

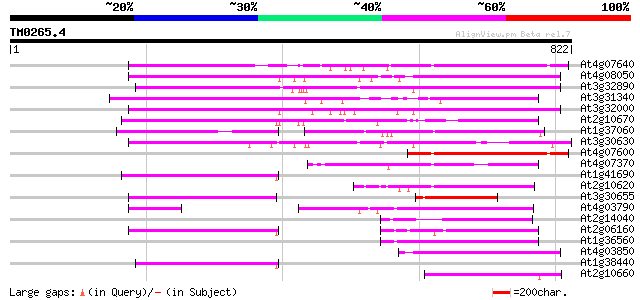
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0265.4
(822 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g07640 putative athila transposon protein 281 1e-75
At4g08050 259 3e-69
At3g32890 Athila ORF 1, putative 253 3e-67
At3g31340 Athila ORF 1, putative 239 6e-63
At3g32000 unknown protein 238 1e-62
At2g10670 pseudogene 219 4e-57
At1g37060 Athila retroelment ORF 1, putative 173 3e-43
At3g30630 hypothetical protein 172 7e-43
At4g07600 151 2e-36
At4g07370 149 5e-36
At1g41690 hypothetical protein 148 1e-35
At2g10620 putative Athila retroelement ORF1 protein 147 2e-35
At3g30655 hypothetical protein, 3' partial 140 3e-33
At4g03790 putative athila-like protein 135 7e-32
At2g14040 putative retroelement pol polyprotein 135 9e-32
At2g06160 putative Athila retroelement ORF1 protein 134 2e-31
At1g36560 hypothetical protein 132 8e-31
At4g03850 putative transposon protein 130 3e-30
At1g38440 hypothetical protein 130 4e-30
At2g10660 putative Athila retroelement ORF1 protein 125 7e-29
>At4g07640 putative athila transposon protein
Length = 866
Score = 281 bits (718), Expect = 1e-75
Identities = 207/713 (29%), Positives = 334/713 (46%), Gaps = 113/713 (15%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I+++ +++ G EDP H++ F R CN +++ VS++ +L LFPFSL D
Sbjct: 38 NFEIKSGLISMIQGNKFYGLPMEDPLDHLDEFDRLCNLTKINGVSADGFKLRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
A W + P SI TW+D + F ++FF A +L+ +I F Q E+ EAWERFK
Sbjct: 98 KAHIWEKNLPHDSIITWDDCKKAFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAARAM 354
+CP H T+A +++T Y G+ R LDTAS+G F + G++L++ +A
Sbjct: 158 GYTNQCPHHGFTKASMLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVDNLAQSDG 217
Query: 355 NSENDRQTRRRMFEFEAYDKLLASNKQISEKVAEIQNHIKVTKLVGASVAKVECVTRGGP 414
N D ++ + + H K K + + ++
Sbjct: 218 NYNED------------------CDRTVRDTADSDDKHRKEIKALNDKLDRI-------- 251
Query: 415 NFSNSCTMTNQGFQGQSSGLFQQVQEQVDVKDGGKKSLEELLESFINRMESNYK------ 468
+ NQ Q F EQ V+DG LEE+ S+IN +S YK
Sbjct: 252 -------LLNQ----QKHVHFLVDDEQFQVQDGEGNQLEEV--SYINNNQSGYKGYNNFK 298
Query: 469 -------IQDAAIKNLKSQFGQLAKQMAK-----------IPQGKFS--------PNALI 502
+ I N + Q +Q + +P+ +F P
Sbjct: 299 TNNPNLSYRSTNIANPQDQVYPPQQQQVQNKPFVPYNQGFVPKQQFQGNYQPPPPPGGKA 358
Query: 503 LSKHENADDVTTRS----GRVLHESKKQVEGEKNDKVEEKDEGVIKKRVSEPV------- 551
L E VT S G L K Q + +++ E + + +P+
Sbjct: 359 LPTREEPKTVTEDSEDQDGEDLSLEKDQADKPHEQPLDQSLEQPLDLSLEQPLDLPLDNV 418
Query: 552 ------------------------KARVLSTPSPELSKIPFPKALANKNLDKQFSKFLEV 587
K +V P P ++PFP DK + F +
Sbjct: 419 TRPTTRPIFPAASATAPKPITVKNKEKVF-VPPPYKPELPFPGRHKKALADKYRAMFAKN 477
Query: 588 FKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKI-PRKR 646
K++ + IP +AL + KF++D++ ++ + EV V+L+ CSAI+Q+KI P+K
Sbjct: 478 IKEVELRIPLVDALALIPDSHKFLKDLIVER--IQEVQGMVVLSHGCSAIIQKKIIPKKL 535
Query: 647 RDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKT 706
DPGSFT+P + L+ LCDLGAS++LM LS+ +RL + + +SL +ADRS++
Sbjct: 536 SDPGSFTLPCSLGPLAFNRCLCDLGASVSLMALSVAKRLGFTQYKSSNISLILADRSVRI 595
Query: 707 PYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVG 766
P+G++++ + + P DFVVL+M+E+ K PLILGRPFLATA A IDV KG++ L +G
Sbjct: 596 PHGLLKNFGITIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLSLG 655
Query: 767 KD-KVRFSVFNPNLQANLNDGFFCDMIQSLSRSSKETPKVSEKDVGLNQALIK 818
KD ++ F V + + + F I+ + + + E + ++ LN AL K
Sbjct: 656 KDFRMTFDVKDAMKKPTIEGQLF--WIKEMDQLADELLEELAEEDHLNSALTK 706
>At4g08050
Length = 1428
Score = 259 bits (663), Expect = 3e-69
Identities = 209/711 (29%), Positives = 332/711 (46%), Gaps = 94/711 (13%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I+++ +++ G EDP H++ F R CN +++ VS + +L LFPFSL D
Sbjct: 38 NFEIKSGLISMIQGNKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
A W + SITTW+D + F ++FF A +L+ +I F Q E+ EAWERFK
Sbjct: 98 KAHIWEKNLSHDSITTWDDYKKAFLSKFFSNARTARLRNEIYGFSQKTGESFCEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAARA- 353
+CP H+ T+A L++T Y G+ R LDTAS+G F + G++L+E +A
Sbjct: 158 GYTNQCPHHSFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSDG 217
Query: 354 -MNSENDRQTRRRMFEFEAYDK-LLASNKQISEKVAEIQNHI---------KVTKLVGAS 402
N + DR R + + K + A N ++ + Q H+ +V G
Sbjct: 218 NYNEDCDRTVRGTADSDDKHRKEIKALNDKLDRILLSQQKHVHFLVDDEQYQVQDGEGNQ 277
Query: 403 VAKVECVT-RGGPN----FSNSCTMTNQGFQGQ--------------------SSGLFQQ 437
+ +V + G N N + Q FQG + + Q
Sbjct: 278 LEEVYLPQQKQGQNKPFVLYNQGFVPKQQFQGNYQPPPPPGFVTQQNQCHAAPDAKMKQM 337
Query: 438 VQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQLAKQMAKIPQGKFS 497
V++ + + + + L ++++ +Y +A ++ L ++ L Q A K +
Sbjct: 338 VKQLLQGQASSSMEIAKKLSELHHKLDCSYNDLNAKVEALNTKVRYLEGQSASTFSPKVT 397
Query: 498 --PNALILSKHENAD------------------DVTTRSGRVLH-ESKKQVE-------- 528
P I + E A D+ TR V E+ QVE
Sbjct: 398 RLPGKSIQNPKEYATAHAITICHDRELPTRHVLDLITRDNDVQEGEASTQVEASVVEFNH 457
Query: 529 --GEKNDKVEEKDEGVIKKRVSEPVKARVLSTPSPELSKIPFP---------KALANKNL 577
G ++ +E K + E + R TP P L +P+ K++A K L
Sbjct: 458 SAGSRHLTQSTSEE---KAAIIERMVKRFKPTPLP-LRALPWTFRKAWMERYKSVAAKQL 513
Query: 578 DKQFSKFLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAI 637
D+ + +P E L +L K +R+++ ++ K+ + A
Sbjct: 514 DE-----------IEAVMPLMEVLNLILDPHKVVRNLILERIKMYHDSDDESDATPSRAA 562
Query: 638 LQRKIPRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSL 697
+R + K DPGSFT+P I + D LCDLGAS+NLMPLSM RRL + P L+L
Sbjct: 563 DKRIVQEKLEDPGSFTLPCSIGEFAFSDCLCDLGASVNLMPLSMARRLEFIQYKPCDLTL 622
Query: 698 QMADRSLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVD 757
+ADRS + +G+++D+ V ++ P DFVVLDME + K PLILGRPFLA+ A IDV
Sbjct: 623 ILADRSSRKHFGMLKDLPVMINGVEVPTDFVVLDMEVEHKDPLILGRPFLASVGAVIDVK 682
Query: 758 KGQLILRVGKD-KVRFSVFNPNLQANLNDGFFCDMIQSLSRSSKETPKVSE 807
+G++ L +GK K++F++ N Q + DG + +SR ET +V E
Sbjct: 683 EGKIGLNLGKHIKLQFNI-NRTPQGSTEDGRTSGNDRVISREEYETERVKE 732
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 253 bits (646), Expect = 3e-67
Identities = 199/690 (28%), Positives = 319/690 (45%), Gaps = 73/690 (10%)
Query: 185 LVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKDAAEEWLNSQP 244
++ +++ G EDP H++ F R N +++ VS + +L LFPFSL D A W + P
Sbjct: 1 MIQGNKFHGLPMEDPLDHLDEFDRLYNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLP 60
Query: 245 QGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFKKLLRKCPQHN 304
SITTW+D + F ++FF A +L+ +I F Q E+ EAWERFK +CP H
Sbjct: 61 HDSITTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTRESFCEAWERFKGYTNQCPHHG 120
Query: 305 LTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAARAMNSENDRQTRR 364
T+A L++T Y G+ R LDT+S+G F + G++L+E +A N D
Sbjct: 121 FTKASLLSTLYRGVLLRIRMLLDTSSNGNFQNKDVEEGWELVENLAQSDGNYNEDCDRNE 180
Query: 365 RMFEFEAYDK-LLASNKQISEKVAEIQNHIKVTKLVGASVAKVECV--TRGG-------- 413
+ D LL+ K + V + Q ++ + G + +V + +GG
Sbjct: 181 IKALNDKLDMILLSQQKHVHFLVDDKQFQVQYGE--GNQLEEVSYIINNQGGYKGYNNFK 238
Query: 414 ---PNFSNSCTMT-----------------NQG------FQG-------------QSSG- 433
PN S T NQG FQG Q+ G
Sbjct: 239 TNNPNLSYRSTNVANPPQQQQGQNKPFVPYNQGFVPKQQFQGNYQPPPPPGFTTQQNQGP 298
Query: 434 ------LFQQVQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQLAKQ 487
+ Q VQ+ ++V+ + + L ++++ +Y +A ++ L ++ L Q
Sbjct: 299 AAPDAEMKQMVQQLLEVQASSSMEIAKKLSELHHKLDCSYNDMNAKVEALNTKVRYLEGQ 358
Query: 488 MAKIPQGKFS--PNALILSKHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKD--EGVI 543
A K + P I + E A T + + H+ + + + D +G
Sbjct: 359 SASTSTPKVTGLPGKSIQNPKEYA---TAHAITICHDRELPTRHVPDFITGDSDVQDGEA 415
Query: 544 KKRVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQF-SKFLEVFKK----LHINIPFS 598
+ + KA ++ P P + K + K+ V K + +P
Sbjct: 416 STQSTSEEKAAIIERMVKRFKPTPLPSSALPWTFRKAWMEKYKSVASKQLDEIEAVMPLI 475
Query: 599 EALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEI 658
E L + K +R+++ ++ K+ + A +R + K DPGSFT+P I
Sbjct: 476 EVLNLIPDPHKDVRNLILERIKMYHDSDDESDATPSRASDKRIVQEKLEDPGSFTLPCSI 535
Query: 659 EGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKV 718
L+ D LCDLGAS++LMPLS+ RRL + P L+L +ADRS + P+G+++D+ V +
Sbjct: 536 GELAFSDCLCDLGASVSLMPLSVARRLEFIQYKPCDLTLILADRSFRKPFGMLKDLPVMI 595
Query: 719 DKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKD-KVRFSVFNP 777
+ P DFVVLDME + K PLILGRP LA+ A IDV +G++ L +GK K++F + N
Sbjct: 596 NGVEVPTDFVVLDMEVEHKDPLILGRPLLASVGAVIDVREGKISLNLGKHIKLQFGI-NK 654
Query: 778 NLQANLNDGFFCDMIQSLSRSSKETPKVSE 807
Q + DG + +SR ET +V E
Sbjct: 655 TPQGSTEDGRTSGNDRVISREGYETERVKE 684
>At3g31340 Athila ORF 1, putative
Length = 781
Score = 239 bits (609), Expect = 6e-63
Identities = 182/652 (27%), Positives = 305/652 (45%), Gaps = 73/652 (11%)
Query: 147 ANRSLRELTSAPMSYDYPGSIVFPEGVGNFKLRPAFINLVSQSQYGGGSSEDPHAHMERF 206
A ++LR+ + + + + P +F+++ + LV Q+Q+ SSE+ H++ F
Sbjct: 58 ARQTLRDHDAPNIDFFRDRNQHLPINRNDFEIKTGLLALVKQNQFFVHSSENHVYHLDNF 117
Query: 207 IRNCNTYRVSNVSSEAIRLSLFPFSLKDAAEEWLNSQPQGSITTWEDLAEKFTTRFFPRA 266
C R++ V + I+ LF FSL D A WL S ++ +WED F ++F ++
Sbjct: 118 EDICRITRMNGVPDDIIKCRLFIFSLADNAHRWLKSLDPINLRSWEDYKAAFLGQYFTQS 177
Query: 267 LLRKLKKDIMNFVQSAEENLYEAWERFKKLLRKCPQHNLTQAELVATFYDGLEYSWRFGL 326
L+ I +F Q E+ EAWERFK R+CP H ++A L++TFY G++ +++ L
Sbjct: 178 RTAILRNKISSFQQGGTESFPEAWERFKDYYRECPHHGFSRATLISTFYQGVDKAYKMAL 237
Query: 327 DTASSGEFDALPPQAGYDLIEKMAARAMNS--ENDRQTRRRMFEFEAYDKLLASNKQISE 384
DTAS+G+F LI+ +AA +N + DR R E + +L +Q+
Sbjct: 238 DTASNGDFMTKTETEATKLIKNLAASNINHNVDYDRSNRGGGGELKQLAELSVKVEQLLR 297
Query: 385 KVAEIQNHIK-VTKLVGASVAKVECVT--RGGPNFSNSCT--MTNQGFQGQS-------- 431
+ ++ N + +K + +C + NF N Q + Q+
Sbjct: 298 RDQKVINFCEDSSKGMVHQEYSGDCSEDLQAEMNFVNGIRYIQLRQAHKVQNFSPTDFRI 357
Query: 432 SGLFQQVQEQVDVKDGGKKSLEEL---LESFINRMESNYKIQDAAIKNLKSQFGQLAK-- 486
G + + +G KK+ ++ ++S N + + + +K L++Q Q+
Sbjct: 358 KGTIRDNSNLQQILEGLKKNAADINVKVDSMYNDLNVKFATLSSHVKTLENQVSQVVSAS 417
Query: 487 -QMAKIPQGKFSPNALILSKHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKK 545
+ A GK P + ++K + V VE DK+ E DE + ++
Sbjct: 418 MRPAGAHSGKEEPREIDVAKQVETNAVV-------------VETLVEDKIVEDDEPLSEE 464
Query: 546 RVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQFSKFLEVFKKLHINIPFSEALEQML 605
P P + K+PFP ++Q S E+
Sbjct: 465 -------------PPPYVPKLPFP------GRERQIQ---------------SRKREEYA 490
Query: 606 IYAKFMRDILSKKRKLSEVDETVM---LTEECSAILQRKIPRKRRDPGSFTIPVEIEGLS 662
+ + R K+ +L++V E + + CSAI IP K DPGSF +P I +
Sbjct: 491 LLVESQRQ--QKEAQLTDVVEVLAGKEMVSTCSAIPPATIPEKLGDPGSFVLPCRIGKSA 548
Query: 663 KVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYV 722
LCDLGA +NLMPLSM +RL + P+ +SL +ADRS++ P G+ E+V V+V +
Sbjct: 549 FERCLCDLGAGVNLMPLSMSKRLGITNFKPSRISLILADRSVRFPVGLAENVHVRVGDFY 608
Query: 723 FPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKDKVRFSV 774
P DFVVL+++++ PL LGRPFL T A IDV + + L++G + F +
Sbjct: 609 IPTDFVVLELDKEPHDPLTLGRPFLNTVGAIIDVRRSTINLQIGDFALEFDM 660
>At3g32000 unknown protein
Length = 839
Score = 238 bits (606), Expect = 1e-62
Identities = 195/701 (27%), Positives = 330/701 (46%), Gaps = 70/701 (9%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I+++ +++ G EDP H++ F R CN +++ VS +L LFPFSL D
Sbjct: 38 NFEIKSGLISMIQGNKFHGLPMEDPLDHLDEFDRLCNLTKINGVSENGFKLRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
A W + P SITT +D + F ++FF +L+ +I F Q E+ EAWERFK
Sbjct: 98 KALIWEMNLPHDSITTRDDCKKAFLSKFFSNDRTARLRNEISGFSQKIGESFCEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAAR-- 352
+CP H T+A L++T Y G+ R LDTAS+G F + G++L++ +A
Sbjct: 158 DYTNQCPHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVDNLAQSDG 217
Query: 353 AMNSENDRQTRRRMFEFEAYDK-LLASNKQISEKVAEIQNHI---------KVTKLVGAS 402
N + DR R + + K + A N ++ + Q H+ +V G
Sbjct: 218 NYNEDCDRTVRGTADSDDKHRKEIKALNDKLDRILLSQQKHVDFLVDDEQYQVQDGEGNQ 277
Query: 403 VAKVECV--TRGGPNFSNSCTMTNQGFQGQSSGLFQQVQEQV---DVKDGGKKSLEELLE 457
+ +V + +GG N+ N +S+ + Q+QV + G K +
Sbjct: 278 LEEVSYINNNQGGYKGYNNFKTNNPNLSYRSTNV-ANPQDQVYPPQQQQGQNKPFVPYNQ 336
Query: 458 SFI--NRMESNYK----------------IQDAAIKNLKSQFGQ--------LAKQMA-- 489
F+ + + NY+ DA +K + Q Q +AK+++
Sbjct: 337 GFVPKQQFQGNYQQPPPPGFAPQQNQGPATPDAEMKQMVQQLLQGQASSSMEIAKKLSEL 396
Query: 490 --KIPQGKFSPNALILS-----KHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGV 542
K+ NA + + ++ +T + +V K + ++ + + + V
Sbjct: 397 HHKLDCSYNDLNAKVEALNTKVRYLEGQSASTSAPKVTGLPGKDSDVQEGEAFTQVEVSV 456
Query: 543 IKKRVSEPVKARVLSTPSPELSKIP------FPKALANKNLDKQFSK-FLEVFK------ 589
++ S + + ST + + I P L ++ L F K ++E +K
Sbjct: 457 VEFNHSAGSRHLIQSTSEEKAAIIERMVKRFKPTPLPSRALPWTFRKSWMERYKSVAAKQ 516
Query: 590 --KLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRR 647
++ +P E L + K +R+++ ++ K+ + A +R + K
Sbjct: 517 PDEIEAVMPLMEVLNLIPDPHKDVRNLILERIKMYHDSDDESDATPSRAADKRIVQEKLE 576
Query: 648 DPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTP 707
DPGSF++P I + D LCDLGA ++LMP S+ RRL + P L+L +ADRS + P
Sbjct: 577 DPGSFSLPCSIGEFAFSDCLCDLGAFVSLMPCSVARRLEFIQYKPCDLTLILADRSSRKP 636
Query: 708 YGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGK 767
+G+++D+ V ++ P DFVVLDME + K PLILGRPFLA+ A IDV +G++ L +GK
Sbjct: 637 FGMLKDLPVMINGVEVPTDFVVLDMEVEHKDPLILGRPFLASVGAVIDVREGKIGLNLGK 696
Query: 768 D-KVRFSVFNPNLQANLNDGFFCDMIQSLSRSSKETPKVSE 807
K++F + N Q + DG + +S ET +V E
Sbjct: 697 HIKLQFGI-NKTPQGSTEDGKTSGNDRVISGEGYETERVKE 736
>At2g10670 pseudogene
Length = 929
Score = 219 bits (559), Expect = 4e-57
Identities = 179/673 (26%), Positives = 301/673 (44%), Gaps = 95/673 (14%)
Query: 165 GSIVFPEGVGNFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIR 224
G ++ P NFK++ I +V +++ G EDP H++ R C +++ VS + +
Sbjct: 60 GIVLRPVQNNNFKIKSGLIAMVQGNKFHGLPMEDPLDHLDELERLCGLTKINGVSEDGFK 119
Query: 225 LSLFPFSLKDAAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEE 284
L LFPFSL D A W + P+ SITTW+D F +FF + +L+ +I F E
Sbjct: 120 LRLFPFSLGDKAHLWEKTLPKNSITTWDDCKMAFLAKFFSNSRTARLRNEISGFTLKQNE 179
Query: 285 NLYEAWERFKKLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYD 344
+ EAWERFK KCP H +QA L+ T Y G+ R LDTAS+G F + G++
Sbjct: 180 SFCEAWERFKGYQTKCPHHGFSQASLLNTLYRGVLPKIRMLLDTASNGNFLKKDIEEGWE 239
Query: 345 LIEKMAARAMNSENDRQTRRRMFEFEAYDKLLASNKQISEKVAEI----QNHI------- 393
L+E +A N N+ R DK K +++K+ +I Q H+
Sbjct: 240 LVENLAQSGGN-YNEDYGRSIYTSSNTDDKHHREMKALNDKLDKIIQMQQKHVHFISEDE 298
Query: 394 --KVTKLVGASVAKVECVTRGGPNFSNSCT-----------------MTNQGFQG----- 429
+V + A++ V GP+ ++ + Q FQG
Sbjct: 299 PFQVQEGENDQCAEIRYVHNQGPSLPSAAAAAEPTKPFVPYNQSLGFVPKQQFQGGYQQQ 358
Query: 430 ----------------QSSGLFQQVQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAA 473
Q+S + +Q+ + + G + + L N+++ ++
Sbjct: 359 QPPPGFTPHQQQAHAPQNSDIMTVLQQLIQGQATGAMEIAKKLSKVNNKVDRQGVELNSK 418
Query: 474 IKNLKSQFGQLAKQMAKIPQGKFSPNALILSKHENADDVTTRSG-RVLHESKKQVEGEKN 532
+++ ++ + +A P +P L +N +T + H+ +
Sbjct: 419 FESMSTRMRYVEGILAS-PSVNNNPCQLPGKAIQNPKQYSTAHAITITHDRELPTRYVPT 477
Query: 533 DKVEE----------KDEGVIKKRVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQFS 582
E+ +D+ + + EP+ + P + K A K D F
Sbjct: 478 SNTEDSVILEGEDFYQDDVLADNPIEEPILTSQPTRPQAPPATPFAKKTEAAKTKDIDFV 537
Query: 583 KFLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKI 642
+K L +PF +F + +++K R L E + +C A++
Sbjct: 538 P--PPYKPL---LPFP---------GRFKKVLVAKYRALLEKHIKDIPLVDCLALIHD-- 581
Query: 643 PRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADR 702
E + L+ + LCDLGAS++LMPLS+ ++L P L+L +ADR
Sbjct: 582 --------------EHKQLTFNNCLCDLGASVSLMPLSVVKKLGFVHYKPCDLTLILADR 627
Query: 703 SLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLI 762
+ KTP+G++EDV V ++ P DFVVL+M+ ++K PLILGRPFLA+ A IDV +G++
Sbjct: 628 TSKTPFGLLEDVPVMINGVEVPTDFVVLEMDGESKDPLILGRPFLASVGAVIDVKQGKIN 687
Query: 763 LRVGKD-KVRFSV 774
L +G+D K++F +
Sbjct: 688 LNLGEDVKMKFDI 700
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 173 bits (439), Expect = 3e-43
Identities = 122/389 (31%), Positives = 201/389 (51%), Gaps = 40/389 (10%)
Query: 432 SGLFQQVQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQLAKQMAKI 491
S L +Q+ + + G L + + N+++ +Y + ++ L S+ + Q
Sbjct: 353 SDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGST 412
Query: 492 PQGKFS-PNALILSKH-ENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIK----- 544
KF+ P+ +S E A +T RSG+ L +K+ D +++ E +
Sbjct: 413 AAPKFTGPSGKSMSNSKEYAHAITLRSGKEL-PTKESPNQNTEDSLDQDGEDFCQNGNSA 471
Query: 545 -KRVSEPV-----------------KARVLST------PSPELSKIPFPKALANKNLDKQ 580
K + EP+ K T P P +PFP + K
Sbjct: 472 EKAIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKY 531
Query: 581 FSKFLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQR 640
+ + K L + +P + L + K+++D+++++ K EV V+L+ ECSAI+Q+
Sbjct: 532 KALLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITERIK--EVQGMVVLSHECSAIIQQ 589
Query: 641 KI-PRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQM 699
KI P+K DPGSFT+P + L+ LCDLGAS++LMPL + ++L + P +SL +
Sbjct: 590 KIIPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLIL 649
Query: 700 ADRSLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKG 759
ADRS++ +G++ED+ V + P DFVVL+M+E+ K PLILGRPFLA ARA IDV KG
Sbjct: 650 ADRSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKG 709
Query: 760 QLILRVGKD-KVRFSVFN----PNLQANL 783
++ L +G+D K+ F + N P ++ N+
Sbjct: 710 KIDLNLGRDLKMTFDITNTMKKPTIEGNI 738
Score = 132 bits (331), Expect = 1e-30
Identities = 78/241 (32%), Positives = 121/241 (49%), Gaps = 23/241 (9%)
Query: 157 APMSYDYPGSIVFPEGVGN-FKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRV 215
AP +++ IV P N F+++ I +V +++ G EDP H++ F R C+ ++
Sbjct: 19 APRNHNQRNGIVPPPVQNNNFEIKSGLIAMVQSNKFHGLPMEDPLDHLDEFDRLCSLTKI 78
Query: 216 SNVSSEAIRLSLFPFSLKDAAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDI 275
+ VS + +L LFPFSL D A +W S PQGSIT+W D + F +FF + +L+ DI
Sbjct: 79 NRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDI 138
Query: 276 MNFVQSAEENLYEAWERFKKLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFD 335
F Q+ E YEAWERFK +CP H + LDTAS+G F
Sbjct: 139 SGFTQTNNETFYEAWERFKGYQTQCPHHEML-------------------LDTASNGNFL 179
Query: 336 ALPPQAGYDLIEKMAAR--AMNSENDRQTRRRMFEFEAYDK-LLASNKQISEKVAEIQNH 392
+ G++++E +A N + DR R E + + + A N ++ + + Q H
Sbjct: 180 NKDVEDGWEVVENLAQSDGNYNEDYDRSIRTSSDSDEKHRREMKAMNDKLDKLLLMQQKH 239
Query: 393 I 393
I
Sbjct: 240 I 240
>At3g30630 hypothetical protein
Length = 785
Score = 172 bits (436), Expect = 7e-43
Identities = 180/710 (25%), Positives = 300/710 (41%), Gaps = 106/710 (14%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NFK++ I+++ +++ G ED H++ F R CN +V+ VS + +L LFPF L D
Sbjct: 38 NFKIKSGLISMIQGNKFHGLPMEDLLDHLDEFDRLCNLTKVNGVSEDGFKLRLFPFFLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
A W + P SI TW D + F +FF A +L +I +F Q E+ EAWERFK
Sbjct: 98 KAHIWEKNMPHDSIITWVDCKKAFLAKFFSNAKTARLINEISSFSQKTGESFCEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMA---- 350
+CP H +A L++T Y G+ R LDTAS+ F + G++LIE +A
Sbjct: 158 GYTNQCPHHGFKKASLLSTLYRGVLPRIRMLLDTASNENFQNKDVEEGWELIENLAQSND 217
Query: 351 ---------ARAMNSENDRQTRRRMFEFEAYDKLLASNKQ-----ISEKVAEIQNHIKVT 396
R +D+ + + DK+L S ++ + ++ ++Q+ +
Sbjct: 218 NYNKDCERTIRGTTDSDDKHRKEIKALNDKQDKILLSQQKHVHFLVDDEQYQVQDG-EGN 276
Query: 397 KLVGASVAKVECVTRGGPNF-----------SNSCTMTNQGFQGQS----SGLFQ----- 436
+L S + +G NF +N +Q + Q SG +Q
Sbjct: 277 QLEEVSYINNQSGYKGYNNFKTNNPNLSYRSTNIANPQDQVYPPQRQQAISGNYQQQPPP 336
Query: 437 ----QVQEQVDVKDGGKKSLEE--LLESFINRMESNYKIQDAAIKNLKSQFGQLAKQMAK 490
Q + D K + + L E + M+ Y +A ++ L ++ L Q A
Sbjct: 337 RFAPQQHQGPPAPDANMKQMAQQLLQEQASSSMDCRYNDLNAKVEALNTKVRYLEGQSAS 396
Query: 491 IPQGKFS--PNALILSKHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEG----VIK 544
K + P I ++ E A T + + H+ + K VE +I+
Sbjct: 397 TSAPKVTGLPGKPIQNQKEYA---TVNAITICHDRELPTRHVKVSVVEFNHSAGSHHLIQ 453
Query: 545 KRVSEPVKARVLSTPSPELSKIPFP-KALANKNLDKQFSKFLEVFKK----LHINIPFSE 599
E K ++ + P P +AL K ++ V +K + + +P E
Sbjct: 454 STSEE--KVAIIERMAKRFKPTPLPSRALPWKFRKAWMERYKSVAEKQLNEIEVVMPLME 511
Query: 600 ALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEIE 659
L + K +R+++ ++ K+ + A +R + K DPGS T+P I
Sbjct: 512 VLNVIHDPHKDVRNLILERIKMYHDSDDECDANPSRAADKRIVQEKLEDPGSCTLPCSIG 571
Query: 660 GLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVD 719
L+ + LCDLGAS++LMPLS+ R + + L+ A
Sbjct: 572 ELAFSNCLCDLGASVSLMPLSVAR-------SGSAYKLRCA------------------- 605
Query: 720 KYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKD-KVRFSVFNPN 778
++E+ K LIL RPFLA+ A IDV + ++ L +G K++F + N
Sbjct: 606 -----------RLKEEHKDHLILERPFLASVGAVIDVREVKINLNLGNHIKLQFDI-NKT 653
Query: 779 LQANLNDGFFCDMIQ------SLSRSSKETPKVSEKDVGLNQALIKLVSE 822
Q + DG + + S ++S KE K S+K + L +V E
Sbjct: 654 PQRSTEDGKTSEDDRVIPGEGSETKSVKELKKRSDKQEKTIEELAHIVEE 703
>At4g07600
Length = 630
Score = 151 bits (381), Expect = 2e-36
Identities = 88/237 (37%), Positives = 148/237 (62%), Gaps = 6/237 (2%)
Query: 584 FLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKI- 642
F + K++ + IP +AL +L KF++D++ ++ + EV V+L+ ECSAI+Q+KI
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVER--IQEVQGMVVLSHECSAIIQKKIV 59
Query: 643 PRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADR 702
P+K DPGSFT+P + ++ LCDLGA ++ MPLS+ +RL + +SL +ADR
Sbjct: 60 PKKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADR 119
Query: 703 SLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLI 762
S++ +G+++++ +++ DFV+L+M+E+ K PLIL RPFLATA A IDV KG++
Sbjct: 120 SVRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKID 179
Query: 763 LRVGKD-KVRFSVFNPNLQANLNDGFFCDMIQSLSRSSKETPKVSEKDVGLNQALIK 818
L +GKD ++ F + + + N+ F I+ + + + E + ++ LN AL K
Sbjct: 180 LNLGKDFRMTFDIKDAMKKPNIEGQLF--WIEEMDQLADELLEELAQEDHLNSALTK 234
>At4g07370
Length = 531
Score = 149 bits (377), Expect = 5e-36
Identities = 106/356 (29%), Positives = 178/356 (49%), Gaps = 46/356 (12%)
Query: 437 QVQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQLAKQMAKIPQGKF 496
QV +++ K + EL + +++ +Y +A ++ L S+ L + A K
Sbjct: 13 QVSSSIEIA----KKISELHQ----KLDCSYNDLNAKVEALNSKVRYLEGESASTSSPKV 64
Query: 497 S--PNALILSKHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKK--RVSEPVK 552
+ P I + E T + ++ DK E E ++ + R + P+
Sbjct: 65 TGLPGKSIQNPKEERPKSVTEDSAYQDGEDFSLNEDQVDKPTEVLEPILDRDTRPTNPLT 124
Query: 553 A------------RVLSTPSPELSKIPFPKALANKNLDKQFSKFLEVFKKLHINIPFSEA 600
+ + P P +PFP + DK + F + KK+ + IP ++A
Sbjct: 125 SSAALKLVAAKNNEIAFIPPPYKPPLPFPGRHKKEVEDKYRAMFAKNIKKVELRIPLADA 184
Query: 601 LEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRK-IPRKRRDPGSFTIPVEIE 659
L + KF++D++ ++ + EV +T +L+ ECSAI+Q + K DPGSFT+P +
Sbjct: 185 LTHIPDSQKFLKDLIMER--IQEVQKTTVLSHECSAIIQENDVSEKLGDPGSFTLPCSLG 242
Query: 660 GLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVD 719
L+ LCDLG S+NLMPLS+ A RS++ P+G++ED+ +K+
Sbjct: 243 SLTFNKCLCDLGPSVNLMPLSV------------------AKRSVRLPHGLLEDLPIKIR 284
Query: 720 KYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKD-KVRFSV 774
P DFVVL+M+E+ K PLILGRPFLAT IDV +G++ L +GK+ +++F +
Sbjct: 285 NVEVPTDFVVLNMDEEPKDPLILGRPFLATVGVIIDVKQGKIDLNLGKNFEMKFDI 340
>At1g41690 hypothetical protein
Length = 371
Score = 148 bits (373), Expect = 1e-35
Identities = 82/233 (35%), Positives = 128/233 (54%), Gaps = 5/233 (2%)
Query: 165 GSIVFPEGVGNFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIR 224
G + P NF+++ + I +V +++ G EDP H++ F R C+ +++ VS + +
Sbjct: 28 GIVPPPVQNNNFEIKSSLIAMVQGNKFHGLPMEDPLDHLDEFERLCSLTKINGVSEDGFK 87
Query: 225 LSLFPFSLKDAAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEE 284
L LFPFSL D A W + PQGSITTW+D + F +FF + +L+ +I F Q E
Sbjct: 88 LRLFPFSLGDKAHLWEKTLPQGSITTWDDCKKAFLAKFFSYSRTARLRNEISGFTQKQSE 147
Query: 285 NLYEAWERFKKLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYD 344
+ EAWERFK KCP H QA L++T Y G+ R LDTAS+G F + G++
Sbjct: 148 SFCEAWERFKGYQTKCPHHGFKQASLLSTLYRGVLPKIRMLLDTASNGNFLKKDVEEGWE 207
Query: 345 LIEKMAARAMNSENDRQTRRRMFEFEAYDKLLASNKQISEKVAEI----QNHI 393
L+EK A++ + N+ R ++ DK K +++K+ +I Q H+
Sbjct: 208 LVEKF-AQSDGNYNEDYDRSIRTSSDSDDKHRRDMKALNDKLDKILQVQQKHV 259
>At2g10620 putative Athila retroelement ORF1 protein
Length = 451
Score = 147 bits (372), Expect = 2e-35
Identities = 99/297 (33%), Positives = 161/297 (53%), Gaps = 38/297 (12%)
Query: 504 SKHENADDVTTRSGRVLHESKKQVEGEKNDKVEEKDEGVIKKRVSEPVKARVLSTPSPEL 563
SK A ++ R G V+ ++ V+ + D KD+ K + +P+ + L P
Sbjct: 123 SKGTKAKEMGKRFGGVI-VTEDSVDHDGEDFSLTKDQA--DKELEQPLD-QSLEQPLDHF 178
Query: 564 SKIPFP-------KALANKNLDKQFSK------------------------FLEVFKKLH 592
++ PFP K + KN +K F F + K++
Sbjct: 179 TRPPFPITAPTAVKPVVVKNKEKVFVPPPYKPHPSFPGRHKKALAHKYRVMFAKNIKEVE 238
Query: 593 INIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKR-RDPGS 651
+ IP +AL ++ KF++D++ ++ + E+ V+L++ECSAI+Q+KI K+ DPGS
Sbjct: 239 LQIPLVDALALIMDSHKFLKDLIVER--IQELQGMVVLSDECSAIIQKKIIHKKLSDPGS 296
Query: 652 FTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGII 711
FT+P + L+ LCDLGAS +LMPLS+ +RL + +SL +ADRS++ P+ +
Sbjct: 297 FTLPCSLGPLAFNKCLCDLGASASLMPLSVTKRLGFTQYKSCNISLILADRSVRIPHDFL 356
Query: 712 EDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKD 768
E++ +++ P DFVVL+M+E+ K LILGRPFL T A I + KG++ L +GKD
Sbjct: 357 ENIPIRIRAVEIPTDFVVLEMDEEPKDHLILGRPFLTTVGAMIYIKKGKINLNLGKD 413
>At3g30655 hypothetical protein, 3' partial
Length = 660
Score = 140 bits (353), Expect = 3e-33
Identities = 75/218 (34%), Positives = 115/218 (52%), Gaps = 2/218 (0%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I +V +++ G EDP H++ F R +++ VS + +L LFPFSL D
Sbjct: 38 NFEIKSGLIAMVQGNKFHGLPMEDPLDHLDEFERLYGLTKINGVSEDGFKLRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
A W + PQ SIT W+D + F +FF + +L+ +I F Q E+ EAWERFK
Sbjct: 98 KAHLWEKTLPQNSITAWDDCKKAFLAKFFSNSRTARLRNEISGFTQKQNESFGEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAAR-- 352
KCP H QA L++T Y G+ R LDTAS+G F + G++L+E A
Sbjct: 158 GYQTKCPHHGFKQASLLSTLYRGVLPKIRILLDTASNGNFLKKDVEEGWELVENFAQSDG 217
Query: 353 AMNSENDRQTRRRMFEFEAYDKLLASNKQISEKVAEIQ 390
N + DR R + +D+ + + EK+ ++Q
Sbjct: 218 NYNEDYDRSIRTSSDSNDKHDREMKALNDKLEKIVQMQ 255
Score = 86.7 bits (213), Expect = 5e-17
Identities = 50/122 (40%), Positives = 82/122 (66%), Gaps = 4/122 (3%)
Query: 595 IPFSEALEQMLIYAKFMRDILSKKRK-LSEVDETVMLTEECSAILQRKI-PRKRRDPGSF 652
+PF +++L+ AK+ R +L K+ K + V ++ +E +AI ++KI P K DPGSF
Sbjct: 540 LPFPGRFKKVLV-AKY-RALLEKQIKDMPLVAYLALIPDEHNAITRQKIVPEKLEDPGSF 597
Query: 653 TIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIE 712
T+P I L+ + LCDLGAS+++MPLS+ R+L + P+ L+L +ADR+ + P+G++E
Sbjct: 598 TLPCSIRQLTFSNCLCDLGASVSIMPLSVARKLGFVQYKPSDLTLILADRTSRRPFGLLE 657
Query: 713 DV 714
DV
Sbjct: 658 DV 659
>At4g03790 putative athila-like protein
Length = 1064
Score = 135 bits (341), Expect = 7e-32
Identities = 108/373 (28%), Positives = 177/373 (46%), Gaps = 33/373 (8%)
Query: 424 NQGFQGQSSGLFQQVQEQVDVKDGGKKSLEELLESFINRMESNYKIQDAAIKNLKSQFGQ 483
NQG + + Q VQ+ + + + + L ++++ NY +A ++ L ++
Sbjct: 161 NQGPTAPDAEMKQMVQQLLQGQASSSMEIPKKLSELHHKLDCNYNDLNAKVEALNTKVRY 220
Query: 484 LAKQMAK--IPQGKFSPNALILSKHENAD----------DVTTRS--GRVLHESKKQVEG 529
L Q A +P+ P I + E A ++ TR ++ ES Q EG
Sbjct: 221 LEGQSASTSVPKVTGLPGKSIQNPKEYATAHAITICHDRELPTRPVLDLIIGESDVQ-EG 279
Query: 530 EKNDKVEE---------------KDEGVIKKRVSEPVKARVLSTPSPELSKIPFPKALAN 574
E ++E + K + E + R TP P + P
Sbjct: 280 EACTQIEVSVIEFNHSAGSHHLIQSTSEEKAAIIERMVKRFKPTPLPSRA---LPWTFRK 336
Query: 575 KNLDKQFSKFLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVMLTEEC 634
+++ S + ++ +P E L + + K +R+++ +K K+ +
Sbjct: 337 AWIERYKSVAAKQLDEIEAVMPLMEVLNLIPDHHKDVRNLILEKIKMYHDSDDESDATPS 396
Query: 635 SAILQRKIPRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGEVTPTM 694
+ +R + K DPGSFT+P I L+ D LCDLGAS++LMPLSM RRL P
Sbjct: 397 RVVDKRIVQEKLEDPGSFTLPCSIGELAFSDFLCDLGASVSLMPLSMGRRLEFIHYKPCD 456
Query: 695 LSLQMADRSLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLATARAKI 754
L+L +ADRS + P+G+++D+ V ++ P DFVVL+ME K PLILGRPFLA+ A I
Sbjct: 457 LTLILADRSSRKPFGMLKDLPVMINGVEVPTDFVVLNMEVKHKDPLILGRPFLASVGAVI 516
Query: 755 DVDKGQLILRVGK 767
D+ +G++ L +GK
Sbjct: 517 DIREGKISLNLGK 529
Score = 65.9 bits (159), Expect = 9e-11
Identities = 29/77 (37%), Positives = 46/77 (59%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I+++ +++ G EDP H++ F R CN +++ VS + +L LFPFSL D
Sbjct: 38 NFEIKSGLISIIQGNKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTW 251
A W + SITTW
Sbjct: 98 KAHIWEKNLSHDSITTW 114
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 135 bits (340), Expect = 9e-32
Identities = 82/223 (36%), Positives = 114/223 (50%), Gaps = 39/223 (17%)
Query: 544 KKRVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQFSKFLEVFKKLHINIPFSEALEQ 603
K+ + E K VL P + K+PFP +K++S F E+ ++L P
Sbjct: 5 KQTLEEKAKKTVLP---PYVPKLPFPGRQRKIQREKEYSLFDEIMRQLQRYSP------- 54
Query: 604 MLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEIEGLSK 663
ECSAILQ IP KR DPGSF +P I +
Sbjct: 55 -----------------------------ECSAILQNVIPVKREDPGSFVLPSRIGEYTF 85
Query: 664 VDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYVF 723
LCDLGA ++LMP S+ +RL TPT +SL + DRS+ P G+ EDV V+V +
Sbjct: 86 DRCLCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYI 145
Query: 724 PIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVG 766
P DFV+++++E+ + LILGRPFL A IDV K ++ LR+G
Sbjct: 146 PTDFVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIG 188
>At2g06160 putative Athila retroelement ORF1 protein
Length = 750
Score = 134 bits (337), Expect = 2e-31
Identities = 73/223 (32%), Positives = 119/223 (52%), Gaps = 5/223 (2%)
Query: 175 NFKLRPAFINLVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKD 234
NF+++ I +V +++ G ED H++ F R C+ +++ VS + + LFPFSL D
Sbjct: 38 NFEIKSGLIAMVQGNKFHGLPMEDSLDHLDEFERLCDLTKINGVSEDGFKFRLFPFSLGD 97
Query: 235 AAEEWLNSQPQGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFK 294
W + PQ SITTW+D + F +FF + +L+ +I F Q E+ EAWERFK
Sbjct: 98 KTHLWEKTLPQNSITTWDDCKKAFFAKFFSNSRTARLRNEISGFTQKQNESFCEAWERFK 157
Query: 295 KLLRKCPQHNLTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAARAM 354
KCP QA L++T Y G+ R LDTAS+G F + G++L+E + A++
Sbjct: 158 GYQTKCPHPGFKQASLLSTLYRGVLPKLRMLLDTASNGNFLNKDVEKGWELVENL-AQSD 216
Query: 355 NSENDRQTRRRMFEFEAYDKLLASNKQISEKVAEI----QNHI 393
+ N+ + E+ DK K +++K+ ++ Q H+
Sbjct: 217 GNYNENYDKSISTSSESDDKHHREIKALNDKIDKLLQVQQKHV 259
Score = 126 bits (316), Expect = 6e-29
Identities = 82/235 (34%), Positives = 139/235 (58%), Gaps = 10/235 (4%)
Query: 544 KKRVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQFSKFLEVFKKLHINIPFSEALEQ 603
K + E + R PSP L+ +P+ A K++ + ++ + ++ +P E L+
Sbjct: 506 KATIIERMVKRFKPAPSPSLA-LPWTFRKAWKSIYESLAE--KQLDEMESVMPLVEVLKL 562
Query: 604 MLIYAKFMRDILSKKRKL---SEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEIEG 660
+ K +R+++ ++ + S+ +E V++ + +R I K DPGSFT+P I
Sbjct: 563 ISDPHKDVRNLILERLNIYQDSDDEEDVIMHQAAG---ERIIQEKLEDPGSFTLPCSISQ 619
Query: 661 LSKVDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDK 720
L+ + LCDLGAS++LMPLS+ R+L + ++L +ADR+ + P+ I+EDV V ++
Sbjct: 620 LTFSNCLCDLGASVSLMPLSVARKLRFVQYKSCDITLILADRTSRRPFSILEDVPVMING 679
Query: 721 YVFPIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGK-DKVRFSV 774
P DFVVL+M+E +K PLILGR FLA+ A IDV G++ L +G+ DK++F +
Sbjct: 680 VEVPTDFVVLEMDEKSKDPLILGRLFLASVGAVIDVRFGKIDLNLGRHDKLQFDI 734
>At1g36560 hypothetical protein
Length = 524
Score = 132 bits (332), Expect = 8e-31
Identities = 80/232 (34%), Positives = 129/232 (55%), Gaps = 4/232 (1%)
Query: 544 KKRVSEPVKARVLSTPSPELSKIPFPKALANKNLDKQFSKFLEVFKKLHINIPFSEALEQ 603
K + E + R TP P + P + + S + ++ +P E L
Sbjct: 20 KAAIIERMVKRFKPTPLPSRA---LPWTFRKAWMARYKSVAAKQLNEIEAVMPLMEVLNL 76
Query: 604 MLIYAKFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEIEGLSK 663
+ K +R+++ + K+ + A +R + K DPGSFT+P + L+
Sbjct: 77 IPNPHKDVRNLILEWIKMYHDSDDESDATPSRAADKRIVQEKLEDPGSFTLPCSLGSLTF 136
Query: 664 VDALCDLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYVF 723
LCDLGAS++LMPLS+ +RL + +SL +AD S++ P+G++ED+ +K+ K
Sbjct: 137 NKCLCDLGASVSLMPLSVAKRLGFNKYKYCNISLILADGSVRQPHGVLEDLPIKIGKVEV 196
Query: 724 PIDFVVLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKD-KVRFSV 774
P DF++L+M+E+ K PLILGRPFLATA A IDV +G++ L +GKD K++F +
Sbjct: 197 PTDFIILNMDEEPKDPLILGRPFLATAGAIIDVKQGKIDLNMGKDFKMKFDI 248
>At4g03850 putative transposon protein
Length = 334
Score = 130 bits (327), Expect = 3e-30
Identities = 87/238 (36%), Positives = 128/238 (53%), Gaps = 11/238 (4%)
Query: 570 KALANKNLDKQFSKFLEVFKKLHINIPFSEALEQMLIYAKFMRDILSKKRKLSEVDETVM 629
K++A K LD+ + +P E L + K +R+++ ++ K+
Sbjct: 5 KSVAAKQLDE-----------IEAVMPLIEVLNLIHDPHKNVRNLILERIKMYHDSNDDS 53
Query: 630 LTEECSAILQRKIPRKRRDPGSFTIPVEIEGLSKVDALCDLGASINLMPLSMFRRLNLGE 689
A +R + K DPGSFT+P I L+ D LCDLGAS++LMPLSM RRL +
Sbjct: 54 DATPSRAADKRIVQEKFGDPGSFTLPCSIGELAFSDCLCDLGASVSLMPLSMARRLEFIQ 113
Query: 690 VTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYVFPIDFVVLDMEEDAKIPLILGRPFLAT 749
P L+L +ADR+ + P+G+++D+ V ++ PIDFVVLDME + K PLILGRPFLA+
Sbjct: 114 YRPCDLTLILADRASRKPFGMLKDLPVMINGVEVPIDFVVLDMEVEHKDPLILGRPFLAS 173
Query: 750 ARAKIDVDKGQLILRVGKDKVRFSVFNPNLQANLNDGFFCDMIQSLSRSSKETPKVSE 807
A IDV +G++ L +GK + N Q + D + + ET KV E
Sbjct: 174 VGAVIDVREGKISLNLGKHIMLQFDINKTPQESKEDEKTSGDDRVIPGEKYETEKVKE 231
>At1g38440 hypothetical protein
Length = 263
Score = 130 bits (326), Expect = 4e-30
Identities = 71/213 (33%), Positives = 115/213 (53%), Gaps = 5/213 (2%)
Query: 185 LVSQSQYGGGSSEDPHAHMERFIRNCNTYRVSNVSSEAIRLSLFPFSLKDAAEEWLNSQP 244
++ ++++ G EDP H++ F R N +++ VS + +L LFPFSL D A W + P
Sbjct: 1 MIQENKFHGLPMEDPLDHLDEFDRLYNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLP 60
Query: 245 QGSITTWEDLAEKFTTRFFPRALLRKLKKDIMNFVQSAEENLYEAWERFKKLLRKCPQHN 304
SITTW+D + F ++FF A +L+ +I F Q E+ EAWERFK +CP H
Sbjct: 61 HDSITTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTSESFCEAWERFKGYTNQCPHHG 120
Query: 305 LTQAELVATFYDGLEYSWRFGLDTASSGEFDALPPQAGYDLIEKMAARAMNSENDRQTRR 364
T+A L++T Y G+ LD AS+G F + G++L+E + A++ + N+ R
Sbjct: 121 FTKASLLSTLYRGVLPCIIMLLDAASNGNFQNKDVEEGWELVENL-AQSYGNYNEDCDRT 179
Query: 365 RMFEFEAYDKLLASNKQISEKVAEI----QNHI 393
++ DK K +++K+ I Q H+
Sbjct: 180 VRGTADSNDKHRKEIKALNDKLDRIFLSQQKHV 212
>At2g10660 putative Athila retroelement ORF1 protein
Length = 1303
Score = 125 bits (315), Expect = 7e-29
Identities = 74/203 (36%), Positives = 113/203 (55%), Gaps = 3/203 (1%)
Query: 609 KFMRDILSKKRKLSEVDETVMLTEECSAILQRKIPRKRRDPGSFTIPVEIEGLSKVDALC 668
K +R+ + ++ K+ + E ++R + K DPGSFT+P I L + LC
Sbjct: 391 KDVRNSILERIKMYQDSEDECDANPSRVTVKRSVQEKLEDPGSFTLPCSIGQLVFSNCLC 450
Query: 669 DLGASINLMPLSMFRRLNLGEVTPTMLSLQMADRSLKTPYGIIEDVMVKVDKYVFPIDFV 728
DLGAS++LMPLS+ R+L + P LSL +ADRS + P+G+++ + + ++ P DFV
Sbjct: 451 DLGASVSLMPLSVARKLEFTQYRPCDLSLILADRSSRKPFGLLKHLPLMINGVEVPTDFV 510
Query: 729 VLDMEEDAKIPLILGRPFLATARAKIDVDKGQLILRVGKDKVRFSVF---NPNLQANLND 785
VL ME + K PLILGRPFLA+ A IDV G++ L +GK V ++ P+
Sbjct: 511 VLGMEVEPKDPLILGRPFLASVGAMIDVKDGRISLNLGKTTVEENIRAQPQPSNSITRQS 570
Query: 786 GFFCDMIQSLSRSSKETPKVSEK 808
F ++ L + S E + EK
Sbjct: 571 TTFTPDLRELKKKSDEQEETIEK 593
Score = 31.2 bits (69), Expect = 2.5
Identities = 20/79 (25%), Positives = 41/79 (51%), Gaps = 4/79 (5%)
Query: 141 EQEVENANRSLRELTSAPMSYDYPGSIVFPEGVGN-FKLRPAFINLVSQSQYGGGSSEDP 199
+++V+ N R+ AP + + IV P N F+++ I +V +++ G EDP
Sbjct: 38 DEQVQPNNIVARD---APRNQNQRNGIVPPPVQNNNFEIQSGLIAMVQSNKFHGLPMEDP 94
Query: 200 HAHMERFIRNCNTYRVSNV 218
+++ F R C+ +++ V
Sbjct: 95 LDYLDEFDRLCSLTKINGV 113
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,974,932
Number of Sequences: 26719
Number of extensions: 710830
Number of successful extensions: 3293
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 3126
Number of HSP's gapped (non-prelim): 184
length of query: 822
length of database: 11,318,596
effective HSP length: 108
effective length of query: 714
effective length of database: 8,432,944
effective search space: 6021122016
effective search space used: 6021122016
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0265.4


