVGHHPTFFTYGS 583
Query: 420 LILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERR 479
L+ LC G +EA + L+S D ++TL A+C+ + + L ++R
Sbjct: 584 LLKGLCKGGHLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRS 643
Query: 480 FMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK----VTARFSYAKMIMGFIKSNRGDIA 535
+P+S TY+ +S LCR G+ L + + + + Y + G K+ +
Sbjct: 644 ILPDSYTYTSLISGLCRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAG 703
Query: 536 ARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEM-MTHGKPHCDIFNSFIDG 594
+M G+ + ++ M L EM +G P+ +N + G
Sbjct: 704 IYFREQMDNLGHTPDIVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHG 763
Query: 595 AMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFND-IRHQVVVS 653
+ ++ + NGI+ + + ++ S + L+ I V V
Sbjct: 764 YSKRKDVSTSFLLYRSIILNGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVEVD 823
Query: 654 TKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEA----- 708
+N +I C + + + A +L M +G++ + + +V L R+ E+
Sbjct: 824 RYTFNMLISKCCANGEINWAFDLVKVMTSLGISLDKDTCDAMVSVLNRNHRFQESRMVLH 883
Query: 709 --------------VNLVNVYEKAGRRLTSF-LGNVLLFHSMISPEVYHSCVDLRREKEG 753
+ L+N + G T+F + ++ H + P V S + K G
Sbjct: 884 EMSKQGISPESRKYIGLINGLCRVGDIKTAFVVKEEMIAHKICPPNVAESAMVRALAKCG 943
Query: 754 E---------FLDSSMLTLIIGAFSG----CLRVSYSIQ--ELEELIAKC-FPVDIYTYN 797
+ F+ L I +F+ C + I+ EL +++ C +D+ +YN
Sbjct: 944 KADEATLLLRFMLKMKLVPTIASFTTLMHLCCKNGNVIEALELRVVMSNCGLKLDLVSYN 1003
Query: 798 LLMRKL-THHDMDKACELFDRMCQRGLEPNRWTYGLMAHG 836
+L+ L DM A EL++ M G N TY + G
Sbjct: 1004 VLITGLCAKGDMALAFELYEEMKGDGFLANATTYKALIRG 1043
Score = 103 bits (258), Expect = 3e-22
Identities = 127/589 (21%), Positives = 234/589 (39%), Gaps = 32/589 (5%)
Query: 145 ILSCARLRPLVFDFLRDFRSCSFPHRARYHDT---LVVGYAIAGKPDIALHLLGRMRFQG 201
I +C R+ L + +R + + R H T LV AGK A + M G
Sbjct: 480 IYNCCRMGCLK-EAIRIYEAMILEGHTRDHFTFNVLVTSLCKAGKVAEAEEFMRCMTSDG 538
Query: 202 LDLDGFGYHILLNSLAENN-CYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAE 260
+ + + L+N + AF V + + + T ++K LCK G L EAE
Sbjct: 539 ILPNTVSFDCLINGYGNSGEGLKAFSVFDEMTKVGHHPTFFTYGSLLKGLCKGGHLREAE 598
Query: 261 AHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEF--GTSLPLENAYGVWIRGL 318
L L + + L+ +C+S +AV L E + LP Y I GL
Sbjct: 599 KFLKSLHAVPAAVDTVMYNTLLTAMCKSGNLAKAVSLFGEMVQRSILPDSYTYTSLISGL 658
Query: 319 VQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPN 378
+ G+ A+ F ++ +P KV Y + + + + K M+ P+
Sbjct: 659 CRKGKTVIAILFAKEAEARGNVLPNKVMYTCFVDGMFKAGQWKAGIYFREQMDNLGHTPD 718
Query: 379 MVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLR 438
+VT NA++ + ++G ++ +L PN Y L+ ++ + R
Sbjct: 719 IVTTNAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHGYSKRKDVSTSFLLYR 778
Query: 439 SSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAG 498
S G PD+ T +L +C ++ +L + R + T++ +S C G
Sbjct: 779 SIILNGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVEVDRYTFNMLISKCCANG 838
Query: 499 RVEDGYLMRGDLDKVTARFSYA-------KMIMGFIKSNRGDIAARLLVEMKEKGYELKR 551
+ + DL KV + M+ +++R + +L EM ++G +
Sbjct: 839 EINWAF----DLVKVMTSLGISLDKDTCDAMVSVLNRNHRFQESRMVLHEMSKQGISPES 894
Query: 552 SSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAM-HANKPDLAREVFEL 610
Y +++ L + + +T F EM+ H ++ S + A+ K D A +
Sbjct: 895 RKYIGLINGLCRVGDIKTAFVVKEEMIAHKICPPNVAESAMVRALAKCGKADEATLLLRF 954
Query: 611 MQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVST---KL----YNRMIVG 663
M + ++ +S +M ++ + +AL +VV+S KL YN +I G
Sbjct: 955 MLKMKLVPTIASFTTLMHLCCKNGNVIEALEL------RVVMSNCGLKLDLVSYNVLITG 1008
Query: 664 LCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLV 712
LC +A EL EM G + Y+ L++ L + + + +++
Sbjct: 1009 LCAKGDMALAFELYEEMKGDGFLANATTYKALIRGLLARETAFSGADII 1057
Score = 103 bits (257), Expect = 4e-22
Identities = 119/574 (20%), Positives = 218/574 (37%), Gaps = 74/574 (12%)
Query: 309 NAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLM 368
+ Y + IR ++ G + ++LE FR GF P N ++G +++ V+ L
Sbjct: 124 SVYDILIRVYLREGMIQDSLEIFRLM-GLYGFNPSVYTCNAILGSVVKSGEDVSVWSFLK 182
Query: 369 DMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDG 428
+M + I P++ T N ++ C G + + L + G +P + Y ++ C G
Sbjct: 183 EMLKRKICPDVATFNILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWYCKKG 242
Query: 429 CPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYS 488
K A +L G D T++ L + LCR +I + + LL +R PN TY+
Sbjct: 243 RFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTYN 302
Query: 489 RFVSALCRAGRVEDGYLMRGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYE 548
++ G+V IA++LL EM G
Sbjct: 303 TLINGFSNEGKVL--------------------------------IASQLLNEMLSFGLS 330
Query: 549 LKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI-FNSFIDGAMHANKPDLAREV 607
++ ++ + N + M G ++ + +DG + DLAR
Sbjct: 331 PNHVTFNALIDGHISEGNFKEALKMFYMMEAKGLTPSEVSYGVLLDGLCKNAEFDLARGF 390
Query: 608 FELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDI-RHQVVVSTKLYNRMIVGLCK 666
+ M+RNG+ + ++ ++ + +A+ N++ + + Y+ +I G CK
Sbjct: 391 YMRMKRNGVCVGRITYTGMIDGLCKNGFLDEAVVLLNEMSKDGIDPDIVTYSALINGFCK 450
Query: 667 SDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFL 726
+ A E+ + +VGL+P+ Y L+ C + EA+ + G F
Sbjct: 451 VGRFKTAKEIVCRIYRVGLSPNGIIYSTLIYNCCRMGCLKEAIRIYEAMILEGHTRDHFT 510
Query: 727 GNVLL--------------------FHSMISPEVYHSCVDLRREKEGEFLDSSML----- 761
NVL+ ++ V C+ GE L + +
Sbjct: 511 FNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFDEMT 570
Query: 762 ------------TLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHH-DM 808
+L+ G G + + + L+ L A VD YN L+ + ++
Sbjct: 571 KVGHHPTFFTYGSLLKGLCKGG-HLREAEKFLKSLHAVPAAVDTVMYNTLLTAMCKSGNL 629
Query: 809 DKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGR 842
KA LF M QR + P+ +TY + G G+
Sbjct: 630 AKAVSLFGEMVQRSILPDSYTYTSLISGLCRKGK 663
Score = 102 bits (254), Expect = 9e-22
Identities = 109/489 (22%), Positives = 192/489 (38%), Gaps = 75/489 (15%)
Query: 413 NYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLL 472
N Y LI +G +++ + R G+ P T + + ++ + + +W L
Sbjct: 122 NPSVYDILIRVYLREGMIQDSLEIFRLMGLYGFNPSVYTCNAILGSVVKSGEDVSVWSFL 181
Query: 473 DFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKS 529
L+R+ P+ +T++ ++ LC G E + ++K +Y ++ + K
Sbjct: 182 KEMLKRKICPDVATFNILINVLCAEGSFEKSSYLMQKMEKSGYAPTIVTYNTVLHWYCKK 241
Query: 530 NRGDIAARLLVEMKEKGYELKRSSYRHVLHCLL-----------------HMDNPRTRFF 572
R A LL MK KG + +Y ++H L M +P +
Sbjct: 242 GRFKAAIELLDHMKSKGVDADVCTYNMLIHDLCRSNRIAKGYLLLRDMRKRMIHPNEVTY 301
Query: 573 NLL------------------EMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQR 613
N L EM++ G P+ FN+ IDG + A ++F +M+
Sbjct: 302 NTLINGFSNEGKVLIASQLLNEMLSFGLSPNHVTFNALIDGHISEGNFKEALKMFYMMEA 361
Query: 614 NGIMTNASSQILVMKSYFRSRRISDALRFFNDI-RHQVVVSTKLYNRMIVGLCKSDKADI 672
G+ + S +++ ++ A F+ + R+ V V Y MI GLCK+ D
Sbjct: 362 KGLTPSEVSYGVLLDGLCKNAEFDLARGFYMRMKRNGVCVGRITYTGMIDGLCKNGFLDE 421
Query: 673 ALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLF 732
A+ L EM K G++P I Y L+ C + R+ A +V + G + + L
Sbjct: 422 AVVLLNEMSKDGIDPDIVTYSALINGFCKVGRFKTAKEIVCRIYRVGLSPNGIIYSTL-- 479
Query: 733 HSMISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVD 792
+Y+ C GCL+ +I+ E +I + D
Sbjct: 480 -------IYNCC-----------------------RMGCLK--EAIRIYEAMILEGHTRD 507
Query: 793 IYTYNLLMRKLTHHDMDKACELFDR-MCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVH 851
+T+N+L+ L E F R M G+ PN ++ + +G+ N G +A
Sbjct: 508 HFTFNVLVTSLCKAGKVAEAEEFMRCMTSDGILPNTVSFDCLINGYGNSGEGLKAFSVFD 567
Query: 852 EMLKKGFNP 860
EM K G +P
Sbjct: 568 EMTKVGHHP 576
Score = 66.2 bits (160), Expect = 7e-11
Identities = 87/406 (21%), Positives = 159/406 (38%), Gaps = 16/406 (3%)
Query: 175 DTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICM 234
+ ++ GY+ GK + LL M Q + Y+ILL+ ++ + ++ I +
Sbjct: 723 NAMIDGYSRMGKIEKTNDLLPEMGNQNGGPNLTTYNILLHGYSKRKDVSTSFLLYRSIIL 782
Query: 235 RGY-ESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFER 293
G +T ++ +C+ LE L + G E+ R + LI C +
Sbjct: 783 NGILPDKLTCHSLVLGICESNMLEIGLKILKAFICRGVEVDRYTFNMLISKCCANGEINW 842
Query: 294 AVELV---SEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNIL 350
A +LV + G SL + + + L + R E+ + +G P +Y L
Sbjct: 843 AFDLVKVMTSLGISLDKDTCDAM-VSVLNRNHRFQES-RMVLHEMSKQGISPESRKYIGL 900
Query: 351 IGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGL 410
I L R +K + + +M I P V +A++ K G D A L + L
Sbjct: 901 INGLCRVGDIKTAFVVKEEMIAHKICPPNVAESAMVRALAKCGKADEATLLLRFMLKMKL 960
Query: 411 SPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWD 470
P ++ L+ C +G EA + S G D +++ L LC + + ++
Sbjct: 961 VPTIASFTTLMHLCCKNGNVIEALELRVVMSNCGLKLDLVSYNVLITGLCAKGDMALAFE 1020
Query: 471 LLDFALERRFMPNSSTYSRFVSALCRAGRVEDG-------YLMRGDLDKVTAR---FSYA 520
L + F+ N++TY + L G L RG + ++
Sbjct: 1021 LYEEMKGDGFLANATTYKALIRGLLARETAFSGADIILKDLLARGFITSMSLSQDSHRNL 1080
Query: 521 KMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDN 566
KM M +K+ + DIA E+ + L ++ + CL+H+ +
Sbjct: 1081 KMAMEKLKALQNDIAVCHREELDLQSNVLSKNQLKLFELCLIHISS 1126
>At1g79540 unknown protein
Length = 780
Score = 141 bits (356), Expect = 1e-33
Identities = 146/613 (23%), Positives = 260/613 (41%), Gaps = 43/613 (7%)
Query: 77 IYNILSSDDDFSAALSALSLPLSETFVLRVLRHGGDDGDILSCLKFFDWAGRQPRFYHTR 136
+ +IL+ AL L LS+ + V++ D+ + +FF WA R+ R +R
Sbjct: 37 VISILAKKKPIEPALEPLVPFLSKNIITSVIK---DEVNRQLGFRFFIWASRRERL-RSR 92
Query: 137 TTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGR 196
+F + +LS L + L + +S + L+ YA G + A+ GR
Sbjct: 93 ESFGLVIDMLSEDNGCDLYWQTLEELKSGGVSVDSYCFCVLISAYAKMGMAEKAVESFGR 152
Query: 197 MRFQGLDLDGFGYHILLNSLAENNCYN--AFDVIANQICMRGYESHMTNVIVIKHLCKQG 254
M+ D F Y+++L + + AF V + + T I++ L K+G
Sbjct: 153 MKEFDCRPDVFTYNVILRVMMREEVFFMLAFAVYNEMLKCNCSPNLYTFGILMDGLYKKG 212
Query: 255 RLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYG 312
R +A+ + + G G +R + LI LC+ + A +L E TS P A+
Sbjct: 213 RTSDAQKMFDDMTGRGISPNRVTYTILISGLCQRGSADDARKLFYEMQTSGNYPDSVAHN 272
Query: 313 VWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNE 372
+ G + GR+ EA E R + +GFV Y+ LI L R R +EL +M +
Sbjct: 273 ALLDGFCKLGRMVEAFELLRLF-EKDGFVLGLRGYSSLIDGLFRARRYTQAFELYANMLK 331
Query: 373 TCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKE 432
I P+++ ++ K G ++ AL+L +S G+SP+ Y +I LC G +E
Sbjct: 332 KNIKPDIILYTILIQGLSKAGKIEDALKLLSSMPSKGISPDTYCYNAVIKALCGRGLLEE 391
Query: 433 AYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVS 492
+ S T FPD T + L ++CR + E ++ + P+ +T++ +
Sbjct: 392 GRSLQLEMSETESFPDACTHTILICSMCRNGLVREAEEIFTEIEKSGCSPSVATFNALID 451
Query: 493 ALCRAGRVEDGYLMRGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRS 552
LC++G +++ L+ ++ + ++ S+ G+ R M E G LK
Sbjct: 452 GLCKSGELKEARLLLHKMEVGRPASLFLRL------SHSGN---RSFDTMVESGSILK-- 500
Query: 553 SYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQ 612
+YR + H T P +N I+G A D A ++ ++Q
Sbjct: 501 AYRDLAHF----------------ADTGSSPDIVSYNVLINGFCRAGDIDGALKLLNVLQ 544
Query: 613 RNGIMTNASSQILVMKSYFRSRRISDALRFF---NDIRHQVVVSTKLYNRMIVGLCKSDK 669
G+ ++ + ++ R R +A + F +D RH S +Y ++ C+ K
Sbjct: 545 LKGLSPDSVTYNTLINGLHRVGREEEAFKLFYAKDDFRH----SPAVYRSLMTWSCRKRK 600
Query: 670 ADIALELCFEMLK 682
+A L + LK
Sbjct: 601 VLVAFNLWMKYLK 613
Score = 88.2 bits (217), Expect = 2e-17
Identities = 110/498 (22%), Positives = 198/498 (39%), Gaps = 62/498 (12%)
Query: 269 SGKELHRSELSF--LIGVLCESNRFE---RAVELVSEFGTSLPLENAYGVWIRGLVQGGR 323
S +E RS SF +I +L E N + + +E + G S+ + V I + G
Sbjct: 84 SRRERLRSRESFGLVIDMLSEDNGCDLYWQTLEELKSGGVSVD-SYCFCVLISAYAKMGM 142
Query: 324 LDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDV-YELLMDMNETCIPPNMVTM 382
++A+E F + ++ + P YN+++ ++RE + + + +M + PN+ T
Sbjct: 143 AEKAVESFGRMKEFD-CRPDVFTYNVILRVMMREEVFFMLAFAVYNEMLKCNCSPNLYTF 201
Query: 383 NAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSG 442
++ K G A ++F+ + G+SPN + Y LI LC G +A ++
Sbjct: 202 GILMDGLYKKGRTSDAQKMFDDMTGRGISPNRVTYTILISGLCQRGSADDARKLFYEMQT 261
Query: 443 TGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVED 502
+G +PD + L + C+ ++ E ++LL + F+ YS + L RA R
Sbjct: 262 SGNYPDSVAHNALLDGFCKLGRMVEAFELLRLFEKDGFVLGLRGYSSLIDGLFRARRYTQ 321
Query: 503 GYLMRGDLDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH 559
+ + ++ K + Y +I G K+ + + A +LL M KG Y V+
Sbjct: 322 AFELYANMLKKNIKPDIILYTILIQGLSKAGKIEDALKLLSSMPSKGISPDTYCYNAVIK 381
Query: 560 CL-----------LHMDNPRTRFFN--------LLEMMTHG-----------------KP 583
L L ++ T F + M +G P
Sbjct: 382 ALCGRGLLEEGRSLQLEMSETESFPDACTHTILICSMCRNGLVREAEEIFTEIEKSGCSP 441
Query: 584 HCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRIS------ 637
FN+ IDG + + AR + M+ + + + S+ +R
Sbjct: 442 SVATFNALIDGLCKSGELKEARLLLHKME-----VGRPASLFLRLSHSGNRSFDTMVESG 496
Query: 638 DALRFFNDIRHQVVVSTK----LYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYE 693
L+ + D+ H + YN +I G C++ D AL+L + GL+P Y
Sbjct: 497 SILKAYRDLAHFADTGSSPDIVSYNVLINGFCRAGDIDGALKLLNVLQLKGLSPDSVTYN 556
Query: 694 VLVQKLCSLKRYYEAVNL 711
L+ L + R EA L
Sbjct: 557 TLINGLHRVGREEEAFKL 574
Score = 77.8 bits (190), Expect = 2e-14
Identities = 105/522 (20%), Positives = 200/522 (38%), Gaps = 67/522 (12%)
Query: 347 YNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS 406
+ ++I L +N ++ L ++ + + ++ + K+GM + A+E F
Sbjct: 95 FGLVIDMLSEDNGCDLYWQTLEELKSGGVSVDSYCFCVLISAYAKMGMAEKAVESFGRMK 154
Query: 407 QFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKID 466
+F P+ Y N + R +
Sbjct: 155 EFDCRPDVFTY---------------------------------------NVILRVMMRE 175
Query: 467 EMWDLLDFALERRFM-----PNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFS 518
E++ +L FA+ + PN T+ + L + GR D M D+ R +
Sbjct: 176 EVFFMLAFAVYNEMLKCNCSPNLYTFGILMDGLYKKGRTSDAQKMFDDMTGRGISPNRVT 235
Query: 519 YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMM 578
Y +I G + D A +L EM+ G ++ +L + F LL +
Sbjct: 236 YTILISGLCQRGSADDARKLFYEMQTSGNYPDSVAHNALLDGFCKLGR-MVEAFELLRLF 294
Query: 579 THGKPHCDI--FNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRI 636
+ ++S IDG A + A E++ M + I + ++++ ++ +I
Sbjct: 295 EKDGFVLGLRGYSSLIDGLFRARRYTQAFELYANMLKKNIKPDIILYTILIQGLSKAGKI 354
Query: 637 SDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVL 695
DAL+ + + + + T YN +I LC + L EM + P + +L
Sbjct: 355 EDALKLLSSMPSKGISPDTYCYNAVIKALCGRGLLEEGRSLQLEMSETESFPDACTHTIL 414
Query: 696 VQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGE- 754
+ +C EA + EK+G + N L+ S E+ + + L + + G
Sbjct: 415 ICSMCRNGLVREAEEIFTEIEKSGCSPSVATFNALIDGLCKSGELKEARLLLHKMEVGRP 474
Query: 755 ---FL------DSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTH 805
FL + S T++ SG + +Y ++L DI +YN+L+
Sbjct: 475 ASLFLRLSHSGNRSFDTMVE---SGSILKAY--RDLAHFADTGSSPDIVSYNVLINGFCR 529
Query: 806 H-DMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEA 846
D+D A +L + + +GL P+ TY + +G GR++EA
Sbjct: 530 AGDIDGALKLLNVLQLKGLSPDSVTYNTLINGLHRVGREEEA 571
Score = 71.6 bits (174), Expect = 2e-12
Identities = 65/270 (24%), Positives = 117/270 (43%), Gaps = 5/270 (1%)
Query: 603 LAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKL-YNRMI 661
LA V+ M + N + ++M ++ R SDA + F+D+ + + ++ Y +I
Sbjct: 181 LAFAVYNEMLKCNCSPNLYTFGILMDGLYKKGRTSDAQKMFDDMTGRGISPNRVTYTILI 240
Query: 662 VGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRR 721
GLC+ AD A +L +EM G P + L+ C L R EA L+ ++EK G
Sbjct: 241 SGLCQRGSADDARKLFYEMQTSGNYPDSVAHNALLDGFCKLGRMVEAFELLRLFEKDGFV 300
Query: 722 LTSFLGNVLLFHSMISPEVYHSCVDLRRE--KEGEFLDSSMLTLIIGAFSGCLRVSYSIQ 779
L G L + Y +L K+ D + T++I S ++ +++
Sbjct: 301 L-GLRGYSSLIDGLFRARRYTQAFELYANMLKKNIKPDIILYTILIQGLSKAGKIEDALK 359
Query: 780 ELEELIAKCFPVDIYTYNLLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGFS 838
L + +K D Y YN +++ L +++ L M + P+ T+ ++
Sbjct: 360 LLSSMPSKGISPDTYCYNAVIKALCGRGLLEEGRSLQLEMSETESFPDACTHTILICSMC 419
Query: 839 NHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
+G EA+ E+ K G +P T N +
Sbjct: 420 RNGLVREAEEIFTEIEKSGCSPSVATFNAL 449
Score = 37.4 bits (85), Expect = 0.037
Identities = 60/226 (26%), Positives = 91/226 (39%), Gaps = 44/226 (19%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSL----AENNCYNAF---D 226
++ L+ G+ AG D AL LL ++ +GL D Y+ L+N L E + F D
Sbjct: 520 YNVLINGFCRAGDIDGALKLLNVLQLKGLSPDSVTYNTLINGLHRVGREEEAFKLFYAKD 579
Query: 227 VIANQICMRGYESHMT------NVIV-----IKHLCKQGRLEEAEAHLNGLVGSGKELHR 275
+ + Y S MT V+V +K+L K L++ A+
Sbjct: 580 DFRHSPAV--YRSLMTWSCRKRKVLVAFNLWMKYLKKISCLDDETAN------------E 625
Query: 276 SELSFLIGVLCESNRFERAVELVSEFGT---SLPLENAYGVWIRGLVQGGRLDEALEFFR 332
E F G ERA+ + E T L L Y +W+ GL Q GR EAL F
Sbjct: 626 IEQCFKEG------ETERALRRLIELDTRKDELTL-GPYTIWLIGLCQSGRFHEALMVFS 678
Query: 333 QKRDSEGFVPCKVRYNILIGRLLRE--NRLKDVYELLMDMNETCIP 376
R+ + V ++ G RE + +V+ +D N +P
Sbjct: 679 VLREKKILVTPPSCVKLIHGLCKREQLDAAIEVFLYTLDNNFKLMP 724
>At5g39710 putative protein
Length = 747
Score = 140 bits (354), Expect = 2e-33
Identities = 123/522 (23%), Positives = 225/522 (42%), Gaps = 27/522 (5%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
++TL+ GY K D LL M +GL+ + Y++++N L + ++
Sbjct: 243 YNTLIDGYCKLRKIDDGFKLLRSMALKGLEPNLISYNVVINGLCREGRMKEVSFVLTEMN 302
Query: 234 MRGYE-SHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
RGY +T +IK CK+G +A ++ G + LI +C++
Sbjct: 303 RRGYSLDEVTYNTLIKGYCKEGNFHQALVMHAEMLRHGLTPSVITYTSLIHSMCKAGNMN 362
Query: 293 RAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNIL 350
RA+E + + P E Y + G Q G ++EA R+ D+ GF P V YN L
Sbjct: 363 RAMEFLDQMRVRGLCPNERTYTTLVDGFSQKGYMNEAYRVLREMNDN-GFSPSVVTYNAL 421
Query: 351 IGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGL 410
I +++D +L DM E + P++V+ + VL FC+ VD AL + + G+
Sbjct: 422 INGHCVTGKMEDAIAVLEDMKEKGLSPDVVSYSTVLSGFCRSYDVDEALRVKREMVEKGI 481
Query: 411 SPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWD 470
P+ + Y LI C KEA + G PD T++ L NA C E +++
Sbjct: 482 KPDTITYSSLIQGFCEQRRTKEACDLYEEMLRVGLPPDEFTYTALINAYCMEGDLEKALQ 541
Query: 471 LLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDL---DKVTARFSYAKMIMGFI 527
L + +E+ +P+ TYS ++ L + R + + L + V + +Y +I
Sbjct: 542 LHNEMVEKGVLPDVVTYSVLINGLNKQSRTREAKRLLLKLFYEESVPSDVTYHTLI---- 597
Query: 528 KSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI 587
N +I + +V + KG+ C+ M + F + H KP
Sbjct: 598 -ENCSNIEFKSVVSL-IKGF------------CMKGMMTEADQVFESMLGKNH-KPDGTA 642
Query: 588 FNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIR 647
+N I G A A +++ M ++G + + + I ++K+ + ++++ +
Sbjct: 643 YNIMIHGHCRAGDIRKAYTLYKEMVKSGFLLHTVTVIALVKALHKEGKVNELNSVIVHVL 702
Query: 648 HQVVVSTKLYNRMIVGL-CKSDKADIALELCFEMLKVGLNPS 688
+S +++V + + D+ L++ EM K G P+
Sbjct: 703 RSCELSEAEQAKVLVEINHREGNMDVVLDVLAEMAKDGFLPN 744
Score = 139 bits (351), Expect = 5e-33
Identities = 130/542 (23%), Positives = 235/542 (42%), Gaps = 27/542 (4%)
Query: 324 LDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENR-LKDVYELLMDMNETCIPPNMVTM 382
+D+AL + + GF+P + YN ++ +R R + + +M E+ + PN+ T
Sbjct: 150 IDKALSIVHLAQ-AHGFMPGVLSYNAVLDATIRSKRNISFAENVFKEMLESQVSPNVFTY 208
Query: 383 NAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSG 442
N ++ FC G +DVAL LF+ G PN + Y LI C + +++LRS +
Sbjct: 209 NILIRGFCFAGNIDVALTLFDKMETKGCLPNVVTYNTLIDGYCKLRKIDDGFKLLRSMAL 268
Query: 443 TGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVED 502
G P+ +++ + N LCRE ++ E+ +L R + + TY+ + C+ G
Sbjct: 269 KGLEPNLISYNVVINGLCREGRMKEVSFVLTEMNRRGYSLDEVTYNTLIKGYCKEGNFHQ 328
Query: 503 GYLMRGDLDK---VTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH 559
+M ++ + + +Y +I K+ + A L +M+ +G +Y ++
Sbjct: 329 ALVMHAEMLRHGLTPSVITYTSLIHSMCKAGNMNRAMEFLDQMRVRGLCPNERTYTTLVD 388
Query: 560 CLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMT 618
+ L EM +G P +N+ I+G K + A V E M+ G+
Sbjct: 389 GFSQKGYMNEAYRVLREMNDNGFSPSVVTYNALINGHCVTGKMEDAIAVLEDMKEKGLSP 448
Query: 619 NASSQILVMKSYFRSRRISDALRFFND-IRHQVVVSTKLYNRMIVGLCKSDKADIALELC 677
+ S V+ + RS + +ALR + + + T Y+ +I G C+ + A +L
Sbjct: 449 DVVSYSTVLSGFCRSYDVDEALRVKREMVEKGIKPDTITYSSLIQGFCEQRRTKEACDLY 508
Query: 678 FEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMIS 737
EML+VGL P Y L+ C +A+ L N + G L +V+ + +I+
Sbjct: 509 EEMLRVGLPPDEFTYTALINAYCMEGDLEKALQLHNEMVEKG-----VLPDVVTYSVLIN 563
Query: 738 PEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYN 797
S R +E + +L + S V+Y LI C ++ +
Sbjct: 564 GLNKQS-----RTREAK----RLLLKLFYEESVPSDVTY-----HTLIENCSNIEFKSVV 609
Query: 798 LLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKK 856
L++ M +A ++F+ M + +P+ Y +M HG G +A EM+K
Sbjct: 610 SLIKGFCMKGMMTEADQVFESMLGKNHKPDGTAYNIMIHGHCRAGDIRKAYTLYKEMVKS 669
Query: 857 GF 858
GF
Sbjct: 670 GF 671
Score = 118 bits (296), Expect = 1e-26
Identities = 109/455 (23%), Positives = 192/455 (41%), Gaps = 49/455 (10%)
Query: 271 KELHRSELS-------FLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQG 321
KE+ S++S LI C + + A+ L + T LP Y I G +
Sbjct: 194 KEMLESQVSPNVFTYNILIRGFCFAGNIDVALTLFDKMETKGCLPNVVTYNTLIDGYCKL 253
Query: 322 GRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVT 381
++D+ + R +G P + YN++I L RE R+K+V +L +MN + VT
Sbjct: 254 RKIDDGFKLLRSMA-LKGLEPNLISYNVVINGLCREGRMKEVSFVLTEMNRRGYSLDEVT 312
Query: 382 MNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSS 441
N ++ +CK G AL + + GL+P+ + Y LI ++C G A L
Sbjct: 313 YNTLIKGYCKEGNFHQALVMHAEMLRHGLTPSVITYTSLIHSMCKAGNMNRAMEFLDQMR 372
Query: 442 GTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVE 501
G P+ RT++TL + ++ ++E + +L + F P+ TY+ ++ C G++E
Sbjct: 373 VRGLCPNERTYTTLVDGFSQKGYMNEAYRVLREMNDNGFSPSVVTYNALINGHCVTGKME 432
Query: 502 DGYLMRGDLDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVL 558
D + D+ + SY+ ++ GF +S D A R+ EM EKG
Sbjct: 433 DAIAVLEDMKEKGLSPDVVSYSTVLSGFCRSYDVDEALRVKREMVEKGI----------- 481
Query: 559 HCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMT 618
KP ++S I G + A +++E M R G+
Sbjct: 482 -----------------------KPDTITYSSLIQGFCEQRRTKEACDLYEEMLRVGLPP 518
Query: 619 NASSQILVMKSYFRSRRISDALRFFND-IRHQVVVSTKLYNRMIVGLCKSDKADIALELC 677
+ + ++ +Y + AL+ N+ + V+ Y+ +I GL K + A L
Sbjct: 519 DEFTYTALINAYCMEGDLEKALQLHNEMVEKGVLPDVVTYSVLINGLNKQSRTREAKRLL 578
Query: 678 FEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLV 712
++ PS Y L++ CS + V+L+
Sbjct: 579 LKLFYEESVPSDVTYHTLIEN-CSNIEFKSVVSLI 612
Score = 110 bits (274), Expect = 4e-24
Identities = 108/513 (21%), Positives = 206/513 (40%), Gaps = 42/513 (8%)
Query: 363 VYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLI- 421
V++ L + + C + V + V+ + +L ++D AL + + G P ++Y ++
Sbjct: 119 VFKSLQETYDLCYSTSSV-FDLVVKSYSRLSLIDKALSIVHLAQAHGFMPGVLSYNAVLD 177
Query: 422 LTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFM 481
T+ A V + + P+ T++ L C ID L D + +
Sbjct: 178 ATIRSKRNISFAENVFKEMLESQVSPNVFTYNILIRGFCFAGNIDVALTLFDKMETKGCL 237
Query: 482 PNSSTYSRFVSALCRAGRVEDGY-LMRGDLDKVTAR--FSYAKMIMGFIKSNRGDIAARL 538
PN TY+ + C+ +++DG+ L+R K SY +I G + R + +
Sbjct: 238 PNVVTYNTLIDGYCKLRKIDDGFKLLRSMALKGLEPNLISYNVVINGLCREGRMKEVSFV 297
Query: 539 LVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMH 597
L EM +GY L +Y ++ N EM+ HG P + S I
Sbjct: 298 LTEMNRRGYSLDEVTYNTLIKGYCKEGNFHQALVMHAEMLRHGLTPSVITYTSLIHSMCK 357
Query: 598 ANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKL 656
A + A E + M+ G+ N + ++ + + +++A R ++ S
Sbjct: 358 AGNMNRAMEFLDQMRVRGLCPNERTYTTLVDGFSQKGYMNEAYRVLREMNDNGFSPSVVT 417
Query: 657 YNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYE 716
YN +I G C + K + A+ + +M + GL+P + Y ++ C EA+ +
Sbjct: 418 YNALINGHCVTGKMEDAIAVLEDMKEKGLSPDVVSYSTVLSGFCRSYDVDEALRVKREMV 477
Query: 717 KAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSY 776
+ G I P D+ + +I F R
Sbjct: 478 EKG----------------IKP------------------DTITYSSLIQGFCEQRRTKE 503
Query: 777 SIQELEELIAKCFPVDIYTYNLLMRKLTHH-DMDKACELFDRMCQRGLEPNRWTYGLMAH 835
+ EE++ P D +TY L+ D++KA +L + M ++G+ P+ TY ++ +
Sbjct: 504 ACDLYEEMLRVGLPPDEFTYTALINAYCMEGDLEKALQLHNEMVEKGVLPDVVTYSVLIN 563
Query: 836 GFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
G + R EAKR + ++ + P + T + +
Sbjct: 564 GLNKQSRTREAKRLLLKLFYEESVPSDVTYHTL 596
Score = 70.9 bits (172), Expect = 3e-12
Identities = 79/349 (22%), Positives = 140/349 (39%), Gaps = 19/349 (5%)
Query: 138 TFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRM 197
T+ ++ + A +FL R R + TLV G++ G + A +L M
Sbjct: 347 TYTSLIHSMCKAGNMNRAMEFLDQMRVRGLCPNERTYTTLVDGFSQKGYMNEAYRVLREM 406
Query: 198 RFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYESHMTNV-IVIKHLCKQGRL 256
G Y+ L+N + + +G + + V+ C+ +
Sbjct: 407 NDNGFSPSVVTYNALINGHCVTGKMEDAIAVLEDMKEKGLSPDVVSYSTVLSGFCRSYDV 466
Query: 257 EEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEF-GTSLPLEN-AYGVW 314
+EA +V G + S LI CE R + A +L E LP + Y
Sbjct: 467 DEALRVKREMVEKGIKPDTITYSSLIQGFCEQRRTKEACDLYEEMLRVGLPPDEFTYTAL 526
Query: 315 IRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETC 374
I G L++AL+ + + +G +P V Y++LI L +++R ++ LL+ +
Sbjct: 527 INAYCMEGDLEKALQLHNEMVE-KGVLPDVVTYSVLINGLNKQSRTREAKRLLLKLFYEE 585
Query: 375 IPPNMVTMNAVL--CF-------------FCKLGMVDVALELFNSRSQFGLSPNYMAYKY 419
P+ VT + ++ C FC GM+ A ++F S P+ AY
Sbjct: 586 SVPSDVTYHTLIENCSNIEFKSVVSLIKGFCMKGMMTEADQVFESMLGKNHKPDGTAYNI 645
Query: 420 LILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEM 468
+I C G ++AY + + +G+ T L AL +E K++E+
Sbjct: 646 MIHGHCRAGDIRKAYTLYKEMVKSGFLLHTVTVIALVKALHKEGKVNEL 694
>At5g65560 putative protein
Length = 915
Score = 140 bits (353), Expect = 3e-33
Identities = 174/777 (22%), Positives = 300/777 (38%), Gaps = 95/777 (12%)
Query: 113 DGDILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPLVFDF-LRDFRSCSFPHRA 171
D D + L F W + PR+ H+ ++ ++ +L +VF L +SC A
Sbjct: 100 DLDPKTALNFSHWISQNPRYKHSVYSYASLLTLLINNGYVGVVFKIRLLMIKSCDSVGDA 159
Query: 172 RYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAF-----D 226
Y V+ D L ++ Y+ LLNSLA + +
Sbjct: 160 LY----VLDLCRKMNKDERFELKYKLIIGC-------YNTLLNSLARFGLVDEMKQVYME 208
Query: 227 VIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLC 286
++ +++C Y + ++ CK G +EEA +++ +V +G + + LI C
Sbjct: 209 MLEDKVCPNIYTYNK----MVNGYCKLGNVEEANQYVSKIVEAGLDPDFFTYTSLIMGYC 264
Query: 287 ESNRFERAVELVSEFGTSLPL------ENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGF 340
+ + A ++ +E +PL E AY I GL R+DEA++ F + +D E F
Sbjct: 265 QRKDLDSAFKVFNE----MPLKGCRRNEVAYTHLIHGLCVARRIDEAMDLFVKMKDDECF 320
Query: 341 VPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALE 400
P Y +LI L R + L+ +M ET I PN+ T ++ C + A E
Sbjct: 321 -PTVRTYTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDSLCSQCKFEKARE 379
Query: 401 LFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALC 460
L + GL PN + Y LI C G ++A V+ P+ RT++ L C
Sbjct: 380 LLGQMLEKGLMPNVITYNALINGYCKRGMIEDAVDVVELMESRKLSPNTRTYNELIKGYC 439
Query: 461 RECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGY---LMRGDLDKVTARF 517
+ + + +L+ LER+ +P+ TY+ + CR+G + Y + D V ++
Sbjct: 440 KS-NVHKAMGVLNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQW 498
Query: 518 SYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEM 577
+Y MI KS R + A L +++KG
Sbjct: 499 TYTSMIDSLCKSKRVEEACDLFDSLEQKGV------------------------------ 528
Query: 578 MTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRIS 637
P+ ++ + IDG A K D A + E M + N+ + ++ ++
Sbjct: 529 ----NPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLK 584
Query: 638 DALRF---FNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEV 694
+A I Q VST +I L K D A +ML G P Y
Sbjct: 585 EATLLEEKMVKIGLQPTVSTDTI--LIHRLLKDGDFDHAYSRFQQMLSSGTKPDAHTYTT 642
Query: 695 LVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLL--FHSMISPEVYHSCVDLRREKE 752
+Q C R +A +++ + G F + L+ + + + R+
Sbjct: 643 FIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTG 702
Query: 753 GEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHDMDKAC 812
E + L+LI L + Y Q+ E P N++ + D
Sbjct: 703 CEPSQHTFLSLI----KHLLEMKYGKQKGSE------PELCAMSNMM-------EFDTVV 745
Query: 813 ELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKR-WVHEMLKKGFNPPENTRNVI 868
EL ++M + + PN +Y + G G A++ + H +G +P E N +
Sbjct: 746 ELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVFNAL 802
Score = 115 bits (287), Expect = 1e-25
Identities = 130/584 (22%), Positives = 233/584 (39%), Gaps = 63/584 (10%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLA-ENNCYNAFDVIANQI 232
+ +L++GY D A + M +G + Y L++ L A D+
Sbjct: 256 YTSLIMGYCQRKDLDSAFKVFNEMPLKGCRRNEVAYTHLIHGLCVARRIDEAMDLFVKMK 315
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
+ + T ++IK LC R EA + + +G + + + LI LC +FE
Sbjct: 316 DDECFPTVRTYTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDSLCSQCKFE 375
Query: 293 RAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFR------------------ 332
+A EL+ + +P Y I G + G +++A++
Sbjct: 376 KARELLGQMLEKGLMPNVITYNALINGYCKRGMIEDAVDVVELMESRKLSPNTRTYNELI 435
Query: 333 ---------------QKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPP 377
K +P V YN LI R Y LL MN+ + P
Sbjct: 436 KGYCKSNVHKAMGVLNKMLERKVLPDVVTYNSLIDGQCRSGNFDSAYRLLSLMNDRGLVP 495
Query: 378 NMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVL 437
+ T +++ CK V+ A +LF+S Q G++PN + Y LI C G EA+ +L
Sbjct: 496 DQWTYTSMIDSLCKSKRVEEACDLFDSLEQKGVNPNVVMYTALIDGYCKAGKVDEAHLML 555
Query: 438 RSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRA 497
P+ TF+ L + LC + K+ E L + ++ P ST + + L +
Sbjct: 556 EKMLSKNCLPNSLTFNALIHGLCADGKLKEATLLEEKMVKIGLQPTVSTDTILIHRLLKD 615
Query: 498 GRVEDGY----LMRGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSS 553
G + Y M K A +Y I + + R A ++ +M+E G +
Sbjct: 616 GDFDHAYSRFQQMLSSGTKPDAH-TYTTFIQTYCREGRLLDAEDMMAKMRENGVSPDLFT 674
Query: 554 YRHVLHCLLHMDNPRTRF-FNLLEMM--THGKPHCDIFNSFIDGAMHANKPDLAREVFEL 610
Y ++ + D +T F F++L+ M T +P F S I + + E+
Sbjct: 675 YSSLIKG--YGDLGQTNFAFDVLKRMRDTGCEPSQHTFLSLI------------KHLLEM 720
Query: 611 MQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKA 670
+ G + ++ M + + + L + H V + K Y ++I+G+C+
Sbjct: 721 --KYGKQKGSEPELCAMSNMMEFDTVVELLE--KMVEHSVTPNAKSYEKLILGICEVGNL 776
Query: 671 DIALELCFEMLK-VGLNPSIECYEVLVQKLCSLKRYYEAVNLVN 713
+A ++ M + G++PS + L+ C LK++ EA +V+
Sbjct: 777 RVAEKVFDHMQRNEGISPSELVFNALLSCCCKLKKHNEAAKVVD 820
Score = 107 bits (267), Expect = 3e-23
Identities = 125/589 (21%), Positives = 231/589 (38%), Gaps = 58/589 (9%)
Query: 133 YHTRTTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALH 192
+ T T+ + + L + + + +++ + L+ K + A
Sbjct: 320 FPTVRTYTVLIKSLCGSERKSEALNLVKEMEETGIKPNIHTYTVLIDSLCSQCKFEKARE 379
Query: 193 LLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYESHMTNVIVIKHLCK 252
LLG+M +GL + Y+ L+N C RG +V+ L +
Sbjct: 380 LLGQMLEKGLMPNVITYNALING----------------YCKRGMIEDAVDVV---ELME 420
Query: 253 QGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTSLPLENAYG 312
+L N L+ K +S + +GVL + ER V LP Y
Sbjct: 421 SRKLSPNTRTYNELI---KGYCKSNVHKAMGVL--NKMLERKV---------LPDVVTYN 466
Query: 313 VWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNE 372
I G + G D A D G VP + Y +I L + R+++ +L + +
Sbjct: 467 SLIDGQCRSGNFDSAYRLLSLMND-RGLVPDQWTYTSMIDSLCKSKRVEEACDLFDSLEQ 525
Query: 373 TCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKE 432
+ PN+V A++ +CK G VD A + PN + + LI LC DG KE
Sbjct: 526 KGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLKE 585
Query: 433 AYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVS 492
A + G P T + L + L ++ D + L P++ TY+ F+
Sbjct: 586 ATLLEEKMVKIGLQPTVSTDTILIHRLLKDGDFDHAYSRFQQMLSSGTKPDAHTYTTFIQ 645
Query: 493 ALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYEL 549
CR GR+ D M + + F+Y+ +I G+ + + A +L M++ G E
Sbjct: 646 TYCREGRLLDAEDMMAKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCEP 705
Query: 550 KRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFE 609
+ ++ ++ LL M + + +P ++ ++ D E+ E
Sbjct: 706 SQHTFLSLIKHLLEMKYGKQK---------GSEPELCAMSNMMEF-------DTVVELLE 749
Query: 610 LMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTK--LYNRMIVGLCKS 667
M + + NA S ++ + A + F+ ++ +S ++N ++ CK
Sbjct: 750 KMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVFNALLSCCCKL 809
Query: 668 DKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYE 716
K + A ++ +M+ VG P +E +VL+ C L + E +V++
Sbjct: 810 KKHNEAAKVVDDMICVGHLPQLESCKVLI---CGLYKKGEKERGTSVFQ 855
Score = 79.7 bits (195), Expect = 6e-15
Identities = 92/465 (19%), Positives = 193/465 (40%), Gaps = 30/465 (6%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
+++L+ G +G D A LL M +GL D + Y +++SL ++ + + +
Sbjct: 465 YNSLIDGQCRSGNFDSAYRLLSLMNDRGLVPDQWTYTSMIDSLCKSKRVEEACDLFDSLE 524
Query: 234 MRGYESHMTN-VIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
+G ++ +I CK G+++EA L ++ + + LI LC + +
Sbjct: 525 QKGVNPNVVMYTALIDGYCKAGKVDEAHLMLEKMLSKNCLPNSLTFNALIHGLCADGKLK 584
Query: 293 RAV---ELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNI 349
A E + + G P + + I L++ G D A F+Q S G P Y
Sbjct: 585 EATLLEEKMVKIGLQ-PTVSTDTILIHRLLKDGDFDHAYSRFQQMLSS-GTKPDAHTYTT 642
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFG 409
I RE RL D +++ M E + P++ T ++++ + LG + A ++ G
Sbjct: 643 FIQTYCREGRLLDAEDMMAKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTG 702
Query: 410 LSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMW 469
P+ + LI L ++ G P+ S + + D +
Sbjct: 703 CEPSQHTFLSLIKHLL----------EMKYGKQKGSEPELCAMSNMM-------EFDTVV 745
Query: 470 DLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK----VTARFSYAKMIMG 525
+LL+ +E PN+ +Y + + +C G + + + + + + ++
Sbjct: 746 ELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDHMQRNEGISPSELVFNALLSC 805
Query: 526 FIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHC 585
K + + AA+++ +M G+ + S + VL C L+ + R ++ + + +
Sbjct: 806 CCKLKKHNEAAKVVDDMICVGHLPQLESCK-VLICGLYKKGEKERGTSVFQNLLQCGYYE 864
Query: 586 D--IFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMK 628
D + IDG + E+F +M++NG ++ + L+++
Sbjct: 865 DELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYSLLIE 909
Score = 42.4 bits (98), Expect = 0.001
Identities = 72/362 (19%), Positives = 131/362 (35%), Gaps = 63/362 (17%)
Query: 99 SETFVLRVLRHGGDDGDILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPL-VFD 157
++T ++ L GD S + +G +P HT TTF+ + C R L D
Sbjct: 604 TDTILIHRLLKDGDFDHAYSRFQQMLSSGTKPDA-HTYTTFIQTY----CREGRLLDAED 658
Query: 158 FLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLA 217
+ R + +L+ GY G+ + A +L RMR G +
Sbjct: 659 MMAKMRENGVSPDLFTYSSLIKGYGDLGQTNFAFDVLKRMRDTGCE-------------- 704
Query: 218 ENNCYNAFDVIANQICMRGYESHMTNVIVIKHL--CKQGRLEEAEAHLNGLVGSGKELHR 275
S T + +IKHL K G+ + +E L
Sbjct: 705 --------------------PSQHTFLSLIKHLLEMKYGKQKGSEPEL------------ 732
Query: 276 SELSFLIGVLCESNRFERAVELVSEF--GTSLPLENAYGVWIRGLVQGGRLDEALEFFRQ 333
+ F+ VEL+ + + P +Y I G+ + G L A + F
Sbjct: 733 -------CAMSNMMEFDTVVELLEKMVEHSVTPNAKSYEKLILGICEVGNLRVAEKVFDH 785
Query: 334 KRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLG 393
+ +EG P ++ +N L+ + + + +++ DM P + + ++C K G
Sbjct: 786 MQRNEGISPSELVFNALLSCCCKLKKHNEAAKVVDDMICVGHLPQLESCKVLICGLYKKG 845
Query: 394 MVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFS 453
+ +F + Q G + +A+K +I + G + Y + G +T+S
Sbjct: 846 EKERGTSVFQNLLQCGYYEDELAWKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYS 905
Query: 454 TL 455
L
Sbjct: 906 LL 907
>At5g12100 unknown protein (At5g12100)
Length = 816
Score = 140 bits (353), Expect = 3e-33
Identities = 142/607 (23%), Positives = 264/607 (43%), Gaps = 28/607 (4%)
Query: 278 LSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFF-RQK 334
L+ L+ L ++ +F + + S P + YG I+ V+ + + LE F R K
Sbjct: 147 LTLLLDHLVKTKQFRVTINVFLNILESDFRPSKFMYGKAIQAAVKLSDVGKGLELFNRMK 206
Query: 335 RDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGM 394
D P YN+LI L + R+ D +L +M + P+++T N ++ +CK G
Sbjct: 207 HDR--IYPSVFIYNVLIDGLCKGKRMNDAEQLFDEMLARRLLPSLITYNTLIDGYCKAGN 264
Query: 395 VDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFST 454
+ + ++ + P+ + + L+ L G ++A VL+ G+ PD TFS
Sbjct: 265 PEKSFKVRERMKADHIEPSLITFNTLLKGLFKAGMVEDAENVLKEMKDLGFVPDAFTFSI 324
Query: 455 LANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRG-DLDK- 512
L + K + + + A++ N+ T S ++ALC+ G++E + G ++ K
Sbjct: 325 LFDGYSSNEKAEAALGVYETAVDSGVKMNAYTCSILLNALCKEGKIEKAEEILGREMAKG 384
Query: 513 -VTARFSYAKMIMGFIKSNRGD-IAARLLVE-MKEKGYELKRSSYRHVLHCLLHMDNPRT 569
V Y MI G+ + +GD + AR+ +E M+++G + +Y ++ +
Sbjct: 385 LVPNEVIYNTMIDGYCR--KGDLVGARMKIEAMEKQGMKPDHLAYNCLIRRFCELGEMEN 442
Query: 570 RFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMK 628
+ +M G P + +N I G + D ++ + M+ NG M N S ++
Sbjct: 443 AEKEVNKMKLKGVSPSVETYNILIGGYGRKYEFDKCFDILKEMEDNGTMPNVVSYGTLIN 502
Query: 629 SYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNP 687
+ ++ +A D+ + V ++YN +I G C K + A EMLK G+
Sbjct: 503 CLCKGSKLLEAQIVKRDMEDRGVSPKVRIYNMLIDGCCSKGKIEDAFRFSKEMLKKGIEL 562
Query: 688 SIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDL 747
++ Y L+ L + EA +L+ + G + F N L+ + V C+ L
Sbjct: 563 NLVTYNTLIDGLSMTGKLSEAEDLLLEISRKGLKPDVFTYNSLISGYGFAGNV-QRCIAL 621
Query: 748 RREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCF-----PVDIYTYN-LLMR 801
E + + ++ T L +S +E EL + F D+ YN +L
Sbjct: 622 YEEMKRSGIKPTLKTY-------HLLISLCTKEGIELTERLFGEMSLKPDLLVYNGVLHC 674
Query: 802 KLTHHDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPP 861
H DM+KA L +M ++ + ++ TY + G G+ E + + EM + P
Sbjct: 675 YAVHGDMEKAFNLQKQMIEKSIGLDKTTYNSLILGQLKVGKLCEVRSLIDEMNAREMEPE 734
Query: 862 ENTRNVI 868
+T N+I
Sbjct: 735 ADTYNII 741
Score = 109 bits (272), Expect = 8e-24
Identities = 131/615 (21%), Positives = 239/615 (38%), Gaps = 50/615 (8%)
Query: 191 LHLLGRMRFQGLDLDGFGYHILLNSLAENNCYN-AFDVIANQICMRGYESHMTNVIVIKH 249
L L RM+ + F Y++L++ L + N A + + R S +T +I
Sbjct: 199 LELFNRMKHDRIYPSVFIYNVLIDGLCKGKRMNDAEQLFDEMLARRLLPSLITYNTLIDG 258
Query: 250 LCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGT--SLPL 307
CK G E++ + E + L+ L ++ E A ++ E +P
Sbjct: 259 YCKAGNPEKSFKVRERMKADHIEPSLITFNTLLKGLFKAGMVEDAENVLKEMKDLGFVPD 318
Query: 308 ENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELL 367
+ + G + + AL + DS G +IL+ L +E +++ E+L
Sbjct: 319 AFTFSILFDGYSSNEKAEAALGVYETAVDS-GVKMNAYTCSILLNALCKEGKIEKAEEIL 377
Query: 368 MDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWD 427
+ PN V N ++ +C+ G + A + + G+ P+++AY LI C
Sbjct: 378 GREMAKGLVPNEVIYNTMIDGYCRKGDLVGARMKIEAMEKQGMKPDHLAYNCLIRRFCEL 437
Query: 428 GCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTY 487
G + A + + G P T++ L R+ + D+ +D+L + MPN +Y
Sbjct: 438 GEMENAEKEVNKMKLKGVSPSVETYNILIGGYGRKYEFDKCFDILKEMEDNGTMPNVVSY 497
Query: 488 SRFVSALCRAGRVEDGYLMRGDLDK--VTARFS-YAKMIMGFIKSNRGDIAARLLVEMKE 544
++ LC+ ++ + +++ D++ V+ + Y +I G + + A R EM +
Sbjct: 498 GTLINCLCKGSKLLEAQIVKRDMEDRGVSPKVRIYNMLIDGCCSKGKIEDAFRFSKEMLK 557
Query: 545 KGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDL 603
KG EL +Y ++ L LLE+ G KP +NS I G A
Sbjct: 558 KGIELNLVTYNTLIDGLSMTGKLSEAEDLLLEISRKGLKPDVFTYNSLISGYGFAGNVQR 617
Query: 604 AREVFELMQRNGIMTNASSQILVM-----------KSYFRSRRISDALRFFNDIRH---- 648
++E M+R+GI + L++ + F + L +N + H
Sbjct: 618 CIALYEEMKRSGIKPTLKTYHLLISLCTKEGIELTERLFGEMSLKPDLLVYNGVLHCYAV 677
Query: 649 -----------------QVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIEC 691
+ + YN +I+G K K L EM + P +
Sbjct: 678 HGDMEKAFNLQKQMIEKSIGLDKTTYNSLILGQLKVGKLCEVRSLIDEMNAREMEPEADT 737
Query: 692 YEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREK 751
Y ++V+ C +K Y A ++ G L +GN E+ + R K
Sbjct: 738 YNIIVKGHCEVKDYMSAYVWYREMQEKGFLLDVCIGN----------ELVSGLKEEWRSK 787
Query: 752 EGEFLDSSMLTLIIG 766
E E + S M ++G
Sbjct: 788 EAEIVISEMNGRMLG 802
Score = 102 bits (255), Expect = 7e-22
Identities = 131/638 (20%), Positives = 257/638 (39%), Gaps = 75/638 (11%)
Query: 247 IKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTSLP 306
++ L +Q R+E A L+ L+ S +S F EL S F S P
Sbjct: 62 LRVLLQQNRIETARGVLSSLLRS-----------------DSTPFASPKELFSAFSLSSP 104
Query: 307 -LENAYGVWIRGLV--QGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDV 363
L++ + + ++ + + EA + F R+ EG P +L+ L++ + +
Sbjct: 105 SLKHDFSYLLLSVLLNESKMISEAADLFFALRN-EGIYPSSDSLTLLLDHLVKTKQFRVT 163
Query: 364 YELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILT 423
+ +++ E+ P+ + KL V LELFN + P+ Y LI
Sbjct: 164 INVFLNILESDFRPSKFMYGKAIQAAVKLSDVGKGLELFNRMKHDRIYPSVFIYNVLIDG 223
Query: 424 LCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPN 483
LC +A ++ P T++TL + C+ ++ + + + P+
Sbjct: 224 LCKGKRMNDAEQLFDEMLARRLLPSLITYNTLIDGYCKAGNPEKSFKVRERMKADHIEPS 283
Query: 484 SSTYSRFVSALCRAGRVEDGYLM---RGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLV 540
T++ + L +AG VED + DL V F+++ + G+ + + + A +
Sbjct: 284 LITFNTLLKGLFKAGMVEDAENVLKEMKDLGFVPDAFTFSILFDGYSSNEKAEAALGVYE 343
Query: 541 EMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHAN 599
+ G ++ + +L+ L L M G P+ I+N+ IDG +
Sbjct: 344 TAVDSGVKMNAYTCSILLNALCKEGKIEKAEEILGREMAKGLVPNEVIYNTMIDG--YCR 401
Query: 600 KPDL--AREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKL 656
K DL AR E M++ G+ + + +++ + + +A + N ++ + V S +
Sbjct: 402 KGDLVGARMKIEAMEKQGMKPDHLAYNCLIRRFCELGEMENAEKEVNKMKLKGVSPSVET 461
Query: 657 YNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYE 716
YN +I G + + D ++ EM G P++ Y L+ LC + EA + E
Sbjct: 462 YNILIGGYGRKYEFDKCFDILKEMEDNGTMPNVVSYGTLINCLCKGSKLLEAQIVKRDME 521
Query: 717 KAGRRLTSFLGNVLLFHSMISPEV--YHSCVDLRREKEGEFLDSSMLTLIIGAFSGCL-- 772
G +SP+V Y+ +D GC
Sbjct: 522 DRG----------------VSPKVRIYNMLID-----------------------GCCSK 542
Query: 773 -RVSYSIQELEELIAKCFPVDIYTYNLLMRKLT-HHDMDKACELFDRMCQRGLEPNRWTY 830
++ + + +E++ K +++ TYN L+ L+ + +A +L + ++GL+P+ +TY
Sbjct: 543 GKIEDAFRFSKEMLKKGIELNLVTYNTLIDGLSMTGKLSEAEDLLLEISRKGLKPDVFTY 602
Query: 831 GLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
+ G+ G EM + G P T +++
Sbjct: 603 NSLISGYGFAGNVQRCIALYEEMKRSGIKPTLKTYHLL 640
Score = 87.8 bits (216), Expect = 2e-17
Identities = 115/528 (21%), Positives = 207/528 (38%), Gaps = 53/528 (10%)
Query: 159 LRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAE 218
L++ + F A L GY+ K + AL + G+ ++ + ILLN+
Sbjct: 307 LKEMKDLGFVPDAFTFSILFDGYSSNEKAEAALGVYETAVDSGVKMNAYTCSILLNA--- 363
Query: 219 NNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSEL 278
LCK+G++E+AE L + G +
Sbjct: 364 -------------------------------LCKEGKIEKAEEILGREMAKGLVPNEVIY 392
Query: 279 SFLIGVLCESNRFERA---VELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKR 335
+ +I C A +E + + G P AY IR + G ++ A E K
Sbjct: 393 NTMIDGYCRKGDLVGARMKIEAMEKQGMK-PDHLAYNCLIRRFCELGEMENA-EKEVNKM 450
Query: 336 DSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMV 395
+G P YNILIG R+ +++L +M + PN+V+ ++ CK +
Sbjct: 451 KLKGVSPSVETYNILIGGYGRKYEFDKCFDILKEMEDNGTMPNVVSYGTLINCLCKGSKL 510
Query: 396 DVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTL 455
A + G+SP Y LI C G ++A+R + G + T++TL
Sbjct: 511 LEAQIVKRDMEDRGVSPKVRIYNMLIDGCCSKGKIEDAFRFSKEMLKKGIELNLVTYNTL 570
Query: 456 ANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK--V 513
+ L K+ E DLL + P+ TY+ +S AG V+ + ++ + +
Sbjct: 571 IDGLSMTGKLSEAEDLLLEISRKGLKPDVFTYNSLISGYGFAGNVQRCIALYEEMKRSGI 630
Query: 514 TARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHC-LLHMDNPRTRFF 572
+++ ++ RL EM K L Y VLHC +H D + F
Sbjct: 631 KPTLKTYHLLISLCTKEGIELTERLFGEMSLKPDLL---VYNGVLHCYAVHGDMEKA--F 685
Query: 573 NLLEMMTHGKPHCD--IFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSY 630
NL + M D +NS I G + K R + + M + A + +++K +
Sbjct: 686 NLQKQMIEKSIGLDKTTYNSLILGQLKVGKLCEVRSLIDEMNAREMEPEADTYNIIVKGH 745
Query: 631 FRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGL---CKSDKADIAL 674
+ A ++ +++ + ++ + N ++ GL +S +A+I +
Sbjct: 746 CEVKDYMSAYVWYREMQEKGFLLDVCIGNELVSGLKEEWRSKEAEIVI 793
>At1g12300 hypothetical protein
Length = 637
Score = 139 bits (351), Expect = 5e-33
Identities = 134/587 (22%), Positives = 234/587 (39%), Gaps = 76/587 (12%)
Query: 323 RLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTM 382
+ D+A++ FR S +P + ++ L + + + V L M I N+ T+
Sbjct: 68 KADDAIDLFRDMIHSRP-LPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYTL 126
Query: 383 NAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSG 442
+ ++ FC+ + +A + G PN + + LI LC +G EA ++
Sbjct: 127 SIMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMVE 186
Query: 443 TGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGR--- 499
G+ PD T +TL N LC K E L+D +E PN+ TY ++ +C++G+
Sbjct: 187 MGHKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQTAL 246
Query: 500 ----------------------VEDGYLMRGDLDK-------------VTARFSYAKMIM 524
+ DG G LD T +Y +I
Sbjct: 247 AMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKGITTNIITYNILIG 306
Query: 525 GFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KP 583
GF + R D A+LL +M ++ ++ ++ + R EM+ G P
Sbjct: 307 GFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLREAEELHKEMIHRGIAP 366
Query: 584 HCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFF 643
+ S IDG N D A ++ +LM G N + +++ Y ++ RI D L F
Sbjct: 367 DTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILINGYCKANRIDDGLELF 426
Query: 644 NDIR-HQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSL 702
+ VV T YN +I G C+ K ++A EL EM+ + P+I Y++L+ LC
Sbjct: 427 RKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPPNIVTYKILLDGLCD- 485
Query: 703 KRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSMLT 762
E+ + ++EK EK LD +
Sbjct: 486 --NGESEKALEIFEKI-------------------------------EKSKMELDIGIYN 512
Query: 763 LIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHH-DMDKACELFDRMCQR 821
+II +V + L K + TYN+++ L + +A LF +M +
Sbjct: 513 IIIHGMCNASKVDDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEED 572
Query: 822 GLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
G P+ WTY ++ G ++ + + E+ + GF+ +T ++
Sbjct: 573 GHAPDGWTYNILIRAHLGDGDATKSVKLIEELKRCGFSVDASTIKMV 619
Score = 129 bits (325), Expect = 5e-30
Identities = 129/589 (21%), Positives = 246/589 (40%), Gaps = 62/589 (10%)
Query: 140 VAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRF 199
+ +FR + +R P V DF R F + A + D+ L L +M
Sbjct: 73 IDLFRDMIHSRPLPTVIDFSRLFSAI----------------AKTKQYDLVLALCKQMEL 116
Query: 200 QGLDLDGFGYHILLNSLAE-NNCYNAFDVIANQICMRGYESH-MTNVIVIKHLCKQGRLE 257
+G+ + + I++N AF + +I GYE + +T +I LC +GR+
Sbjct: 117 KGIAHNLYTLSIMINCFCRCRKLCLAFSAMG-KIIKLGYEPNTITFSTLINGLCLEGRVS 175
Query: 258 EAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVS---EFGTSLPLENAYGVW 314
EA ++ +V G + ++ L+ LC S + A+ L+ E+G P YG
Sbjct: 176 EALELVDRMVEMGHKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQ-PNAVTYGPV 234
Query: 315 IRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETC 374
+ + + G+ A+E R K + V+Y+I+I L + L + + L +M
Sbjct: 235 LNVMCKSGQTALAMELLR-KMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKG 293
Query: 375 IPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAY 434
I N++T N ++ FC G D +L + ++PN + + LI + +G +EA
Sbjct: 294 ITTNIITYNILIGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLREAE 353
Query: 435 RVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSAL 494
+ + G PD T+++L + C+E +D+ ++D + + PN T++ ++
Sbjct: 354 ELHKEMIHRGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILINGY 413
Query: 495 CRAGRVEDGYLMRGDLD---KVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKR 551
C+A R++DG + + V +Y +I GF + + ++A L EM +
Sbjct: 414 CKANRIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKV---- 469
Query: 552 SSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELM 611
P+ + +DG + + A E+FE +
Sbjct: 470 ------------------------------PPNIVTYKILLDGLCDNGESEKALEIFEKI 499
Query: 612 QRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKA 670
+++ + + +++ + ++ DA F + + V K YN MI GLCK
Sbjct: 500 EKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPGVKTYNIMIGGLCKKGPL 559
Query: 671 DIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAG 719
A L +M + G P Y +L++ ++V L+ ++ G
Sbjct: 560 SEAELLFRKMEEDGHAPDGWTYNILIRAHLGDGDATKSVKLIEELKRCG 608
Score = 115 bits (289), Expect = 8e-26
Identities = 95/386 (24%), Positives = 177/386 (45%), Gaps = 11/386 (2%)
Query: 184 AGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCY-NAFDVIANQICMRGYESHM- 241
+G+ +A+ LL +M + + LD Y I+++ L ++ NAF++ N++ M+G +++
Sbjct: 241 SGQTALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLF-NEMEMKGITTNII 299
Query: 242 TNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEF 301
T I+I C GR ++ L ++ + S LI + + A EL E
Sbjct: 300 TYNILIGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLREAEELHKEM 359
Query: 302 ---GTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLREN 358
G + P Y I G + LD+A + S+G P +NILI + N
Sbjct: 360 IHRGIA-PDTITYTSLIDGFCKENHLDKANQMV-DLMVSKGCDPNIRTFNILINGYCKAN 417
Query: 359 RLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYK 418
R+ D EL M+ + + VT N ++ FC+LG ++VA ELF + PN + YK
Sbjct: 418 RIDDGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPPNIVTYK 477
Query: 419 YLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALER 478
L+ LC +G ++A + + D ++ + + +C K+D+ WDL +
Sbjct: 478 ILLDGLCDNGESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLK 537
Query: 479 RFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKSNRGDIA 535
P TY+ + LC+ G + + L+ +++ ++Y +I + +
Sbjct: 538 GVKPGVKTYNIMIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNILIRAHLGDGDATKS 597
Query: 536 ARLLVEMKEKGYELKRSSYRHVLHCL 561
+L+ E+K G+ + S+ + V+ L
Sbjct: 598 VKLIEELKRCGFSVDASTIKMVIDML 623
Score = 77.4 bits (189), Expect = 3e-14
Identities = 89/410 (21%), Positives = 172/410 (41%), Gaps = 15/410 (3%)
Query: 462 ECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLD-KVTARFSYA 520
+ K D+ DL + R +P +SR SA+ + + + + ++ K A Y
Sbjct: 66 DIKADDAIDLFRDMIHSRPLPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYT 125
Query: 521 KMIM--GFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH--CLLHMDNPRTRFFNLLE 576
IM F + + +A + ++ + GYE ++ +++ CL + + +
Sbjct: 126 LSIMINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMV 185
Query: 577 MMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRI 636
M H KP N+ ++G + K A + + M G NA + V+ +S +
Sbjct: 186 EMGH-KPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQT 244
Query: 637 SDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVL 695
+ A+ + + + + Y+ +I GLCK D A L EM G+ +I Y +L
Sbjct: 245 ALAMELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKGITTNIITYNIL 304
Query: 696 VQKLCSLKRYYEAVNLVN--VYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREK-- 751
+ C+ R+ + L+ + K + +F +L S + +L +E
Sbjct: 305 IGGFCNAGRWDDGAKLLRDMIKRKINPNVVTFS---VLIDSFVKEGKLREAEELHKEMIH 361
Query: 752 EGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDK 810
G D+ T +I F + + Q ++ +++K +I T+N+L+ + +D
Sbjct: 362 RGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILINGYCKANRIDD 421
Query: 811 ACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNP 860
ELF +M RG+ + TY + GF G+ + AK EM+ + P
Sbjct: 422 GLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPP 471
>At1g64100 hypothetical protein
Length = 806
Score = 138 bits (347), Expect = 2e-32
Identities = 129/537 (24%), Positives = 227/537 (42%), Gaps = 23/537 (4%)
Query: 186 KPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYESHMTNVI 245
+PD+A+ L +M + + L+ + ++IL+ + + + ++ G++ +
Sbjct: 121 RPDVAISLYRKMEIRRIPLNIYSFNILIKCFCDCHKLSFSLSTFGKLTKLGFQPDVVTFN 180
Query: 246 VIKH-LCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS 304
+ H LC + R+ EA A +V +G FL V F++ VE+
Sbjct: 181 TLLHGLCLEDRISEALALFGYMVETG---------FLEAVAL----FDQMVEI-----GL 222
Query: 305 LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVY 364
P+ + I GL GR+ EA K +G V Y ++ + + K
Sbjct: 223 TPVVITFNTLINGLCLEGRVLEAAALVN-KMVGKGLHIDVVTYGTIVNGMCKMGDTKSAL 281
Query: 365 ELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTL 424
LL M ET I P++V +A++ CK G A LF+ + G++PN Y +I
Sbjct: 282 NLLSKMEETHIKPDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEKGIAPNVFTYNCMIDGF 341
Query: 425 CWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNS 484
C G +A R+LR PD TF+ L +A +E K+ E L D L R P++
Sbjct: 342 CSFGRWSDAQRLLRDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEMLHRCIFPDT 401
Query: 485 STYSRFVSALCRAGRVEDGYLMRGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKE 544
TY+ + C+ R +D M DL ++ +I + ++ R D +LL E+
Sbjct: 402 VTYNSMIYGFCKHNRFDDAKHM-FDLMASPDVVTFNTIIDVYCRAKRVDEGMQLLREISR 460
Query: 545 KGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGK-PHCDIFNSFIDGAMHANKPDL 603
+G ++Y ++H +DN EM++HG P N + G K +
Sbjct: 461 RGLVANTTTYNTLIHGFCEVDNLNAAQDLFQEMISHGVCPDTITCNILLYGFCENEKLEE 520
Query: 604 AREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIR-HQVVVSTKLYNRMIV 662
A E+FE++Q + I + + +++ + ++ +A F + H V + YN MI
Sbjct: 521 ALELFEVIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGVEPDVQTYNVMIS 580
Query: 663 GLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAG 719
G C A L +M G P Y L++ +++ L++ G
Sbjct: 581 GFCGKSAISDANVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIELISEMRSNG 637
Score = 130 bits (328), Expect = 2e-30
Identities = 147/588 (25%), Positives = 243/588 (41%), Gaps = 69/588 (11%)
Query: 281 LIGVLCESNRFERAVELVSEFGTS-LPLE-NAYGVWIRGLVQGGRLDEALEFFRQKRDSE 338
+IGV NR + A+ L + +PL ++ + I+ +L +L F K
Sbjct: 112 VIGVFVRMNRPDVAISLYRKMEIRRIPLNIYSFNILIKCFCDCHKLSFSLSTFG-KLTKL 170
Query: 339 GFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVA 398
GF P V +N L+ L E+R+ + L M ET G ++ A
Sbjct: 171 GFQPDVVTFNTLLHGLCLEDRISEALALFGYMVET-------------------GFLE-A 210
Query: 399 LELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANA 458
+ LF+ + GL+P + + LI LC +G EA ++ G G D T+ T+ N
Sbjct: 211 VALFDQMVEIGLTPVVITFNTLINGLCLEGRVLEAAALVNKMVGKGLHIDVVTYGTIVNG 270
Query: 459 LCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDG-YLMRGDLDKVTAR- 516
+C+ +LL E P+ YS + LC+ G D YL L+K A
Sbjct: 271 MCKMGDTKSALNLLSKMEETHIKPDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEKGIAPN 330
Query: 517 -FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLL 575
F+Y MI GF R A RLL +M E+ ++ ++ + + L
Sbjct: 331 VFTYNCMIDGFCSFGRWSDAQRLLRDMIEREINPDVLTFNALISASVK-EGKLFEAEKLC 389
Query: 576 EMMTHGK--PHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRS 633
+ M H P +NS I G N+ D A+ +F+LM ++T + ++ Y R+
Sbjct: 390 DEMLHRCIFPDTVTYNSMIYGFCKHNRFDDAKHMFDLMASPDVVTFNT----IIDVYCRA 445
Query: 634 RRISDALRFFNDI-RHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECY 692
+R+ + ++ +I R +V +T YN +I G C+ D + A +L EM+ G+ P
Sbjct: 446 KRVDEGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQDLFQEMISHGVCPDTITC 505
Query: 693 EVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKE 752
+L+ C ++ EA+ L V + + L + N+++ H M + K
Sbjct: 506 NILLYGFCENEKLEEALELFEVIQMSKIDLDTVAYNIII-HGM-----------CKGSKV 553
Query: 753 GEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDKA 811
E D L S I +E D+ TYN+++ + A
Sbjct: 554 DEAWD--------------LFCSLPIHGVEP--------DVQTYNVMISGFCGKSAISDA 591
Query: 812 CELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFN 859
LF +M G EP+ TY + G G D++ + EM GF+
Sbjct: 592 NVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIELISEMRSNGFS 639
Score = 115 bits (289), Expect = 8e-26
Identities = 119/567 (20%), Positives = 224/567 (38%), Gaps = 62/567 (10%)
Query: 324 LDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMN 383
LD+A++FF S F V N +IG +R NR L M IP N+ + N
Sbjct: 87 LDDAIDFFDYMVRSRPFYTA-VDCNKVIGVFVRMNRPDVAISLYRKMEIRRIPLNIYSFN 145
Query: 384 AVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGT 443
++ FC + +L F ++ G P+ + + L+ LC + EA + T
Sbjct: 146 ILIKCFCDCHKLSFSLSTFGKLTKLGFQPDVVTFNTLLHGLCLEDRISEALALFGYMVET 205
Query: 444 GYF---------------PDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYS 488
G+ P TF+TL N LC E ++ E L++ + + + TY
Sbjct: 206 GFLEAVALFDQMVEIGLTPVVITFNTLINGLCLEGRVLEAAALVNKMVGKGLHIDVVTYG 265
Query: 489 RFVSALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEK 545
V+ +C+ G + + +++ + Y+ +I K A L EM EK
Sbjct: 266 TIVNGMCKMGDTKSALNLLSKMEETHIKPDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEK 325
Query: 546 GYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAR 605
G P+ +N IDG + A+
Sbjct: 326 GIA----------------------------------PNVFTYNCMIDGFCSFGRWSDAQ 351
Query: 606 EVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVV-STKLYNRMIVGL 664
+ M I + + ++ + + ++ +A + +++ H+ + T YN MI G
Sbjct: 352 RLLRDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEMLHRCIFPDTVTYNSMIYGF 411
Query: 665 CKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTS 724
CK ++ D A + F+++ +P + + ++ C KR E + L+ + G +
Sbjct: 412 CKHNRFDDAKHM-FDLMA---SPDVVTFNTIIDVYCRAKRVDEGMQLLREISRRGLVANT 467
Query: 725 FLGNVLLFHSMISPEVYHSCVDLRRE--KEGEFLDSSMLTLIIGAFSGCLRVSYSIQELE 782
N L+ H + ++ DL +E G D+ +++ F ++ +++ E
Sbjct: 468 TTYNTLI-HGFCEVDNLNAAQDLFQEMISHGVCPDTITCNILLYGFCENEKLEEALELFE 526
Query: 783 ELIAKCFPVDIYTYNLLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHG 841
+ +D YN+++ + +D+A +LF + G+EP+ TY +M GF
Sbjct: 527 VIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGVEPDVQTYNVMISGFCGKS 586
Query: 842 RKDEAKRWVHEMLKKGFNPPENTRNVI 868
+A H+M G P +T N +
Sbjct: 587 AISDANVLFHKMKDNGHEPDNSTYNTL 613
Score = 89.4 bits (220), Expect = 8e-18
Identities = 111/506 (21%), Positives = 199/506 (38%), Gaps = 64/506 (12%)
Query: 175 DTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICM 234
+TL+ G + G+ A L+ +M +GL +D Y ++N + C A +
Sbjct: 230 NTLINGLCLEGRVLEAAALVNKMVGKGLHIDVVTYGTIVNGM----CKMGDTKSALNLLS 285
Query: 235 RGYESHMTNVIV-----IKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESN 289
+ E+H+ +V I LCK G H S+ +L + E
Sbjct: 286 KMEETHIKPDVVIYSAIIDRLCKDG-------------------HHSDAQYLFSEMLEKG 326
Query: 290 RFERAVELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNI 349
P Y I G GR +A R + E P + +N
Sbjct: 327 --------------IAPNVFTYNCMIDGFCSFGRWSDAQRLLRDMIERE-INPDVLTFNA 371
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFG 409
LI ++E +L + +L +M CI P+ VT N+++ FCK D A +F+ +
Sbjct: 372 LISASVKEGKLFEAEKLCDEMLHRCIFPDTVTYNSMIYGFCKHNRFDDAKHMFDLMA--- 428
Query: 410 LSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMW 469
SP+ + + +I C E ++LR S G + T++TL + C ++
Sbjct: 429 -SPDVVTFNTIIDVYCRAKRVDEGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQ 487
Query: 470 DLLDFALERRFMPNSSTYSRFVSALCRAGRVEDG-------YLMRGDLDKVTARFSYAKM 522
DL + P++ T + + C ++E+ + + DLD V +Y +
Sbjct: 488 DLFQEMISHGVCPDTITCNILLYGFCENEKLEEALELFEVIQMSKIDLDTV----AYNII 543
Query: 523 IMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVL--HCLLHMDNPRTRFFNLLEMMTH 580
I G K ++ D A L + G E +Y ++ C + F+ ++ H
Sbjct: 544 IHGMCKGSKVDEAWDLFCSLPIHGVEPDVQTYNVMISGFCGKSAISDANVLFHKMKDNGH 603
Query: 581 GKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDAL 640
+P +N+ I G + A + D + E+ M+ NG +A + + + R+SD
Sbjct: 604 -EPDNSTYNTLIRGCLKAGEIDKSIELISEMRSNGFSGDAFTIKMAEEIIC---RVSDEE 659
Query: 641 RFFNDIRHQVVVSTKLYNRMIVGLCK 666
N +R ++ T R +V L +
Sbjct: 660 IIENYLRPKINGETSSIPRYVVELAE 685
Score = 82.8 bits (203), Expect = 8e-16
Identities = 74/331 (22%), Positives = 139/331 (41%), Gaps = 8/331 (2%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLN-SLAENNCYNAFDVIANQI 232
++ ++ G+ G+ A LL M + ++ D ++ L++ S+ E + A + +
Sbjct: 334 YNCMIDGFCSFGRWSDAQRLLRDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEML 393
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
+ +T +I CK R ++A+ H+ L+ S + + +I V C + R +
Sbjct: 394 HRCIFPDTVTYNSMIYGFCKHNRFDDAK-HMFDLMASPDVV---TFNTIIDVYCRAKRVD 449
Query: 293 RAVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNIL 350
++L+ E + N Y I G + L+ A + F Q+ S G P + NIL
Sbjct: 450 EGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQDLF-QEMISHGVCPDTITCNIL 508
Query: 351 IGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGL 410
+ +L++ EL + + I + V N ++ CK VD A +LF S G+
Sbjct: 509 LYGFCENEKLEEALELFEVIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGV 568
Query: 411 SPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWD 470
P+ Y +I C +A + G+ PD T++TL + +ID+ +
Sbjct: 569 EPDVQTYNVMISGFCGKSAISDANVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIE 628
Query: 471 LLDFALERRFMPNSSTYSRFVSALCRAGRVE 501
L+ F ++ T +CR E
Sbjct: 629 LISEMRSNGFSGDAFTIKMAEEIICRVSDEE 659
Score = 72.4 bits (176), Expect = 1e-12
Identities = 90/376 (23%), Positives = 158/376 (41%), Gaps = 69/376 (18%)
Query: 516 RFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLL 575
R + ++I G + + +G + L ++ +G ELK S H L + FF
Sbjct: 43 RLNSRRLIHGRV-AEKGTKSLPSLTQVTFEGEELKLKSGSHYFKSL----DDAIDFF--- 94
Query: 576 EMMTHGKPHCDIF--NSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRS 633
+ M +P N I + N+PD+A ++ M+ I N S +++K +
Sbjct: 95 DYMVRSRPFYTAVDCNKVIGVFVRMNRPDVAISLYRKMEIRRIPLNIYSFNILIKCFCDC 154
Query: 634 RRISDALRFFNDIRH---QVVVSTKLYNRMIVGLCKSDKADIALELCF------------ 678
++S +L F + Q V T +N ++ GLC D+ AL L
Sbjct: 155 HKLSFSLSTFGKLTKLGFQPDVVT--FNTLLHGLCLEDRISEALALFGYMVETGFLEAVA 212
Query: 679 ---EMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSM 735
+M+++GL P + + L+ LC R EA LVN G + +V+ + ++
Sbjct: 213 LFDQMVEIGLTPVVITFNTLINGLCLEGRVLEAAALVNKMVGKGLHI-----DVVTYGTI 267
Query: 736 ISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYT 795
++ K G+ S L L+ ++EE K D+
Sbjct: 268 VNGMC----------KMGD--TKSALNLL--------------SKMEETHIK---PDVVI 298
Query: 796 YNLLMRKLT---HHDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHE 852
Y+ ++ +L HH A LF M ++G+ PN +TY M GF + GR +A+R + +
Sbjct: 299 YSAIIDRLCKDGHHS--DAQYLFSEMLEKGIAPNVFTYNCMIDGFCSFGRWSDAQRLLRD 356
Query: 853 MLKKGFNPPENTRNVI 868
M+++ NP T N +
Sbjct: 357 MIEREINPDVLTFNAL 372
Score = 38.5 bits (88), Expect = 0.016
Identities = 49/209 (23%), Positives = 81/209 (38%), Gaps = 10/209 (4%)
Query: 138 TFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRM 197
TF I + A+ LR+ ++TL+ G+ + A L M
Sbjct: 434 TFNTIIDVYCRAKRVDEGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQDLFQEM 493
Query: 198 RFQGLDLDGFGYHILLNSLAEN----NCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQ 253
G+ D +ILL EN F+VI Q+ ++ N I+I +CK
Sbjct: 494 ISHGVCPDTITCNILLYGFCENEKLEEALELFEVI--QMSKIDLDTVAYN-IIIHGMCKG 550
Query: 254 GRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAY 311
+++EA L G E + +I C + A L + + P + Y
Sbjct: 551 SKVDEAWDLFCSLPIHGVEPDVQTYNVMISGFCGKSAISDANVLFHKMKDNGHEPDNSTY 610
Query: 312 GVWIRGLVQGGRLDEALEFFRQKRDSEGF 340
IRG ++ G +D+++E + R S GF
Sbjct: 611 NTLIRGCLKAGEIDKSIELISEMR-SNGF 638
>At2g02150 hypothetical protein
Length = 1107
Score = 137 bits (344), Expect = 3e-32
Identities = 102/423 (24%), Positives = 191/423 (45%), Gaps = 7/423 (1%)
Query: 306 PLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYE 365
P Y + I + + G ++ A F + + G VP V YN +I + RL D
Sbjct: 128 PTVFTYNIMIDCMCKEGDVEAARGLFEEMK-FRGLVPDTVTYNSMIDGFGKVGRLDDTVC 186
Query: 366 LLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLC 425
+M + C P+++T NA++ FCK G + + LE + GL PN ++Y L+ C
Sbjct: 187 FFEEMKDMCCEPDVITYNALINCFCKFGKLPIGLEFYREMKGNGLKPNVVSYSTLVDAFC 246
Query: 426 WDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSS 485
+G ++A + G P+ T+++L +A C+ + + + L + L+ N
Sbjct: 247 KEGMMQQAIKFYVDMRRVGLVPNEYTYTSLIDANCKIGNLSDAFRLGNEMLQVGVEWNVV 306
Query: 486 TYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKSNRGDIAARLLVEM 542
TY+ + LC A R+++ + G +D + SY +I GF+K+ D A LL E+
Sbjct: 307 TYTALIDGLCDAERMKEAEELFGKMDTAGVIPNLASYNALIHGFVKAKNMDRALELLNEL 366
Query: 543 KEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKP 601
K +G + Y + L ++ + EM G K + I+ + +D + P
Sbjct: 367 KGRGIKPDLLLYGTFIWGLCSLEKIEAAKVVMNEMKECGIKANSLIYTTLMDAYFKSGNP 426
Query: 602 DLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ--VVVSTKLYNR 659
+ + M+ I + +++ +++ +S A+ +FN I + + + ++
Sbjct: 427 TEGLHLLDEMKELDIEVTVVTFCVLIDGLCKNKLVSKAVDYFNRISNDFGLQANAAIFTA 486
Query: 660 MIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAG 719
MI GLCK ++ + A L +M++ GL P Y L+ EA+ L + + G
Sbjct: 487 MIDGLCKDNQVEAATTLFEQMVQKGLVPDRTAYTSLMDGNFKQGNVLEALALRDKMAEIG 546
Query: 720 RRL 722
+L
Sbjct: 547 MKL 549
Score = 119 bits (298), Expect = 7e-27
Identities = 124/550 (22%), Positives = 227/550 (40%), Gaps = 23/550 (4%)
Query: 281 LIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSE 338
L VL + E A++ S+ P + + + G+ D+ FF+ +
Sbjct: 66 LFSVLIDLGMLEEAIQCFSKMKRFRVFPKTRSCNGLLHRFAKLGKTDDVKRFFKDMIGA- 124
Query: 339 GFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVA 398
G P YNI+I + +E ++ L +M + P+ VT N+++ F K+G +D
Sbjct: 125 GARPTVFTYNIMIDCMCKEGDVEAARGLFEEMKFRGLVPDTVTYNSMIDGFGKVGRLDDT 184
Query: 399 LELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANA 458
+ F P+ + Y LI C G R G G P+ ++STL +A
Sbjct: 185 VCFFEEMKDMCCEPDVITYNALINCFCKFGKLPIGLEFYREMKGNGLKPNVVSYSTLVDA 244
Query: 459 LCRECKIDEMWDLLDFALERR---FMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTA 515
C+E M + F ++ R +PN TY+ + A C+ G + D + + ++ +V
Sbjct: 245 FCKE---GMMQQAIKFYVDMRRVGLVPNEYTYTSLIDANCKIGNLSDAFRLGNEMLQVGV 301
Query: 516 RF---SYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFF 572
+ +Y +I G + R A L +M G +SY ++H + N
Sbjct: 302 EWNVVTYTALIDGLCDAERMKEAEELFGKMDTAGVIPNLASYNALIHGFVKAKNMDRALE 361
Query: 573 NLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYF 631
L E+ G KP ++ +FI G K + A+ V M+ GI N+ +M +YF
Sbjct: 362 LLNELKGRGIKPDLLLYGTFIWGLCSLEKIEAAKVVMNEMKECGIKANSLIYTTLMDAYF 421
Query: 632 RSRRISDALRFFNDIRH-QVVVSTKLYNRMIVGLCKSDKADIALELCFEMLK--VGLNPS 688
+S ++ L ++++ + V+ + +I GLCK+ A++ F + GL +
Sbjct: 422 KSGNPTEGLHLLDEMKELDIEVTVVTFCVLIDGLCKNKLVSKAVDY-FNRISNDFGLQAN 480
Query: 689 IECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLR 748
+ ++ LC + A L + G L+ + V +
Sbjct: 481 AAIFTAMIDGLCKDNQVEAATTLFEQMVQKGLVPDRTAYTSLMDGNFKQGNVLEALA--L 538
Query: 749 REKEGEF---LDSSMLTLIIGAFSGCLRVSYSIQELEELIAK-CFPVDIYTYNLLMRKLT 804
R+K E LD T ++ S C ++ + LEE+I + P ++ ++L +
Sbjct: 539 RDKMAEIGMKLDLLAYTSLVWGLSHCNQLQKARSFLEEMIGEGIHPDEVLCISVLKKHYE 598
Query: 805 HHDMDKACEL 814
+D+A EL
Sbjct: 599 LGCIDEAVEL 608
Score = 110 bits (274), Expect = 4e-24
Identities = 106/526 (20%), Positives = 218/526 (41%), Gaps = 12/526 (2%)
Query: 347 YNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS 406
++ L L+ L++ + M + P + N +L F KLG D F
Sbjct: 63 FDALFSVLIDLGMLEEAIQCFSKMKRFRVFPKTRSCNGLLHRFAKLGKTDDVKRFFKDMI 122
Query: 407 QFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKID 466
G P Y +I +C +G + A + G PD T++++ + + ++D
Sbjct: 123 GAGARPTVFTYNIMIDCMCKEGDVEAARGLFEEMKFRGLVPDTVTYNSMIDGFGKVGRLD 182
Query: 467 EMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYL----MRGDLDKVTARFSYAKM 522
+ + + P+ TY+ ++ C+ G++ G M+G+ K SY+ +
Sbjct: 183 DTVCFFEEMKDMCCEPDVITYNALINCFCKFGKLPIGLEFYREMKGNGLKPNV-VSYSTL 241
Query: 523 IMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGK 582
+ F K A + V+M+ G +Y ++ + N F EM+ G
Sbjct: 242 VDAFCKEGMMQQAIKFYVDMRRVGLVPNEYTYTSLIDANCKIGNLSDAFRLGNEMLQVGV 301
Query: 583 P-HCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALR 641
+ + + IDG A + A E+F M G++ N +S ++ + +++ + AL
Sbjct: 302 EWNVVTYTALIDGLCDAERMKEAEELFGKMDTAGVIPNLASYNALIHGFVKAKNMDRALE 361
Query: 642 FFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLC 700
N+++ + + LY I GLC +K + A + EM + G+ + Y L+
Sbjct: 362 LLNELKGRGIKPDLLLYGTFIWGLCSLEKIEAAKVVMNEMKECGIKANSLIYTTLMDAYF 421
Query: 701 SLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEF---LD 757
E ++L++ ++ +T VL+ + ++ VD +F +
Sbjct: 422 KSGNPTEGLHLLDEMKELDIEVTVVTFCVLI-DGLCKNKLVSKAVDYFNRISNDFGLQAN 480
Query: 758 SSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMR-KLTHHDMDKACELFD 816
+++ T +I +V + E+++ K D Y LM ++ +A L D
Sbjct: 481 AAIFTAMIDGLCKDNQVEAATTLFEQMVQKGLVPDRTAYTSLMDGNFKQGNVLEALALRD 540
Query: 817 RMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPE 862
+M + G++ + Y + G S+ + +A+ ++ EM+ +G +P E
Sbjct: 541 KMAEIGMKLDLLAYTSLVWGLSHCNQLQKARSFLEEMIGEGIHPDE 586
Score = 80.9 bits (198), Expect = 3e-15
Identities = 83/398 (20%), Positives = 163/398 (40%), Gaps = 11/398 (2%)
Query: 156 FDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNS 215
F+ ++D C P Y+ L+ + GK I L M+ GL + Y L+++
Sbjct: 188 FEEMKDM--CCEPDVITYN-ALINCFCKFGKLPIGLEFYREMKGNGLKPNVVSYSTLVDA 244
Query: 216 LA-ENNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELH 274
E A + + + T +I CK G L +A N ++ G E +
Sbjct: 245 FCKEGMMQQAIKFYVDMRRVGLVPNEYTYTSLIDANCKIGNLSDAFRLGNEMLQVGVEWN 304
Query: 275 RSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFR 332
+ LI LC++ R + A EL + T+ +P +Y I G V+ +D ALE
Sbjct: 305 VVTYTALIDGLCDAERMKEAEELFGKMDTAGVIPNLASYNALIHGFVKAKNMDRALELLN 364
Query: 333 QKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKL 392
+ + G P + Y I L +++ ++ +M E I N + ++ + K
Sbjct: 365 ELK-GRGIKPDLLLYGTFIWGLCSLEKIEAAKVVMNEMKECGIKANSLIYTTLMDAYFKS 423
Query: 393 GMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVL-RSSSGTGYFPDRRT 451
G L L + + + + + LI LC + +A R S+ G +
Sbjct: 424 GNPTEGLHLLDEMKELDIEVTVVTFCVLIDGLCKNKLVSKAVDYFNRISNDFGLQANAAI 483
Query: 452 FSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLD 511
F+ + + LC++ +++ L + +++ +P+ + Y+ + + G V + +R +
Sbjct: 484 FTAMIDGLCKDNQVEAATTLFEQMVQKGLVPDRTAYTSLMDGNFKQGNVLEALALRDKMA 543
Query: 512 KVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKG 546
++ + +Y ++ G N+ A L EM +G
Sbjct: 544 EIGMKLDLLAYTSLVWGLSHCNQLQKARSFLEEMIGEG 581
Score = 47.0 bits (110), Expect = 5e-05
Identities = 58/246 (23%), Positives = 96/246 (38%), Gaps = 38/246 (15%)
Query: 159 LRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAE 218
+ + + C + + TL+ Y +G P LHLL M+ +LD
Sbjct: 398 MNEMKECGIKANSLIYTTLMDAYFKSGNPTEGLHLLDEMK----ELD------------- 440
Query: 219 NNCYNAFDVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGS-GKELHRSE 277
+V C ++I LCK + +A + N + G + + +
Sbjct: 441 ------IEVTVVTFC-----------VLIDGLCKNKLVSKAVDYFNRISNDFGLQANAAI 483
Query: 278 LSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKR 335
+ +I LC+ N+ E A L + +P AY + G + G + EAL R K
Sbjct: 484 FTAMIDGLCKDNQVEAATTLFEQMVQKGLVPDRTAYTSLMDGNFKQGNVLEALA-LRDKM 542
Query: 336 DSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMV 395
G + Y L+ L N+L+ L +M I P+ V +VL +LG +
Sbjct: 543 AEIGMKLDLLAYTSLVWGLSHCNQLQKARSFLEEMIGEGIHPDEVLCISVLKKHYELGCI 602
Query: 396 DVALEL 401
D A+EL
Sbjct: 603 DEAVEL 608
Score = 40.8 bits (94), Expect = 0.003
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 1/77 (1%)
Query: 793 IYTYNLLMRKLTHH-DMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVH 851
++TYN+++ + D++ A LF+ M RGL P+ TY M GF GR D+ +
Sbjct: 130 VFTYNIMIDCMCKEGDVEAARGLFEEMKFRGLVPDTVTYNSMIDGFGKVGRLDDTVCFFE 189
Query: 852 EMLKKGFNPPENTRNVI 868
EM P T N +
Sbjct: 190 EMKDMCCEPDVITYNAL 206
Score = 31.6 bits (70), Expect = 2.0
Identities = 15/61 (24%), Positives = 32/61 (51%)
Query: 808 MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNV 867
+++A + F +M + + P + + H F+ G+ D+ KR+ +M+ G P T N+
Sbjct: 76 LEEAIQCFSKMKRFRVFPKTRSCNGLLHRFAKLGKTDDVKRFFKDMIGAGARPTVFTYNI 135
Query: 868 I 868
+
Sbjct: 136 M 136
>At5g59900 putative protein
Length = 907
Score = 135 bits (341), Expect = 8e-32
Identities = 141/648 (21%), Positives = 258/648 (39%), Gaps = 80/648 (12%)
Query: 226 DVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVL 285
+++ +C+R S +++ L K+G++EEA + +V G + + LI L
Sbjct: 318 EMMDEMLCLRFSPSEAAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNALIDSL 377
Query: 286 CESNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPC 343
C+ +F A L G P + Y + I + G+LD AL F + D+ G
Sbjct: 378 CKGRKFHEAELLFDRMGKIGLRPNDVTYSILIDMFCRRGKLDTALSFLGEMVDT-GLKLS 436
Query: 344 KVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFN 403
YN LI + + + +M + P +VT +++ +C G ++ AL L++
Sbjct: 437 VYPYNSLINGHCKFGDISAAEGFMAEMINKKLEPTVVTYTSLMGGYCSKGKINKALRLYH 496
Query: 404 SRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCREC 463
+ G++P+ + L+ L G ++A ++ + P+R T++ + C E
Sbjct: 497 EMTGKGIAPSIYTFTTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYNVMIEGYCEEG 556
Query: 464 KIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTARFS---YA 520
+ + ++ L E+ +P++ +Y + LC G+ + + L K + Y
Sbjct: 557 DMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCLTGQASEAKVFVDGLHKGNCELNEICYT 616
Query: 521 KMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLH-CLLHMDNPRTRFFNLL-EMM 578
++ GF + + + A + EM ++G +L Y ++ L H D R FF LL EM
Sbjct: 617 GLLHGFCREGKLEEALSVCQEMVQRGVDLDLVCYGVLIDGSLKHKD--RKLFFGLLKEMH 674
Query: 579 THG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRIS 637
G KP I+ S ID A +++LM G + N +
Sbjct: 675 DRGLKPDDVIYTSMIDAKSKTGDFKEAFGIWDLMINEGCVPNEVT--------------- 719
Query: 638 DALRFFNDIRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQ 697
Y +I GLCK+ + A LC +M V P+ Y +
Sbjct: 720 -------------------YTAVINGLCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLD 760
Query: 698 KLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLD 757
L + V++ E L L N ++
Sbjct: 761 ILTKGE-----VDMQKAVELHNAILKGLLANTATYN------------------------ 791
Query: 758 SSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTH-HDMDKACELFD 816
++I F R+ + + + +I D TY ++ +L +D+ KA EL++
Sbjct: 792 -----MLIRGFCRQGRIEEASELITRMIGDGVSPDCITYTTMINELCRRNDVKKAIELWN 846
Query: 817 RMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENT 864
M ++G+ P+R Y + HG G +A +EML++G P T
Sbjct: 847 SMTEKGIRPDRVAYNTLIHGCCVAGEMGKATELRNEMLRQGLIPNNKT 894
Score = 116 bits (290), Expect = 6e-26
Identities = 151/634 (23%), Positives = 261/634 (40%), Gaps = 59/634 (9%)
Query: 278 LSFLIGVLCESNRFERAVELVSEFGTSLPLENAY---GVWIRGLVQGGRLDEALEFFRQK 334
LS L+ L + F A+EL ++ + + Y GV IR L + L A E
Sbjct: 195 LSALLHGLVKFRHFGLAMELFNDMVSVGIRPDVYIYTGV-IRSLCELKDLSRAKEMIAHM 253
Query: 335 RDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGM 394
++ G V YN+LI L ++ ++ + + D+ + P++VT ++ CK+
Sbjct: 254 -EATGCDVNIVPYNVLIDGLCKKQKVWEAVGIKKDLAGKDLKPDVVTYCTLVYGLCKVQE 312
Query: 395 VDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFST 454
++ LE+ + SP+ A L+ L G +EA +++ G P+ ++
Sbjct: 313 FEIGLEMMDEMLCLRFSPSEAAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNA 372
Query: 455 LANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVT 514
L ++LC+ K E L D + PN TYS + CR G+++ G++
Sbjct: 373 LIDSLCKGRKFHEAELLFDRMGKIGLRPNDVTYSILIDMFCRRGKLDTALSFLGEMVDTG 432
Query: 515 ARFS---YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVL--HCLLHMDNPRT 569
+ S Y +I G K A + EM K E +Y ++ +C N
Sbjct: 433 LKLSVYPYNSLINGHCKFGDISAAEGFMAEMINKKLEPTVVTYTSLMGGYCSKGKINKAL 492
Query: 570 RFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMK 628
R ++ EM G P F + + G A A ++F M + N + ++++
Sbjct: 493 RLYH--EMTGKGIAPSIYTFTTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYNVMIE 550
Query: 629 SYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNP 687
Y +S A F ++ + +V T Y +I GLC + +A A + K
Sbjct: 551 GYCEEGDMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCLTGQASEAKVFVDGLHKGNCEL 610
Query: 688 SIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSM------------ 735
+ CY L+ C + EA+++ + G L VL+ S+
Sbjct: 611 NEICYTGLLHGFCREGKLEEALSVCQEMVQRGVDLDLVCYGVLIDGSLKHKDRKLFFGLL 670
Query: 736 -------ISPE--VYHSCVDLRREKEGEFLDS-SMLTLIIGAFSGCL--RVSYS------ 777
+ P+ +Y S +D + K G+F ++ + L+I GC+ V+Y+
Sbjct: 671 KEMHDRGLKPDDVIYTSMID-AKSKTGDFKEAFGIWDLMIN--EGCVPNEVTYTAVINGL 727
Query: 778 -----IQELEELIAKCFPV----DIYTYNLLMRKLT--HHDMDKACELFDRMCQRGLEPN 826
+ E E L +K PV + TY + LT DM KA EL + + +GL N
Sbjct: 728 CKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLDILTKGEVDMQKAVELHNAIL-KGLLAN 786
Query: 827 RWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNP 860
TY ++ GF GR +EA + M+ G +P
Sbjct: 787 TATYNMLIRGFCRQGRIEEASELITRMIGDGVSP 820
Score = 105 bits (262), Expect = 1e-22
Identities = 132/596 (22%), Positives = 226/596 (37%), Gaps = 64/596 (10%)
Query: 305 LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVY 364
L +++ + I+ V+ R+ + + F+ +P + L+ L++
Sbjct: 153 LSSSSSFDLLIQHYVRSRRVLDGVLVFKMMITKVSLLPEVRTLSALLHGLVKFRHFGLAM 212
Query: 365 ELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTL 424
EL DM I P++ V+ C+L + A E+ G N + Y LI L
Sbjct: 213 ELFNDMVSVGIRPDVYIYTGVIRSLCELKDLSRAKEMIAHMEATGCDVNIVPYNVLIDGL 272
Query: 425 CWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALC------------------------ 460
C EA + + +G PD T+ TL LC
Sbjct: 273 CKKQKVWEAVGIKKDLAGKDLKPDVVTYCTLVYGLCKVQEFEIGLEMMDEMLCLRFSPSE 332
Query: 461 -----------RECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGD 509
+ KI+E +L+ ++ PN Y+ + +LC+ + + L+
Sbjct: 333 AAVSSLVEGLRKRGKIEEALNLVKRVVDFGVSPNLFVYNALIDSLCKGRKFHEAELLFDR 392
Query: 510 LDKVTAR---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVL--HCLLHM 564
+ K+ R +Y+ +I F + + D A L EM + G +L Y ++ HC
Sbjct: 393 MGKIGLRPNDVTYSILIDMFCRRGKLDTALSFLGEMVDTGLKLSVYPYNSLINGHCKFGD 452
Query: 565 DNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQ 623
+ F + EM+ +P + S + G K + A ++ M GI + +
Sbjct: 453 ISAAEGF--MAEMINKKLEPTVVTYTSLMGGYCSKGKINKALRLYHEMTGKGIAPSIYTF 510
Query: 624 ILVMKSYFRSRRISDALRFFNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEMLK 682
++ FR+ I DA++ FN++ V ++ YN MI G C+ A E EM +
Sbjct: 511 TTLLSGLFRAGLIRDAVKLFNEMAEWNVKPNRVTYNVMIEGYCEEGDMSKAFEFLKEMTE 570
Query: 683 VGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSM------- 735
G+ P Y L+ LC + EA V+ K L LL H
Sbjct: 571 KGIVPDTYSYRPLIHGLCLTGQASEAKVFVDGLHKGNCELNEICYTGLL-HGFCREGKLE 629
Query: 736 ----ISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPV 791
+ E+ VDL G +D S+ F G L+ E+ + K P
Sbjct: 630 EALSVCQEMVQRGVDLDLVCYGVLIDGSLKHKDRKLFFGLLK------EMHDRGLK--PD 681
Query: 792 DIYTYNLLMRKLTHHDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAK 847
D+ +++ K D +A ++D M G PN TY + +G G +EA+
Sbjct: 682 DVIYTSMIDAKSKTGDFKEAFGIWDLMINEGCVPNEVTYTAVINGLCKAGFVNEAE 737
Score = 99.4 bits (246), Expect = 8e-21
Identities = 114/527 (21%), Positives = 217/527 (40%), Gaps = 52/527 (9%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFD-VIANQI 232
+ L+ + GK D AL LG M GL L + Y+ L+N + +A + +A I
Sbjct: 405 YSILIDMFCRRGKLDTALSFLGEMVDTGLKLSVYPYNSLINGHCKFGDISAAEGFMAEMI 464
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
+ + +T ++ C +G++ +A + + G G + L+ L +
Sbjct: 465 NKKLEPTVVTYTSLMGGYCSKGKINKALRLYHEMTGKGIAPSIYTFTTLLSGLFRAGLIR 524
Query: 293 RAVELVSEFG--TSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNIL 350
AV+L +E P Y V I G + G + +A EF ++ + +G VP Y L
Sbjct: 525 DAVKLFNEMAEWNVKPNRVTYNVMIEGYCEEGDMSKAFEFLKEMTE-KGIVPDTYSYRPL 583
Query: 351 IGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGL 410
I L + + + +++ N + +L FC+ G ++ AL + Q G+
Sbjct: 584 IHGLCLTGQASEAKVFVDGLHKGNCELNEICYTGLLHGFCREGKLEEALSVCQEMVQRGV 643
Query: 411 SPNYMAYKYLILTLCWDGCPKEAYR-----VLRSSSGTGYFPDRRTFSTLANALCRECKI 465
+ + Y LI DG K R +L+ G PD ++++ +A +
Sbjct: 644 DLDLVCYGVLI-----DGSLKHKDRKLFFGLLKEMHDRGLKPDDVIYTSMIDAKSKTGDF 698
Query: 466 DEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTA---RFSYAKM 522
E + + D + +PN TY+ ++ LC+AG V + ++ + V++ + +Y
Sbjct: 699 KEAFGIWDLMINEGCVPNEVTYTAVINGLCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCF 758
Query: 523 IMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGK 582
+ DI + V+M +K EL + + +L T +N+L
Sbjct: 759 L---------DILTKGEVDM-QKAVELHNAILKGLL--------ANTATYNML------- 793
Query: 583 PHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRF 642
I G + + A E+ M +G+ + + ++ R + A+
Sbjct: 794 ---------IRGFCRQGRIEEASELITRMIGDGVSPDCITYTTMINELCRRNDVKKAIEL 844
Query: 643 FNDIRHQVVVSTKL-YNRMIVGLCKSDKADIALELCFEMLKVGLNPS 688
+N + + + ++ YN +I G C + + A EL EML+ GL P+
Sbjct: 845 WNSMTEKGIRPDRVAYNTLIHGCCVAGEMGKATELRNEMLRQGLIPN 891
Score = 84.7 bits (208), Expect = 2e-16
Identities = 92/358 (25%), Positives = 147/358 (40%), Gaps = 29/358 (8%)
Query: 168 PHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDV 227
P+R Y+ ++ GY G A L M +G+ D + Y L++ L + V
Sbjct: 540 PNRVTYN-VMIEGYCEEGDMSKAFEFLKEMTEKGIVPDTYSYRPLIHGLCLTGQASEAKV 598
Query: 228 IA----------NQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSE 277
N+IC G ++ C++G+LEEA + +V G +L
Sbjct: 599 FVDGLHKGNCELNEICYTG---------LLHGFCREGKLEEALSVCQEMVQRGVDLDLVC 649
Query: 278 LSFLI-GVLCESNR--FERAVELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQK 334
LI G L +R F ++ + + G P + Y I + G EA +
Sbjct: 650 YGVLIDGSLKHKDRKLFFGLLKEMHDRGLK-PDDVIYTSMIDAKSKTGDFKEAFGIW-DL 707
Query: 335 RDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGM 394
+EG VP +V Y +I L + + + L M PN VT L K G
Sbjct: 708 MINEGCVPNEVTYTAVINGLCKAGFVNEAEVLCSKMQPVSSVPNQVTYGCFLDILTK-GE 766
Query: 395 VDV--ALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTF 452
VD+ A+EL N+ + GL N Y LI C G +EA ++ G G PD T+
Sbjct: 767 VDMQKAVELHNAILK-GLLANTATYNMLIRGFCRQGRIEEASELITRMIGDGVSPDCITY 825
Query: 453 STLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDL 510
+T+ N LCR + + +L + E+ P+ Y+ + C AG + +R ++
Sbjct: 826 TTMINELCRRNDVKKAIELWNSMTEKGIRPDRVAYNTLIHGCCVAGEMGKATELRNEM 883
Score = 73.9 bits (180), Expect = 4e-13
Identities = 68/282 (24%), Positives = 129/282 (45%), Gaps = 6/282 (2%)
Query: 177 LVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRG 236
L+ G+ GK + AL + M +G+DLD Y +L++ ++ F + ++ RG
Sbjct: 618 LLHGFCREGKLEEALSVCQEMVQRGVDLDLVCYGVLIDGSLKHKDRKLFFGLLKEMHDRG 677
Query: 237 YE-SHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAV 295
+ + +I K G +EA + ++ G + + +I LC++ A
Sbjct: 678 LKPDDVIYTSMIDAKSKTGDFKEAFGIWDLMINEGCVPNEVTYTAVINGLCKAGFVNEAE 737
Query: 296 ELVSEFG--TSLPLENAYGVWIRGLVQGG-RLDEALEFFRQKRDSEGFVPCKVRYNILIG 352
L S+ +S+P + YG ++ L +G + +A+E +G + YN+LI
Sbjct: 738 VLCSKMQPVSSVPNQVTYGCFLDILTKGEVDMQKAVEL--HNAILKGLLANTATYNMLIR 795
Query: 353 RLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSP 412
R+ R+++ EL+ M + P+ +T ++ C+ V A+EL+NS ++ G+ P
Sbjct: 796 GFCRQGRIEEASELITRMIGDGVSPDCITYTTMINELCRRNDVKKAIELWNSMTEKGIRP 855
Query: 413 NYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFST 454
+ +AY LI C G +A + G P+ +T T
Sbjct: 856 DRVAYNTLIHGCCVAGEMGKATELRNEMLRQGLIPNNKTSRT 897
>At1g63130 unknown protein
Length = 630
Score = 134 bits (336), Expect = 3e-31
Identities = 135/562 (24%), Positives = 231/562 (41%), Gaps = 45/562 (8%)
Query: 212 LLNSLAENNCYNAFDVIANQICMRGYESHM-TNVIVIKHLCKQGRLEEAEAHLNGLVGSG 270
LL+++A+ N ++ + Q+ G ++ T I+I C++ +L A A L ++ G
Sbjct: 87 LLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSILINCFCRRSQLSLALAVLAKMMKLG 146
Query: 271 KELHRSELSFLIGVLCESNRFERAVELVS---EFGTSLPLENAYGVWIRGLVQGGRLDEA 327
E L+ L+ C NR AV LV E G P + I GL + R EA
Sbjct: 147 YEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQ-PDSFTFNTLIHGLFRHNRASEA 205
Query: 328 LEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLC 387
+ + +G P V Y I++ L + + LL M + I P +V N ++
Sbjct: 206 VALV-DRMVVKGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIID 264
Query: 388 FFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFP 447
C V+ AL LF G+ PN + Y LI LC G +A R+L P
Sbjct: 265 ALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINP 324
Query: 448 DRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDG---Y 504
+ TFS L +A +E K+ E L D ++R P+ TYS ++ C R+++ +
Sbjct: 325 NVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMF 384
Query: 505 LMRGDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHM 564
+ D +Y +I GF K+ R D L EM ++G +Y ++H
Sbjct: 385 ELMISKDCFPNVVTYNTLIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIH----- 439
Query: 565 DNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQI 624
G A + D A+ VF+ M +G++ + +
Sbjct: 440 -----------------------------GFFQARECDNAQIVFKQMVSDGVLPDIMTYS 470
Query: 625 LVMKSYFRSRRISDALRFFNDI-RHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKV 683
+++ + ++ AL F + R ++ YN MI G+CK+ K + +L +
Sbjct: 471 ILLDGLCNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLK 530
Query: 684 GLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHS 743
G+ P++ Y ++ C EA L ++ G S N L+ + + S
Sbjct: 531 GVKPNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTYNTLIRAHLRDGDKAAS 590
Query: 744 CVDLRREKEGEFL-DSSMLTLI 764
+R + F+ D+S + L+
Sbjct: 591 AELIREMRSCRFVGDASTIGLV 612
Score = 133 bits (334), Expect = 5e-31
Identities = 124/543 (22%), Positives = 235/543 (42%), Gaps = 29/543 (5%)
Query: 345 VRYN---ILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALEL 401
VRY+ I I RL + +L D L DM ++ P++V + +L K+ D+ + L
Sbjct: 45 VRYDYRKISINRL-NDLKLDDAVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISL 103
Query: 402 FNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCR 461
G+S N Y LI C A VL GY PD T ++L N C
Sbjct: 104 GEQMQNLGISHNLYTYSILINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCH 163
Query: 462 ECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR----- 516
+I + L+ +E + P+S T++ + L R R + + +D++ +
Sbjct: 164 GNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGLFRHNRASEAVAL---VDRMVVKGCQPD 220
Query: 517 -FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLL 575
+Y ++ G K D+A LL +M++ E Y ++ L + N
Sbjct: 221 LVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFT 280
Query: 576 EMMTHG-KPHCDIFNSFIDGAMHANK-PDLAREVFELMQRNGIMTNASSQILVMKSYFRS 633
EM G +P+ +NS I + + D +R + ++++R I N + ++ ++ +
Sbjct: 281 EMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERK-INPNVVTFSALIDAFVKE 339
Query: 634 RRISDALRFFND-IRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECY 692
++ +A + +++ I+ + Y+ +I G C D+ D A + M+ P++ Y
Sbjct: 340 GKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTY 399
Query: 693 EVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMI-----SPEVYHSCVDL 747
L++ C KR E + L + G +GN + + ++I + E ++ +
Sbjct: 400 NTLIKGFCKAKRVDEGMELFREMSQRG-----LVGNTVTYTTLIHGFFQARECDNAQIVF 454
Query: 748 RRE-KEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHH 806
++ +G D ++++ +V ++ E L DIYTYN+++ +
Sbjct: 455 KQMVSDGVLPDIMTYSILLDGLCNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKA 514
Query: 807 D-MDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTR 865
++ +LF + +G++PN TY M GF G K+EA EM ++G P T
Sbjct: 515 GKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTY 574
Query: 866 NVI 868
N +
Sbjct: 575 NTL 577
Score = 131 bits (330), Expect = 1e-30
Identities = 128/552 (23%), Positives = 228/552 (41%), Gaps = 17/552 (3%)
Query: 143 FRILSCARLRPLVFD-----FLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRM 197
+R +S RL L D F +S FP + L+ A K D+ + L +M
Sbjct: 49 YRKISINRLNDLKLDDAVNLFGDMVKSRPFPSIVEF-SKLLSAIAKMNKFDLVISLGEQM 107
Query: 198 RFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYESHMTNV-IVIKHLCKQGRL 256
+ G+ + + Y IL+N + + + ++ GYE + + ++ C R+
Sbjct: 108 QNLGISHNLYTYSILINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNRI 167
Query: 257 EEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVW 314
+A + + +V G + + LI L NR AV LV P YG+
Sbjct: 168 SDAVSLVGQMVEMGYQPDSFTFNTLIHGLFRHNRASEAVALVDRMVVKGCQPDLVTYGIV 227
Query: 315 IRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETC 374
+ GL + G +D AL + K + P V YN +I L + D L +M+
Sbjct: 228 VNGLCKRGDIDLALSLLK-KMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKG 286
Query: 375 IPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAY 434
I PN+VT N+++ C G A L + + ++PN + + LI +G EA
Sbjct: 287 IRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAE 346
Query: 435 RVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSAL 494
++ PD T+S+L N C ++DE + + + + PN TY+ +
Sbjct: 347 KLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGF 406
Query: 495 CRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKR 551
C+A RV++G + ++ + V +Y +I GF ++ D A + +M G
Sbjct: 407 CKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSDGVLPDI 466
Query: 552 SSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI--FNSFIDGAMHANKPDLAREVFE 609
+Y +L L + T + E + K DI +N I+G A K + ++F
Sbjct: 467 MTYSILLDGLCNNGKVETALV-VFEYLQRSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFC 525
Query: 610 LMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSD 668
+ G+ N + +M + R +A F +++ + + + YN +I +
Sbjct: 526 SLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTYNTLIRAHLRDG 585
Query: 669 KADIALELCFEM 680
+ EL EM
Sbjct: 586 DKAASAELIREM 597
Score = 70.5 bits (171), Expect = 4e-12
Identities = 69/292 (23%), Positives = 125/292 (42%), Gaps = 16/292 (5%)
Query: 177 LVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENN----CYNAFDVIANQI 232
L+ + GK A L M + +D D F Y L+N ++ + F+++ ++
Sbjct: 332 LIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKD 391
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEA-----EAHLNGLVGSGKELHRSELSFLIGVLCE 287
C ++ T +IK CK R++E E GLVG+ F C+
Sbjct: 392 CFPNVVTYNT---LIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECD 448
Query: 288 SNRFERAVELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRY 347
+ + ++VS+ LP Y + + GL G+++ AL F + S+ P Y
Sbjct: 449 NAQIVFK-QMVSD--GVLPDIMTYSILLDGLCNNGKVETALVVFEYLQRSK-MEPDIYTY 504
Query: 348 NILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQ 407
NI+I + + +++D ++L ++ + PN+VT ++ FC+ G+ + A LF +
Sbjct: 505 NIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKE 564
Query: 408 FGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANAL 459
G P+ Y LI DG + ++R + D T + N L
Sbjct: 565 EGPLPDSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIGLVTNML 616
>At5g64320 putative protein
Length = 730
Score = 133 bits (334), Expect = 5e-31
Identities = 125/491 (25%), Positives = 197/491 (39%), Gaps = 15/491 (3%)
Query: 135 TRTTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLL 194
T ++ + IL + + D S P ++ + + D AL LL
Sbjct: 181 TFKSYNVVLEILVSGNCHKVAANVFYDMLSRKIPPTLFTFGVVMKAFCAVNEIDSALSLL 240
Query: 195 GRMRFQGLDLDGFGYHILLNSLAENNCYN-AFDVIANQICMRGYESHMTNVIVIKHLCKQ 253
M G + Y L++SL++ N N A ++ M T VI LCK
Sbjct: 241 RDMTKHGCVPNSVIYQTLIHSLSKCNRVNEALQLLEEMFLMGCVPDAETFNDVILGLCKF 300
Query: 254 GRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTSLPLENAYGV 313
R+ EA +N ++ G +L+ LC+ R + A +L + P +
Sbjct: 301 DRINEAAKMVNRMLIRGFAPDDITYGYLMNGLCKIGRVDAAKDLF--YRIPKPEIVIFNT 358
Query: 314 WIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNET 373
I G V GRLD+A S G VP YN LI +E + E+L DM
Sbjct: 359 LIHGFVTHGRLDDAKAVLSDMVTSYGIVPDVCTYNSLIYGYWKEGLVGLALEVLHDMRNK 418
Query: 374 CIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEA 433
PN+ + ++ FCKLG +D A + N S GL PN + + LI C + EA
Sbjct: 419 GCKPNVYSYTILVDGFCKLGKIDEAYNVLNEMSADGLKPNTVGFNCLISAFCKEHRIPEA 478
Query: 434 YRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSA 493
+ R G PD TF++L + LC +I LL + + N+ TY+ ++A
Sbjct: 479 VEIFREMPRKGCKPDVYTFNSLISGLCEVDEIKHALWLLRDMISEGVVANTVTYNTLINA 538
Query: 494 LCRAGRVEDGYLMRGD-------LDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKG 546
R G +++ + + LD++T Y +I G ++ D A L +M G
Sbjct: 539 FLRRGEIKEARKLVNEMVFQGSPLDEIT----YNSLIKGLCRAGEVDKARSLFEKMLRDG 594
Query: 547 YELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGK-PHCDIFNSFIDGAMHANKPDLAR 605
+ S +++ L EM+ G P FNS I+G A + +
Sbjct: 595 HAPSNISCNILINGLCRSGMVEEAVEFQKEMVLRGSTPDIVTFNSLINGLCRAGRIEDGL 654
Query: 606 EVFELMQRNGI 616
+F +Q GI
Sbjct: 655 TMFRKLQAEGI 665
Score = 132 bits (331), Expect = 1e-30
Identities = 126/551 (22%), Positives = 235/551 (41%), Gaps = 17/551 (3%)
Query: 327 ALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVL 386
++E F G+ Y +LIG+L K + LL+ M + I +++
Sbjct: 94 SMELFSWTGSQNGYRHSFDVYQVLIGKLGANGEFKTIDRLLIQMKDEGIVFKESLFISIM 153
Query: 387 CFFCKLGMVDVALELF-NSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGY 445
+ K G L R+ + P + +Y ++ L C K A V
Sbjct: 154 RDYDKAGFPGQTTRLMLEMRNVYSCEPTFKSYNVVLEILVSGNCHKVAANVFYDMLSRKI 213
Query: 446 FPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYL 505
P TF + A C +ID LL + +PNS Y + +L + RV +
Sbjct: 214 PPTLFTFGVVMKAFCAVNEIDSALSLLRDMTKHGCVPNSVIYQTLIHSLSKCNRVNEALQ 273
Query: 506 MRGD---LDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLL 562
+ + + V ++ +I+G K +R + AA+++ M +G+ +Y ++++ L
Sbjct: 274 LLEEMFLMGCVPDAETFNDVILGLCKFDRINEAAKMVNRMLIRGFAPDDITYGYLMNGLC 333
Query: 563 ---HMDNPRTRFFNLLEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVF-ELMQRNGIMT 618
+D + F+ + KP IFN+ I G + + D A+ V +++ GI+
Sbjct: 334 KIGRVDAAKDLFYRI------PKPEIVIFNTLIHGFVTHGRLDDAKAVLSDMVTSYGIVP 387
Query: 619 NASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELC 677
+ + ++ Y++ + AL +D+R++ + Y ++ G CK K D A +
Sbjct: 388 DVCTYNSLIYGYWKEGLVGLALEVLHDMRNKGCKPNVYSYTILVDGFCKLGKIDEAYNVL 447
Query: 678 FEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMIS 737
EM GL P+ + L+ C R EAV + + G + + N L+
Sbjct: 448 NEMSADGLKPNTVGFNCLISAFCKEHRIPEAVEIFREMPRKGCKPDVYTFNSLISGLCEV 507
Query: 738 PEVYHSCVDLR-REKEGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTY 796
E+ H+ LR EG ++ +I AF + + + + E++ + P+D TY
Sbjct: 508 DEIKHALWLLRDMISEGVVANTVTYNTLINAFLRRGEIKEARKLVNEMVFQGSPLDEITY 567
Query: 797 NLLMRKLTH-HDMDKACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLK 855
N L++ L ++DKA LF++M + G P+ + ++ +G G +EA + EM+
Sbjct: 568 NSLIKGLCRAGEVDKARSLFEKMLRDGHAPSNISCNILINGLCRSGMVEEAVEFQKEMVL 627
Query: 856 KGFNPPENTRN 866
+G P T N
Sbjct: 628 RGSTPDIVTFN 638
Score = 127 bits (318), Expect = 4e-29
Identities = 128/527 (24%), Positives = 210/527 (39%), Gaps = 41/527 (7%)
Query: 347 YNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS 406
YN+++ L+ N K + DM IPP + T V+ FC + +D AL L +
Sbjct: 185 YNVVLEILVSGNCHKVAANVFYDMLSRKIPPTLFTFGVVMKAFCAVNEIDSALSLLRDMT 244
Query: 407 QFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKID 466
+ G PN + Y+ LI +L EA ++L G PD TF+ + LC+ +I+
Sbjct: 245 KHGCVPNSVIYQTLIHSLSKCNRVNEALQLLEEMFLMGCVPDAETFNDVILGLCKFDRIN 304
Query: 467 EMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVE---------------------DGYL 505
E +++ L R F P+ TY ++ LC+ GRV+ G++
Sbjct: 305 EAAKMVNRMLIRGFAPDDITYGYLMNGLCKIGRVDAAKDLFYRIPKPEIVIFNTLIHGFV 364
Query: 506 MRGDLDKVTARFS--------------YAKMIMGFIKSNRGDIAARLLVEMKEKGYELKR 551
G LD A S Y +I G+ K +A +L +M+ KG +
Sbjct: 365 THGRLDDAKAVLSDMVTSYGIVPDVCTYNSLIYGYWKEGLVGLALEVLHDMRNKGCKPNV 424
Query: 552 SSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDGAMHANKPDLAREVFEL 610
SY ++ + + L EM G KP+ FN I ++ A E+F
Sbjct: 425 YSYTILVDGFCKLGKIDEAYNVLNEMSADGLKPNTVGFNCLISAFCKEHRIPEAVEIFRE 484
Query: 611 MQRNGIMTNASSQILVMKSYFRSRRISDALRFFND-IRHQVVVSTKLYNRMIVGLCKSDK 669
M R G + + ++ I AL D I VV +T YN +I + +
Sbjct: 485 MPRKGCKPDVYTFNSLISGLCEVDEIKHALWLLRDMISEGVVANTVTYNTLINAFLRRGE 544
Query: 670 ADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNV 729
A +L EM+ G Y L++ LC +A +L + G ++ N+
Sbjct: 545 IKEARKLVNEMVFQGSPLDEITYNSLIKGLCRAGEVDKARSLFEKMLRDGHAPSNISCNI 604
Query: 730 LLFHSMISPEVYHSCVDLRREK--EGEFLDSSMLTLIIGAFSGCLRVSYSIQELEELIAK 787
L+ + + + V+ ++E G D +I R+ + +L A+
Sbjct: 605 LI-NGLCRSGMVEEAVEFQKEMVLRGSTPDIVTFNSLINGLCRAGRIEDGLTMFRKLQAE 663
Query: 788 CFPVDIYTYNLLMRKLTHHD-MDKACELFDRMCQRGLEPNRWTYGLM 833
P D T+N LM L + AC L D + G PN T+ ++
Sbjct: 664 GIPPDTVTFNTLMSWLCKGGFVYDACLLLDEGIEDGFVPNHRTWSIL 710
Score = 122 bits (306), Expect = 9e-28
Identities = 155/687 (22%), Positives = 263/687 (37%), Gaps = 120/687 (17%)
Query: 115 DILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPLVFDFLRDFRSCSFPHRARYH 174
++ + ++ F W G Q + H+ + + L + L + +
Sbjct: 90 NVSTSMELFSWTGSQNGYRHSFDVYQVLIGKLGANGEFKTIDRLLIQMKDEGIVFKESLF 149
Query: 175 DTLVVGYAIAGKPDIALHLLGRMR-FQGLDLDGFGYHILLNSLAENNCYN-AFDVIANQI 232
+++ Y AG P L+ MR + Y+++L L NC+ A +V + +
Sbjct: 150 ISIMRDYDKAGFPGQTTRLMLEMRNVYSCEPTFKSYNVVLEILVSGNCHKVAANVFYDML 209
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSG---------------------- 270
+ + T +V+K C ++ A + L + G
Sbjct: 210 SRKIPPTLFTFGVVMKAFCAVNEIDSALSLLRDMTKHGCVPNSVIYQTLIHSLSKCNRVN 269
Query: 271 KELHRSELSFLIGV-------------LCESNRFERAVELVSEFGTS--LPLENAYGVWI 315
+ L E FL+G LC+ +R A ++V+ P + YG +
Sbjct: 270 EALQLLEEMFLMGCVPDAETFNDVILGLCKFDRINEAAKMVNRMLIRGFAPDDITYGYLM 329
Query: 316 RGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETC- 374
GL + GR+D A + F + E + +N LI + RL D +L DM +
Sbjct: 330 NGLCKIGRVDAAKDLFYRIPKPEIVI-----FNTLIHGFVTHGRLDDAKAVLSDMVTSYG 384
Query: 375 IPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAY 434
I P++ T N+++ + K G+V +ALE+ + G PN +Y L+ C G EAY
Sbjct: 385 IVPDVCTYNSLIYGYWKEGLVGLALEVLHDMRNKGCKPNVYSYTILVDGFCKLGKIDEAY 444
Query: 435 RVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSAL 494
VL S G P+ F+ L +A C+E +I E ++ + P+ T++ +S L
Sbjct: 445 NVLNEMSADGLKPNTVGFNCLISAFCKEHRIPEAVEIFREMPRKGCKPDVYTFNSLISGL 504
Query: 495 CRAGRVEDG-YLMRGDLDK--VTARFSYAKMIMGFIKSNRGDIA-ARLLVEMKEKGYELK 550
C ++ +L+R + + V +Y +I F++ RG+I AR LV
Sbjct: 505 CEVDEIKHALWLLRDMISEGVVANTVTYNTLINAFLR--RGEIKEARKLVN--------- 553
Query: 551 RSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI-FNSFIDGAMHANKPDLAREVFE 609
EM+ G P +I +NS I G A + D AR +FE
Sbjct: 554 -------------------------EMVFQGSPLDEITYNSLIKGLCRAGEVDKARSLFE 588
Query: 610 LMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIVGLCKSDK 669
M R+G +A S I N +I GLC+S
Sbjct: 589 KMLRDG---HAPSNISC-------------------------------NILINGLCRSGM 614
Query: 670 ADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNV 729
+ A+E EM+ G P I + L+ LC R + + + + G + N
Sbjct: 615 VEEAVEFQKEMVLRGSTPDIVTFNSLINGLCRAGRIEDGLTMFRKLQAEGIPPDTVTFNT 674
Query: 730 LLFHSMISPEVYHSCVDLRREKEGEFL 756
L+ VY +C+ L E F+
Sbjct: 675 LMSWLCKGGFVYDACLLLDEGIEDGFV 701
Score = 96.3 bits (238), Expect = 7e-20
Identities = 77/317 (24%), Positives = 142/317 (44%), Gaps = 6/317 (1%)
Query: 154 LVFDFLRDFRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILL 213
L + L D R+ + LV G+ GK D A ++L M GL + G++ L+
Sbjct: 407 LALEVLHDMRNKGCKPNVYSYTILVDGFCKLGKIDEAYNVLNEMSADGLKPNTVGFNCLI 466
Query: 214 NSLAENNCYNAFDVIANQICMRGYESHM-TNVIVIKHLCKQGRLEEAEAHLNGLVGSGKE 272
++ + + I ++ +G + + T +I LC+ ++ A L ++ G
Sbjct: 467 SAFCKEHRIPEAVEIFREMPRKGCKPDVYTFNSLISGLCEVDEIKHALWLLRDMISEGVV 526
Query: 273 LHRSELSFLIGVLCESNRFERAVELVSEF---GTSLPLENAYGVWIRGLVQGGRLDEALE 329
+ + LI + A +LV+E G+ L E Y I+GL + G +D+A
Sbjct: 527 ANTVTYNTLINAFLRRGEIKEARKLVNEMVFQGSPLD-EITYNSLIKGLCRAGEVDKARS 585
Query: 330 FFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFF 389
F +K +G P + NILI L R +++ E +M P++VT N+++
Sbjct: 586 LF-EKMLRDGHAPSNISCNILINGLCRSGMVEEAVEFQKEMVLRGSTPDIVTFNSLINGL 644
Query: 390 CKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDR 449
C+ G ++ L +F G+ P+ + + L+ LC G +A +L G+ P+
Sbjct: 645 CRAGRIEDGLTMFRKLQAEGIPPDTVTFNTLMSWLCKGGFVYDACLLLDEGIEDGFVPNH 704
Query: 450 RTFSTLANALCRECKID 466
RT+S L ++ + +D
Sbjct: 705 RTWSILLQSIIPQETLD 721
Score = 91.7 bits (226), Expect = 2e-18
Identities = 78/324 (24%), Positives = 144/324 (44%), Gaps = 4/324 (1%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
+++L+ GY G +AL +L MR +G + + Y IL++ + + + N++
Sbjct: 392 YNSLIYGYWKEGLVGLALEVLHDMRNKGCKPNVYSYTILVDGFCKLGKIDEAYNVLNEMS 451
Query: 234 MRGYESHMTNV-IVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
G + + +I CK+ R+ EA + G + + LI LCE + +
Sbjct: 452 ADGLKPNTVGFNCLISAFCKEHRIPEAVEIFREMPRKGCKPDVYTFNSLISGLCEVDEIK 511
Query: 293 RAVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNIL 350
A+ L+ + + + N Y I ++ G + EA + + +G ++ YN L
Sbjct: 512 HALWLLRDMISEGVVANTVTYNTLINAFLRRGEIKEARKLVNEMV-FQGSPLDEITYNSL 570
Query: 351 IGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGL 410
I L R + L M P+ ++ N ++ C+ GMV+ A+E G
Sbjct: 571 IKGLCRAGEVDKARSLFEKMLRDGHAPSNISCNILINGLCRSGMVEEAVEFQKEMVLRGS 630
Query: 411 SPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWD 470
+P+ + + LI LC G ++ + R G PD TF+TL + LC+ + +
Sbjct: 631 TPDIVTFNSLINGLCRAGRIEDGLTMFRKLQAEGIPPDTVTFNTLMSWLCKGGFVYDACL 690
Query: 471 LLDFALERRFMPNSSTYSRFVSAL 494
LLD +E F+PN T+S + ++
Sbjct: 691 LLDEGIEDGFVPNHRTWSILLQSI 714
>At3g22470 hypothetical protein
Length = 619
Score = 132 bits (331), Expect = 1e-30
Identities = 134/525 (25%), Positives = 217/525 (40%), Gaps = 21/525 (4%)
Query: 209 YHILLNSLAENNCYNAFDVIANQICMRGYESHM-TNVIVIKHLCKQGRLEEAEAHLNGLV 267
++ L +++A Y+ + + G E M T I+I C++ +L A + L
Sbjct: 73 FNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYTMTIMINCYCRKKKLLFAFSVLGRAW 132
Query: 268 GSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTSLPLENAYGV--WIRGLVQGGRLD 325
G E S L+ C R AV LV + V I GL GR+
Sbjct: 133 KLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMVEMKQRPDLVTVSTLINGLCLKGRVS 192
Query: 326 EALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAV 385
EAL + GF P +V Y ++ RL + +L M E I ++V + V
Sbjct: 193 EALVLI-DRMVEYGFQPDEVTYGPVLNRLCKSGNSALALDLFRKMEERNIKASVVQYSIV 251
Query: 386 LCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGY 445
+ CK G D AL LFN G+ + + Y LI LC DG + ++LR G
Sbjct: 252 IDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGLCNDGKWDDGAKMLREMIGRNI 311
Query: 446 FPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCR------AGR 499
PD TFS L + +E K+ E +L + + R P++ TY+ + C+ A +
Sbjct: 312 IPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHEANQ 371
Query: 500 VEDGYLMRG-DLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVL 558
+ D + +G + D VT Y+ +I + K+ R D RL E+ KG +Y ++
Sbjct: 372 MFDLMVSKGCEPDIVT----YSILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLV 427
Query: 559 --HCLLHMDNPRTRFFNLLEMMTHGKPHCDI-FNSFIDGAMHANKPDLAREVFELMQRNG 615
C N F EM++ G P + + +DG + + A E+FE MQ++
Sbjct: 428 LGFCQSGKLNAAKELFQ--EMVSRGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSR 485
Query: 616 IMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIAL 674
+ +++ + ++ DA F + + V YN MI GLCK A
Sbjct: 486 MTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEAD 545
Query: 675 ELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAG 719
L +M + G P Y +L++ +V L+ + G
Sbjct: 546 MLFRKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELIEEMKVCG 590
Score = 128 bits (322), Expect = 1e-29
Identities = 130/573 (22%), Positives = 234/573 (40%), Gaps = 57/573 (9%)
Query: 281 LIGVLCESNRFERAVELVSEFGTSL-------PLENAYGVWIRGLVQGGRLDEALEFFRQ 333
+I L NR + E GTSL + +Y +R + ++++A++ F
Sbjct: 1 MIQRLIPLNRKASNFTQILEKGTSLLHYSSITEAKLSYKERLRNGIVDIKVNDAIDLFES 60
Query: 334 KRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLG 393
S +P + +N L + R + V M I +M TM ++ +C+
Sbjct: 61 MIQSRP-LPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYTMTIMINCYCRKK 119
Query: 394 MVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFS 453
+ A + + G P+ + + L+ C +G EA ++ PD T S
Sbjct: 120 KLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMVEMKQRPDLVTVS 179
Query: 454 TLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGR-------------- 499
TL N LC + ++ E L+D +E F P+ TY ++ LC++G
Sbjct: 180 TLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSALALDLFRKMEER 239
Query: 500 -----------VEDGYLMRGDLDKVTARFS-------------YAKMIMGFIKSNRGDIA 535
V D G D + F+ Y+ +I G + D
Sbjct: 240 NIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGLCNDGKWDDG 299
Query: 536 ARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCDIFNSFIDG 594
A++L EM + ++ ++ + EM+T G P +NS IDG
Sbjct: 300 AKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTITYNSLIDG 359
Query: 595 AMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ-VVVS 653
N A ++F+LM G + + +++ SY +++R+ D +R F +I + ++ +
Sbjct: 360 FCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREISSKGLIPN 419
Query: 654 TKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVN 713
T YN +++G C+S K + A EL EM+ G+ PS+ Y +L+ LC +A+ +
Sbjct: 420 TITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELNKALEIFE 479
Query: 714 VYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEGEFLDSSMLTLIIGAFSGCLR 773
+K+ L + N+++ H M + L + + ++T G L
Sbjct: 480 KMQKSRMTLGIGIYNIII-HGMCNASKVDDAWSLFCSLSDKGVKPDVVT--YNVMIGGLC 536
Query: 774 VSYSIQELEELIAK-----CFPVDIYTYNLLMR 801
S+ E + L K C P D +TYN+L+R
Sbjct: 537 KKGSLSEADMLFRKMKEDGCTP-DDFTYNILIR 568
Score = 117 bits (293), Expect = 3e-26
Identities = 115/538 (21%), Positives = 222/538 (40%), Gaps = 19/538 (3%)
Query: 344 KVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFN 403
K+ Y + + + ++ D +L M ++ P + N + + D+ L
Sbjct: 35 KLSYKERLRNGIVDIKVNDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCK 94
Query: 404 SRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCREC 463
G+ + +I C A+ VL + GY PD TFSTL N C E
Sbjct: 95 GMELNGIEHDMYTMTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEG 154
Query: 464 KIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDL-------DKVTAR 516
++ E L+D +E + P+ T S ++ LC GRV + ++ + D+VT
Sbjct: 155 RVSEAVALVDRMVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVT-- 212
Query: 517 FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDN--PRTRFFNL 574
Y ++ KS +A L +M+E+ + Y V+ L + FN
Sbjct: 213 --YGPVLNRLCKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNE 270
Query: 575 LEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSR 634
+EM K ++S I G + K D ++ M I+ + + ++ + +
Sbjct: 271 MEMKGI-KADVVTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEG 329
Query: 635 RISDALRFFND-IRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYE 693
++ +A +N+ I + T YN +I G CK + A ++ M+ G P I Y
Sbjct: 330 KLLEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYS 389
Query: 694 VLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRREKEG 753
+L+ C KR + + L G + N L+ S ++ ++ +L +E
Sbjct: 390 ILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKL-NAAKELFQEMVS 448
Query: 754 EFLDSSMLT--LIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDK 810
+ S++T +++ ++ +++ E++ + I YN+++ + + +D
Sbjct: 449 RGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDD 508
Query: 811 ACELFDRMCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
A LF + +G++P+ TY +M G G EA +M + G P + T N++
Sbjct: 509 AWSLFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNIL 566
Score = 112 bits (281), Expect = 7e-25
Identities = 119/553 (21%), Positives = 231/553 (41%), Gaps = 19/553 (3%)
Query: 107 LRHGGDDGDILSCLKFFDWAGRQPRFYHTRTTFVAIFRILSCARLRPLVFDFLRDFRSCS 166
LR+G D + + F+ + Q R T F + ++ + LV F +
Sbjct: 42 LRNGIVDIKVNDAIDLFE-SMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNG 100
Query: 167 FPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLA-ENNCYNAF 225
H ++ Y K A +LGR G + D + L+N E A
Sbjct: 101 IEHDMYTMTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAV 160
Query: 226 DVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVL 285
++ + M+ +T +I LC +GR+ EA ++ +V G + ++ L
Sbjct: 161 ALVDRMVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRL 220
Query: 286 CESNRFERAVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPC 343
C+S A++L + + Y + I L + G D+AL F + + +G
Sbjct: 221 CKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEM-EMKGIKAD 279
Query: 344 KVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFN 403
V Y+ LIG L + + D ++L +M I P++VT +A++ F K G + A EL+N
Sbjct: 280 VVTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYN 339
Query: 404 SRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCREC 463
G++P+ + Y LI C + C EA ++ G PD T+S L N+ C+
Sbjct: 340 EMITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAK 399
Query: 464 KIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVE------DGYLMRGDLDKVTARF 517
++D+ L + +PN+ TY+ V C++G++ + RG V
Sbjct: 400 RVDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVV--- 456
Query: 518 SYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLH---MDNPRTRFFNL 574
+Y ++ G + + A + +M++ L Y ++H + + +D+ + F +L
Sbjct: 457 TYGILLDGLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSL 516
Query: 575 LEMMTHGKPHCDIFNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSR 634
+ KP +N I G A +F M+ +G + + ++++++
Sbjct: 517 SDKGV--KPDVVTYNVMIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILIRAHLGGS 574
Query: 635 RISDALRFFNDIR 647
+ ++ +++
Sbjct: 575 GLISSVELIEEMK 587
Score = 52.4 bits (124), Expect = 1e-06
Identities = 55/217 (25%), Positives = 94/217 (42%), Gaps = 6/217 (2%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQIC 233
+ L+ Y A + D + L + +GL + Y+ L+ ++ NA + ++
Sbjct: 388 YSILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMV 447
Query: 234 MRGYE-SHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFE 292
RG S +T I++ LC G L +A + S L + +I +C +++ +
Sbjct: 448 SRGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVD 507
Query: 293 RAVEL---VSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNI 349
A L +S+ G P Y V I GL + G L EA FR+ ++ +G P YNI
Sbjct: 508 DAWSLFCSLSDKGVK-PDVVTYNVMIGGLCKKGSLSEADMLFRKMKE-DGCTPDDFTYNI 565
Query: 350 LIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVL 386
LI L + L EL+ +M + T+ V+
Sbjct: 566 LIRAHLGGSGLISSVELIEEMKVCGFSADSSTIKMVI 602
>At1g62930 unknown protein
Length = 659
Score = 131 bits (329), Expect = 2e-30
Identities = 117/531 (22%), Positives = 226/531 (42%), Gaps = 25/531 (4%)
Query: 354 LLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPN 413
+L + +L D +L +M ++ P++V N +L K+ D+ + L +S +
Sbjct: 55 VLLDLKLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYD 114
Query: 414 YMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLD 473
+Y LI C A VL GY PD T S+L N C +I E L+D
Sbjct: 115 LYSYNILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVD 174
Query: 474 FALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR------FSYAKMIMGFI 527
+ PN+ T++ + L + + + +D++ AR F+Y ++ G
Sbjct: 175 QMFVMEYQPNTVTFNTLIHGLFLHNKASEAVAL---IDRMVARGCQPDLFTYGTVVNGLC 231
Query: 528 KSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHG-KPHCD 586
K D+A LL +M++ E Y ++ L + N EM G +P+
Sbjct: 232 KRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVV 291
Query: 587 IFNSFIDGAMHANK-PDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFND 645
+NS I + + D +R + ++++R I N + ++ ++ + ++ +A + +++
Sbjct: 292 TYNSLIRCLCNYGRWSDASRLLSDMIERK-INPNVVTFSALIDAFVKEGKLVEAEKLYDE 350
Query: 646 -IRHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKR 704
I+ + Y+ +I G C D+ D A + M+ P++ Y L++ C KR
Sbjct: 351 MIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKR 410
Query: 705 YYEAVNLVNVYEKAGRRLTSFLGNVLLFHSMISPEVYHSCVDLRRE------KEGEFLDS 758
E + L + G +GN + ++++I D+ ++ +G D
Sbjct: 411 VEEGMELFREMSQRG-----LVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDI 465
Query: 759 SMLTLIIGAFSGCLRVSYSIQELEELIAKCFPVDIYTYNLLMRKLTHHD-MDKACELFDR 817
++++ ++ ++ E L DIYTYN+++ + ++ +LF
Sbjct: 466 ITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCS 525
Query: 818 MCQRGLEPNRWTYGLMAHGFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
+ +G++PN Y M GF G K+EA EM + G P T N +
Sbjct: 526 LSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTL 576
Score = 117 bits (293), Expect = 3e-26
Identities = 113/528 (21%), Positives = 212/528 (39%), Gaps = 42/528 (7%)
Query: 177 LVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRG 236
L+ A K D+ + L RM+ + D + Y+IL+N + + ++ G
Sbjct: 86 LLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILINCFCRRSQLPLALAVLGKMMKLG 145
Query: 237 YESHMTNV-IVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAV 295
YE + + ++ C R+ EA A ++ + + + + LI L N+ AV
Sbjct: 146 YEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVMEYQPNTVTFNTLIHGLFLHNKASEAV 205
Query: 296 ELVSEFGT--SLPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGR 353
L+ P YG + GL + G +D AL + K + V Y +I
Sbjct: 206 ALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLK-KMEKGKIEADVVIYTTIIDA 264
Query: 354 LLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPN 413
L + D L +M+ I PN+VT N+++ C G A L + + ++PN
Sbjct: 265 LCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPN 324
Query: 414 YMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLD 473
+ + LI +G EA ++ PD T+S+L N C ++DE + +
Sbjct: 325 VVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFE 384
Query: 474 FALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDK---VTARFSYAKMIMGFIKSN 530
+ + PN TY+ + C+A RVE+G + ++ + V +Y +I G ++
Sbjct: 385 LMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAG 444
Query: 531 RGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDIFNS 590
D+A ++ +M G P ++
Sbjct: 445 DCDMAQKIFKKMVSDGV----------------------------------PPDIITYSI 470
Query: 591 FIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQ- 649
+DG K + A VFE +Q++ + + + ++++ ++ ++ D F + +
Sbjct: 471 LLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKG 530
Query: 650 VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQ 697
V + +Y MI G C+ + A L EM + G P+ Y L++
Sbjct: 531 VKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIR 578
Score = 111 bits (277), Expect = 2e-24
Identities = 104/399 (26%), Positives = 177/399 (44%), Gaps = 17/399 (4%)
Query: 175 DTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICM 234
+TL+ G + K A+ L+ RM +G D F Y ++N L + + + ++
Sbjct: 189 NTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEK 248
Query: 235 RGYESHMT-NVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFER 293
E+ + +I LC + +A + G + + LI LC R+
Sbjct: 249 GKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSD 308
Query: 294 AVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQ--KRDSEGFVPCKVRYNI 349
A L+S+ N + I V+ G+L EA + + + KR + P Y+
Sbjct: 309 ASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSID---PDIFTYSS 365
Query: 350 LIGRLLRENRL---KDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS 406
LI +RL K ++EL++ ++ C P N+VT N ++ FCK V+ +ELF S
Sbjct: 366 LINGFCMHDRLDEAKHMFELMI--SKDCFP-NVVTYNTLIKGFCKAKRVEEGMELFREMS 422
Query: 407 QFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKID 466
Q GL N + Y LI L G A ++ + G PD T+S L + LC+ K++
Sbjct: 423 QRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLE 482
Query: 467 EMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMI 523
+ + ++ + + P+ TY+ + +C+AG+VEDG+ + L + Y MI
Sbjct: 483 KALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMI 542
Query: 524 MGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLL 562
GF + + A L EMKE G +Y ++ L
Sbjct: 543 SGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARL 581
Score = 109 bits (272), Expect = 8e-24
Identities = 125/581 (21%), Positives = 232/581 (39%), Gaps = 54/581 (9%)
Query: 162 FRSCSFPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGF-GYHILLNSLAENN 220
F + S+ +R + +++ K D A+ L G M Q L ++ LL+++A+
Sbjct: 40 FSAASYDYREKLSRNVLLDL----KLDDAVDLFGEM-VQSRPLPSIVEFNKLLSAIAK-- 92
Query: 221 CYNAFDVIAN-----QICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHR 275
N FD++ + Q Y+ + N++ I C++ +L A A L ++ G E
Sbjct: 93 -MNKFDLVISLGERMQNLRISYDLYSYNIL-INCFCRRSQLPLALAVLGKMMKLGYEPDI 150
Query: 276 SELSFLIGVLCESNRFERAVELVSEFGTSLPLENAYGVWIRGLVQGGRLDEALEFFRQKR 335
LS L+ C R AV LV + F +
Sbjct: 151 VTLSSLLNGYCHGKRISEAVALVDQM----------------------------FVME-- 180
Query: 336 DSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMV 395
+ P V +N LI L N+ + L+ M P++ T V+ CK G +
Sbjct: 181 ----YQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDI 236
Query: 396 DVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTL 455
D+AL L + + + + Y +I LC +A + G P+ T+++L
Sbjct: 237 DLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSL 296
Query: 456 ANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTA 515
LC + + LL +ER+ PN T+S + A + G++ + + ++ K +
Sbjct: 297 IRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSI 356
Query: 516 R---FSYAKMIMGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFF 572
F+Y+ +I GF +R D A + M K +Y ++
Sbjct: 357 DPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGME 416
Query: 573 NLLEMMTHGKPHCDI-FNSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYF 631
EM G + +N+ I G A D+A+++F+ M +G+ + + +++
Sbjct: 417 LFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLC 476
Query: 632 RSRRISDALRFFNDI-RHQVVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIE 690
+ ++ AL F + + ++ YN MI G+CK+ K + +L + G+ P++
Sbjct: 477 KYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVI 536
Query: 691 CYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRLTSFLGNVLL 731
Y ++ C EA L ++ G S N L+
Sbjct: 537 IYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLI 577
Score = 108 bits (269), Expect = 2e-23
Identities = 90/391 (23%), Positives = 173/391 (44%), Gaps = 7/391 (1%)
Query: 347 YNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRS 406
YNILI R ++L +L M + P++VT++++L +C + A+ L +
Sbjct: 118 YNILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMF 177
Query: 407 QFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKID 466
PN + + LI L EA ++ G PD T+ T+ N LC+ ID
Sbjct: 178 VMEYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDID 237
Query: 467 EMWDLLDFALERRFMPNSSTYSRFVSALCRAGRVEDGYLMRGDLDKVTAR---FSYAKMI 523
LL + + + Y+ + ALC V D + ++D R +Y +I
Sbjct: 238 LALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLI 297
Query: 524 MGFIKSNRGDIAARLLVEMKEKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKP 583
R A+RLL +M E+ ++ ++ + + L + M
Sbjct: 298 RCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFV-KEGKLVEAEKLYDEMIKRSI 356
Query: 584 HCDIF--NSFIDGAMHANKPDLAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALR 641
DIF +S I+G ++ D A+ +FELM N + ++K + +++R+ + +
Sbjct: 357 DPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGME 416
Query: 642 FFNDIRHQ-VVVSTKLYNRMIVGLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLC 700
F ++ + +V +T YN +I GL ++ D+A ++ +M+ G+ P I Y +L+ LC
Sbjct: 417 LFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLC 476
Query: 701 SLKRYYEAVNLVNVYEKAGRRLTSFLGNVLL 731
+ +A+ + +K+ + N+++
Sbjct: 477 KYGKLEKALVVFEYLQKSKMEPDIYTYNIMI 507
Score = 88.6 bits (218), Expect = 1e-17
Identities = 84/363 (23%), Positives = 149/363 (40%), Gaps = 39/363 (10%)
Query: 174 HDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAE----NNCYNAFDVIA 229
+ T+V G G D+AL LL +M ++ D Y ++++L N+ N F +
Sbjct: 223 YGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMD 282
Query: 230 NQ---------------ICMRGYESHMTNVI-----------------VIKHLCKQGRLE 257
N+ +C G S + ++ +I K+G+L
Sbjct: 283 NKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLV 342
Query: 258 EAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTS--LPLENAYGVWI 315
EAE + ++ + S LI C +R + A + + P Y I
Sbjct: 343 EAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLI 402
Query: 316 RGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDMNETCI 375
+G + R++E +E FR+ G V V YN LI L + ++ M +
Sbjct: 403 KGFCKAKRVEEGMELFREM-SQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGV 461
Query: 376 PPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCPKEAYR 435
PP+++T + +L CK G ++ AL +F + + P+ Y +I +C G ++ +
Sbjct: 462 PPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWD 521
Query: 436 VLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRFVSALC 495
+ S S G P+ ++T+ + CR+ +E L E +PNS TY+ + A
Sbjct: 522 LFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARL 581
Query: 496 RAG 498
R G
Sbjct: 582 RDG 584
Score = 74.3 bits (181), Expect = 3e-13
Identities = 71/303 (23%), Positives = 136/303 (44%), Gaps = 21/303 (6%)
Query: 177 LVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENN----CYNAFDVIANQI 232
L+ + GK A L M + +D D F Y L+N ++ + F+++ ++
Sbjct: 331 LIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKD 390
Query: 233 CMRGYESHMTNVIVIKHLCKQGRLEEA-----EAHLNGLVGSGKELHRSELSFLIGVLCE 287
C ++ T +IK CK R+EE E GLVG+ + LI L +
Sbjct: 391 CFPNVVTYNT---LIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNT-----LIQGLFQ 442
Query: 288 SNRFERAVELVSEFGTS--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKV 345
+ + A ++ + + P Y + + GL + G+L++AL F + S+ P
Sbjct: 443 AGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSK-MEPDIY 501
Query: 346 RYNILIGRLLRENRLKDVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSR 405
YNI+I + + +++D ++L ++ + PN++ ++ FC+ G+ + A LF
Sbjct: 502 TYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREM 561
Query: 406 SQFGLSPNYMAYKYLILTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLA-NALCRECK 464
+ G PN Y LI DG + +++ G+ D T S L+ NA+ ++
Sbjct: 562 KEDGTLPNSGTYNTLIRARLRDGDKAASAELIKEMRSCGFVGDASTISMLSGNAVLKQSI 621
Query: 465 IDE 467
+++
Sbjct: 622 LEK 624
Score = 52.0 bits (123), Expect = 1e-06
Identities = 52/207 (25%), Positives = 89/207 (42%), Gaps = 5/207 (2%)
Query: 167 FPHRARYHDTLVVGYAIAGKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAE-NNCYNAF 225
FP+ Y+ TL+ G+ A + + + L M +GL + Y+ L+ L + +C A
Sbjct: 392 FPNVVTYN-TLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQ 450
Query: 226 DVIANQICMRGYESHMTNVIVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVL 285
+ + +T I++ LCK G+LE+A L S E + +I +
Sbjct: 451 KIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGM 510
Query: 286 CESNRFERAVELVSEFGTSLPLENA--YGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPC 343
C++ + E +L N Y I G + G +EA FR+ ++ +G +P
Sbjct: 511 CKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKE-DGTLPN 569
Query: 344 KVRYNILIGRLLRENRLKDVYELLMDM 370
YN LI LR+ EL+ +M
Sbjct: 570 SGTYNTLIRARLRDGDKAASAELIKEM 596
>At5g14770 putative protein
Length = 938
Score = 130 bits (326), Expect = 4e-30
Identities = 163/753 (21%), Positives = 306/753 (39%), Gaps = 47/753 (6%)
Query: 139 FVAIFRI-LSCARLRPLVFDFLRDFRS-CSFP--HRARYHDTLVVGYAIAGKPDIALHLL 194
F +FR+ LSC RL + R + C+F +R ++L+ + + G + L+
Sbjct: 61 FHTLFRLYLSCERL----YGAARTLSAMCTFGVVPDSRLWNSLIHQFNVNGLVHDQVSLI 116
Query: 195 -GRMRFQGLDLDGFGYHILLNSLAENNCYN-AFDVIANQICMRGYESHMTNVIVIKHLCK 252
+M G+ D F ++L++S + + A ++ N++ ++ T VI LC+
Sbjct: 117 YSKMIACGVSPDVFALNVLIHSFCKVGRLSFAISLLRNRVISIDTVTYNT---VISGLCE 173
Query: 253 QGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGT------SLP 306
G +EA L+ +V G + LI C+ F RA LV E ++
Sbjct: 174 HGLADEAYQFLSEMVKMGILPDTVSYNTLIDGFCKVGNFVRAKALVDEISELNLITHTIL 233
Query: 307 LENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYEL 366
L + Y + A+E + GF P V ++ +I RL + ++ + L
Sbjct: 234 LSSYYNL------------HAIEEAYRDMVMSGFDPDVVTFSSIINRLCKGGKVLEGGLL 281
Query: 367 LMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCW 426
L +M E + PN VT ++ K + AL L++ G+ + + Y L+ L
Sbjct: 282 LREMEEMSVYPNHVTYTTLVDSLFKANIYRHALALYSQMVVRGIPVDLVVYTVLMDGLFK 341
Query: 427 DGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSST 486
G +EA + + P+ T++ L + LC+ + ++ LE+ +PN T
Sbjct: 342 AGDLREAEKTFKMLLEDNQVPNVVTYTALVDGLCKAGDLSSAEFIITQMLEKSVIPNVVT 401
Query: 487 YSRFVSALCRAGRVEDGY-LMR--GDLDKVTARFSYAKMIMGFIKSNRGDIAARLLVEMK 543
YS ++ + G +E+ L+R D + V F+Y +I G K+ + ++A L EM+
Sbjct: 402 YSSMINGYVKKGMLEEAVSLLRKMEDQNVVPNGFTYGTVIDGLFKAGKEEMAIELSKEMR 461
Query: 544 EKGYELKRSSYRHVLHCLLHMDNPRTRFFNLLEMMTHGKPHCDI-FNSFIDGAMHANKPD 602
G E +++ L + + + +M++ G I + S ID +
Sbjct: 462 LIGVEENNYILDALVNHLKRIGRIKEVKGLVKDMVSKGVTLDQINYTSLIDVFFKGGDEE 521
Query: 603 LAREVFELMQRNGIMTNASSQILVMKSYFRSRRISDALRFFNDIRHQVVVSTKLYNRMIV 662
A E MQ G+ + S +++ + ++ + + +N M+
Sbjct: 522 AALAWAEEMQERGMPWDVVSYNVLISGMLKFGKVGADWAYKGMREKGIEPDIATFNIMMN 581
Query: 663 GLCKSDKADIALELCFEMLKVGLNPSIECYEVLVQKLCSLKRYYEAVNLVNVYEKAGRRL 722
K ++ L+L +M G+ PS+ ++V LC + EA++++N L
Sbjct: 582 SQRKQGDSEGILKLWDKMKSCGIKPSLMSCNIVVGMLCENGKMEEAIHILN-----QMML 636
Query: 723 TSFLGNVLLFHSMISPEVYHSCVDLRREKE------GEFLDSSMLTLIIGAFSGCLRVSY 776
N+ + + H D + G L + +I
Sbjct: 637 MEIHPNLTTYRIFLDTSSKHKRADAIFKTHETLLSYGIKLSRQVYNTLIATLCKLGMTKK 696
Query: 777 SIQELEELIAKCFPVDIYTYNLLMR-KLTHHDMDKACELFDRMCQRGLEPNRWTYGLMAH 835
+ + ++ A+ F D T+N LM + KA + M + G+ PN TY +
Sbjct: 697 AAMVMGDMEARGFIPDTVTFNSLMHGYFVGSHVRKALSTYSVMMEAGISPNVATYNTIIR 756
Query: 836 GFSNHGRKDEAKRWVHEMLKKGFNPPENTRNVI 868
G S+ G E +W+ EM +G P + T N +
Sbjct: 757 GLSDAGLIKEVDKWLSEMKSRGMRPDDFTYNAL 789
Score = 68.9 bits (167), Expect = 1e-11
Identities = 71/306 (23%), Positives = 130/306 (42%), Gaps = 10/306 (3%)
Query: 197 MRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYE-SHMTNVIVIKHLCKQGR 255
MR +G++ D ++I++NS + + +++ G + S M+ IV+ LC+ G+
Sbjct: 564 MREKGIEPDIATFNIMMNSQRKQGDSEGILKLWDKMKSCGIKPSLMSCNIVVGMLCENGK 623
Query: 256 LEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGTSLPL-----ENA 310
+EEA LN ++ E+H + ++ I L S++ +RA + T L
Sbjct: 624 MEEAIHILNQMMLM--EIHPNLTTYRI-FLDTSSKHKRADAIFKTHETLLSYGIKLSRQV 680
Query: 311 YGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLKDVYELLMDM 370
Y I L + G +A ++ GF+P V +N L+ + ++ M
Sbjct: 681 YNTLIATLCKLGMTKKAAMVMGDM-EARGFIPDTVTFNSLMHGYFVGSHVRKALSTYSVM 739
Query: 371 NETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLILTLCWDGCP 430
E I PN+ T N ++ G++ + + G+ P+ Y LI G
Sbjct: 740 MEAGISPNVATYNTIIRGLSDAGLIKEVDKWLSEMKSRGMRPDDFTYNALISGQAKIGNM 799
Query: 431 KEAYRVLRSSSGTGYFPDRRTFSTLANALCRECKIDEMWDLLDFALERRFMPNSSTYSRF 490
K + + G P T++ L + K+ + +LL +R PN+STY
Sbjct: 800 KGSMTIYCEMIADGLVPKTSTYNVLISEFANVGKMLQARELLKEMGKRGVSPNTSTYCTM 859
Query: 491 VSALCR 496
+S LC+
Sbjct: 860 ISGLCK 865
Score = 64.7 bits (156), Expect = 2e-10
Identities = 62/282 (21%), Positives = 115/282 (39%), Gaps = 4/282 (1%)
Query: 185 GKPDIALHLLGRMRFQGLDLDGFGYHILLNSLAENNCYNAFDVIANQICMRGYESHMTNV 244
G + L L +M+ G+ +I++ L EN I NQ+ + ++T
Sbjct: 587 GDSEGILKLWDKMKSCGIKPSLMSCNIVVGMLCENGKMEEAIHILNQMMLMEIHPNLTTY 646
Query: 245 -IVIKHLCKQGRLEEAEAHLNGLVGSGKELHRSELSFLIGVLCESNRFERAVELVSEFGT 303
I + K R + L+ G +L R + LI LC+ ++A ++ +
Sbjct: 647 RIFLDTSSKHKRADAIFKTHETLLSYGIKLSRQVYNTLIATLCKLGMTKKAAMVMGDMEA 706
Query: 304 S--LPLENAYGVWIRGLVQGGRLDEALEFFRQKRDSEGFVPCKVRYNILIGRLLRENRLK 361
+P + + G G + +AL + ++ G P YN +I L +K
Sbjct: 707 RGFIPDTVTFNSLMHGYFVGSHVRKALSTYSVMMEA-GISPNVATYNTIIRGLSDAGLIK 765
Query: 362 DVYELLMDMNETCIPPNMVTMNAVLCFFCKLGMVDVALELFNSRSQFGLSPNYMAYKYLI 421
+V + L +M + P+ T NA++ K+G + ++ ++ GL P Y LI
Sbjct: 766 EVDKWLSEMKSRGMRPDDFTYNALISGQAKIGNMKGSMTIYCEMIADGLVPKTSTYNVLI 825
Query: 422 LTLCWDGCPKEAYRVLRSSSGTGYFPDRRTFSTLANALCREC 463
G +A +L+ G P+ T+ T+ + LC+ C
Sbjct: 826 SEFANVGKMLQARELLKEMGKRGVSPNTSTYCTMISGLCKLC 867
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,063,483
Number of Sequences: 26719
Number of extensions: 888746
Number of successful extensions: 12146
Number of sequences better than 10.0: 447
Number of HSP's better than 10.0 without gapping: 306
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 3088
Number of HSP's gapped (non-prelim): 3275
length of query: 868
length of database: 11,318,596
effective HSP length: 108
effective length of query: 760
effective length of database: 8,432,944
effective search space: 6409037440
effective search space used: 6409037440
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0266.19


