

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0265.3
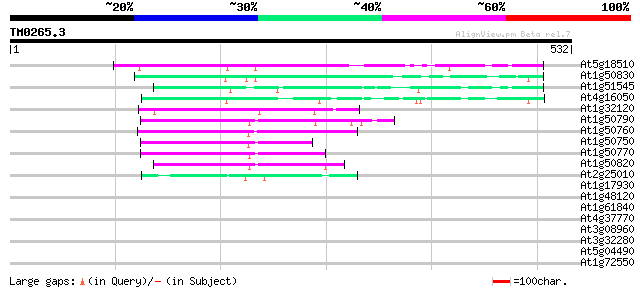
(532 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g18510 putative protein 99 6e-21
At1g50830 hypothetical protein 94 2e-19
At1g51545 unknown protein 93 4e-19
At4g16050 hypothetical protein 71 2e-12
At1g32120 hypothetical protein 67 2e-11
At1g50790 hypothetical protein 66 5e-11
At1g50760 hypothetical protein 66 5e-11
At1g50750 hypothetical protein 64 2e-10
At1g50770 hypothetical protein 60 2e-09
At1g50820 hypothetical protein 58 1e-08
At2g25010 unknown protein 48 1e-05
At1g17930 unknown protein 41 0.001
At1g48120 serine/threonine phosphatase PP7, putative 37 0.027
At1g61840 hypothetical protein 30 3.3
At4g37770 1-aminocyclopropane-1-carboxylate synthase - like pro... 30 4.4
At3g08960 hypothetical protein 30 4.4
At3g32280 hypothetical protein 29 5.7
At5g04490 unknown protein 29 7.4
At1g72550 putative phenylalanyl-tRNA synthetase beta-subunit; PheHB 29 7.4
>At5g18510 putative protein
Length = 702
Score = 99.0 bits (245), Expect = 6e-21
Identities = 119/434 (27%), Positives = 177/434 (40%), Gaps = 65/434 (14%)
Query: 99 PFLKDPKRTFRSAPPNPSAGEGA-----YLRWLDRVEASKFGHWKVTGIFDLIQLSRSPI 153
P L D +R S+ N S+G A +L WL +++A WK GIF+ I+ S I
Sbjct: 32 PRLDDQQRLSDSSLHNTSSGFWAADHHFFLSWLGKMQALYEPIWKKAGIFEAIKASTYKI 91
Query: 154 TYNPAMLLSSLFFWERSTNSFHVPFGTLSPTLLDVAAITGLWVVGDDYHSSA-------A 206
+ + +LS W T SF P+G + TL DV + G V+G S +
Sbjct: 92 IKDTSSILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPVFSPLECSEMRDS 151
Query: 207 PTNPIAIPTDNIAFSKFIKDHYVEN---GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPAK 262
+ D++ +K + + G EH AFL+ WLS +VF K R I
Sbjct: 152 AEKLEKVRRDSLGKAKRVSQRSWTSSFMGRGGQMEHEAFLVLWLSLFVFPGKFCRSISTN 211
Query: 263 LLPLANLLHEGRIIAMARLVLGNLYQMINEAIADIRDPKVASLNAAGPFWLLQLWMNAVF 322
++P+A L G IA+A VL LY+ ++ R +N F L+Q+W F
Sbjct: 212 VIPIAVRLARGERIALAPAVLAFLYKDLDRICDFSRGKCAGKVNLKSLFKLVQVWTWERF 271
Query: 323 EPSLPAKNPPPTVSNTRIEAFRLEALTPPYDASTFEFDFKKYFTLFFELKRFRSSFAPYH 382
SN R +A + P K L F++ +R PY
Sbjct: 272 -------------SNIRPKAKEIPKGEPRIAQWDGLQQISKNVKLSFDVFEWR----PYT 314
Query: 383 KPSYGPRWLRNSYPNLPGSENLSQHQVE-LWQTI---------LSPRVLTINFASNDFTL 432
KP L+N P L + E +W T+ R + +++ + + +
Sbjct: 315 KP------LKNWNP-------LRFYVDEAMWLTVDDSVDDAFASFARCVKVSYLAGNGFV 361
Query: 433 CGYNPQLVSRQFRLSQDLPNTLFDKSLILYPGTITKRSVFDTTIGYYNEEELLGLSPFSF 492
Y P V+RQF LSQDLP L+ IT++ +D Y N E L L S
Sbjct: 362 EDYFPYRVARQFGLSQDLP------GLVTRRRKITEKDAWD---DYSNSLEGLNLYLPSQ 412
Query: 493 TPSFYVTQAFKAWW 506
YVT ++ WW
Sbjct: 413 LDRGYVTARYQDWW 426
>At1g50830 hypothetical protein
Length = 768
Score = 93.6 bits (231), Expect = 2e-19
Identities = 101/403 (25%), Positives = 161/403 (39%), Gaps = 46/403 (11%)
Query: 119 EGAYLRWLDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPF 178
+ ++ WL ++EA W+ GIF+ I++S IT NP+++LS W T SF P+
Sbjct: 79 DAPFVSWLAKMEALHAQTWRKAGIFEAIKVSTYSITKNPSLILSVSEKWCPETKSFVFPW 138
Query: 179 GTLSPTLLDVAAITGLWVVGDDYHS--SAAPTNPIAIPTDNIAFSKF--IKDHYVEN--- 231
G + TL DV + G V+G + + T +N+ D V
Sbjct: 139 GEATITLEDVMVLLGFSVLGSPVFAPLETSETRDSVKKLENVRIQHMNSSTDRRVSQKSW 198
Query: 232 -----GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPAKLLPLANLLHEGRIIAMARLVLGN 285
G D EHVAFL+ WLS +VF KS R I + P+A L G IA+A +L
Sbjct: 199 VSTFLGRGGDMEHVAFLVLWLSLFVFPVKSRRNISNHVFPIAVRLARGERIALAPAILAI 258
Query: 286 LYQMINEAIADIRDPKVASLNAAGPFWLLQLWMNAVFEPSLPAKNPPPTVSNTRIEAFRL 345
LY+ ++ R+ V + F L+Q+W F P + P + L
Sbjct: 259 LYRDLDRIHEVSREDCVDKFHLESLFKLVQVWTWERFRNIRPKASDIPKGEPRIAQWHGL 318
Query: 346 EALTPPYDASTFEFDFKKYFTLFFELKRFRSSFAPYHKPSYGPRWLRNSYPNLPGSENLS 405
+ +F+++ Y + +++ P+ W + E++
Sbjct: 319 HRRSKDAWFCFDDFEWRPY-------TKALNNWNPFRFYLEEAIW-------VTVDESID 364
Query: 406 QHQVELWQTILSPRVLTINFASNDFTLCGYNPQLVSRQFRLSQDLPNTLFDKSLILYPGT 465
R +T++ D + Y P V+RQF L QDLP T
Sbjct: 365 DEFASF------ARCVTVSQIVGDGFVEDYFPNRVARQFGLDQDLPGL----------AT 408
Query: 466 ITKRSVFDTTIGYYNEEELLGLSPF--SFTPSFYVTQAFKAWW 506
+ S YN + L+GL+ + S VT ++ WW
Sbjct: 409 CQRNSTEKEAWNDYN-KSLIGLNLYMPSRLDQGSVTARYRVWW 450
>At1g51545 unknown protein
Length = 629
Score = 92.8 bits (229), Expect = 4e-19
Identities = 99/393 (25%), Positives = 157/393 (39%), Gaps = 66/393 (16%)
Query: 137 WKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTLLDVAAITGLWV 196
W+ GIF+ I+ S I N ++LL+ + W T SF P+G + TL DV + G V
Sbjct: 9 WRKAGIFEAIKASMYKIRKNQSLLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSV 68
Query: 197 VGDDYHSSAAPT-----------------NPIAIPTDNIAFSKFIKDHYVENGEVSDAEH 239
G + + + N+ S F+ G EH
Sbjct: 69 QGSPVFAPLESSEMRDSVEKLEKARLENRGQDGLVRQNLWVSSFL-------GRGDQMEH 121
Query: 240 VAFLLYWLSAYVF---CTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQMINEAIAD 296
AFL +WLS +VF C +S I K+LP+A L G IA A VL LY+ + + A
Sbjct: 122 EAFLAFWLSQFVFPDMCRRS--ISTKVLPMAVRLARGERIAFAPAVLARLYRDLGQIQAS 179
Query: 297 IRDPKVASLNAAGPFWLLQLWMNAVFEPSLPAKNPPPTVSNTRIEAFRLEALTPPYDAST 356
R+ ++ F L+QLW F+ + P P RI + + + +
Sbjct: 180 AREKSTPNVTLKSLFKLVQLWAWERFKSTSPKARVIPK-GEPRISRWHSQT-SKNVRLNL 237
Query: 357 FEFDFKKYFTLFFELKRFRSSFAPYHKPSY---GPRWLRNSYPNLPGSENLSQHQVELWQ 413
+FD++ PY KP PR+ + +NL V +
Sbjct: 238 VDFDWR-----------------PYTKPLQIWNPPRFYPEEAMWMTVDDNLDDGFVSFAR 280
Query: 414 TILSPRVLTINFASNDFTLCGYNPQLVSRQFRLSQDLPNTLFDKSLILYPGTITKRSVFD 473
+ +++ + + Y P V+ QF L+QDLP + D S + T++ +D
Sbjct: 281 CMRVSQLVGVGIVED------YYPNRVAMQFGLAQDLPGLVTDHS------SFTEKEAWD 328
Query: 474 TTIGYYNEEELLGLSPFSFTPSFYVTQAFKAWW 506
GY + L L S + VT+ ++ WW
Sbjct: 329 ---GYNKSLDGLMLYIPSRVATTSVTERYRDWW 358
>At4g16050 hypothetical protein
Length = 900
Score = 70.9 bits (172), Expect = 2e-12
Identities = 94/398 (23%), Positives = 156/398 (38%), Gaps = 66/398 (16%)
Query: 126 LDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTL 185
+D +A W +GIF+ I+ S I NP+++LS W TN+F P+G + TL
Sbjct: 330 IDGFQALHKPTWLKSGIFEAIKASTYRIHKNPSLILSLAQNWCPETNTFVFPWGEATITL 389
Query: 186 LDVAAITGLWVVGDDYHSS--AAPTNPIAIPTDNIAFSKFIKDHYVENGEVSDAEHVAFL 243
DV + G + G +S ++ ++ ++ + + EH AFL
Sbjct: 390 EDVNVLLGFSISGSSVFASLQSSEMKEAVEKLQKRCQGSMKQESWISSFVDDEMEHEAFL 449
Query: 244 LYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQMINE--AIADIRDPK 301
+ WLS +VF K G IA A VL NLY + +A I++
Sbjct: 450 VLWLSKFVFPDK-------------FCTRGERIAFAPAVLANLYNDLGHICVLASIQNVL 496
Query: 302 VASLNAAGPFWLLQLWMNAVFEPSLPAKNPPPTVSNTRIEAFRLEALTPPYDASTFEFDF 361
+SL F L+Q+W+ F+ P P RI + S + F
Sbjct: 497 ASSL-----FKLVQVWIWERFKSIRPEAKVIPR-GQPRIAQW-----------SGLKQRF 539
Query: 362 KKYFTLFFELKRFRSSFAPYHKP--SYGP--------RWLRNSYPNLPGSENLSQHQVEL 411
K + F + PY +P ++ P +W+R +L G + V
Sbjct: 540 KNVGLIIF---HGNFDWRPYSEPLENWNPPRFYVEEAKWVRID-ESLDGDYDDDDEFVSF 595
Query: 412 WQTILSPRVLTINFASNDFTLCGYNPQLVSRQFRLSQDLPNTLFDKSLILYPGTITKRSV 471
+ + +++ I N Y P V+ QF L+QD+P GT +R+
Sbjct: 596 ARCVRVSKLVGIGVVEN------YYPNRVAMQFGLAQDVP----------VLGTNHRRNF 639
Query: 472 FDTTIGYYNEEELLGLSPF--SFTPSFYVTQAFKAWWS 507
+ + L+GL + S + VT ++ WW+
Sbjct: 640 TEEEAWDDYNKPLVGLKLYFPSRVATASVTTRYRDWWA 677
>At1g32120 hypothetical protein
Length = 1206
Score = 67.0 bits (162), Expect = 2e-11
Identities = 56/226 (24%), Positives = 98/226 (42%), Gaps = 19/226 (8%)
Query: 123 LRWLDRVEASKFGH---WKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFG 179
L W++ V H WK +G++D I SR I + ++++ + W TN+F P+G
Sbjct: 82 LNWIEWVNVMAKSHATVWKKSGVYDAILASRYQIKRHDDLIVALVEKWCIETNTFVFPWG 141
Query: 180 TLSPTLLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIKDHYVENGEVS---- 235
+ TL D+ + GL V G++ + + ++I+ + VS
Sbjct: 142 EATLTLEDMIVLGGLSVTGNNALAPVKRDGMKEVEEKMKEAKRYIEVSLEKKCCVSMWMK 201
Query: 236 -------DAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLY- 287
+ EH AF++ WLS +VF + KL P A L +G +A+A VL +Y
Sbjct: 202 EMMNSGNEIEHEAFMVSWLSRFVFTNSGDVLREKLFPAAVQLAKGVRLALAPAVLARIYG 261
Query: 288 --QMINEAIADIRDPKVASLNAAGPFWLLQLWMNAVFEPSLPAKNP 331
++ E + + + + + PF +Q+W F P P
Sbjct: 262 DLGVLKEFLTGYSEKETVVVKS--PFQFVQVWALERFMALQPPGQP 305
>At1g50790 hypothetical protein
Length = 812
Score = 65.9 bits (159), Expect = 5e-11
Identities = 65/278 (23%), Positives = 116/278 (41%), Gaps = 43/278 (15%)
Query: 125 WLDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPT 184
W ++ A W+ GIF+ I S I N +++ W TN+F +G + T
Sbjct: 67 WARKMSALHEPIWRKAGIFEAILASTYKIFKNTDLVMGIAEKWCPDTNTFVFSWGEATIT 126
Query: 185 LLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIKD------------HYVENG 232
L DV + G V+G ++ + + + K KD ++++G
Sbjct: 127 LEDVMVLLGFSVLGSPVFATLDSSGKEIMAKLGKEWLKIKKDKGTFVTQIAWIERFMDSG 186
Query: 233 EVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQ---- 288
+ + EH+AFL+ WLS +VF T+ I + P+A L G +A+A VL +LY
Sbjct: 187 D--ELEHLAFLVLWLSYFVFPTRYYHIYEAIWPIAIHLSNGTKMALAPAVLAHLYADLSL 244
Query: 289 MINEAIADIRDPKVASLNAAGPFWLLQLWMNAVF-----EPSLPAKNPP----------- 332
+ N A P ++ + L+ +W+ F +P+L K P
Sbjct: 245 LKNHITALSESPIKVEIDLSSLCKLVNVWIWERFRALQSKPNLLLKGEPRLALWNDLKQR 304
Query: 333 -----PTVSNTRIEAFRLEALTPPYDASTFEFDFKKYF 365
++N++I+ F PY + +DF +++
Sbjct: 305 TSNAKRILNNSKIDGFE----WCPYTKTVKNWDFPQFY 338
>At1g50760 hypothetical protein
Length = 649
Score = 65.9 bits (159), Expect = 5e-11
Identities = 54/223 (24%), Positives = 93/223 (41%), Gaps = 16/223 (7%)
Query: 122 YLRWLDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTL 181
+ RW ++ A W+ GIF+ + S I N ++L W T +F +G
Sbjct: 53 FKRWARKMSALHEPIWRKAGIFEAVNASTYKIHKNTDLVLGVAEKWSPDTKTFVFSWGEA 112
Query: 182 SPTLLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIK-------------DHY 228
+ TL DV ++G V+G ++ + + A+ + D +
Sbjct: 113 TITLEDVMVLSGFSVLGSPAFATLDSSGKKIMERLKNAWQTIKREGKVNLLTQVAWMDRF 172
Query: 229 VENGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQ 288
+ G + EHVAFL+ WL +V ++ + + P+A L G IA+A VL +LY
Sbjct: 173 MNCG--GELEHVAFLVLWLGYFVLPSRFHHLCETIFPIAVNLSSGTRIALAPAVLAHLYA 230
Query: 289 MINEAIADI-RDPKVASLNAAGPFWLLQLWMNAVFEPSLPAKN 330
++ I + P + + F L+Q+W F P N
Sbjct: 231 DLSLLKTHISKSPPRVEIRLSALFMLVQVWTWERFRELQPKPN 273
>At1g50750 hypothetical protein
Length = 816
Score = 63.9 bits (154), Expect = 2e-10
Identities = 46/175 (26%), Positives = 80/175 (45%), Gaps = 14/175 (8%)
Query: 125 WLDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPT 184
W + A W+ GIF+ + S I NP ++L W T +F P+G + T
Sbjct: 42 WAIEMAALHEPTWREAGIFEAVMASIYRIPKNPDLILGIAEKWCPYTKTFVFPWGETAVT 101
Query: 185 LLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIK------------DHYVENG 232
L DV ++G V+G ++ + + + K K + ++++G
Sbjct: 102 LEDVMVLSGFSVLGSPVFATLDSSGKEVKAKLDKEWKKIKKAKVNFVTQVAWMERFMDSG 161
Query: 233 EVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLY 287
+ + EHVAFL+ WL+ +VF ++ + + P+ L G IA+A VL +LY
Sbjct: 162 D--ELEHVAFLVLWLNYFVFPSRLYHLYKAVFPIVVHLSTGTRIALALAVLAHLY 214
>At1g50770 hypothetical protein
Length = 632
Score = 60.5 bits (145), Expect = 2e-09
Identities = 46/186 (24%), Positives = 80/186 (42%), Gaps = 13/186 (6%)
Query: 125 WLDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPT 184
W ++ A W+ GIF+ + S I N ++L W T +F P+G + T
Sbjct: 66 WARKMAALHEPIWRKAGIFEAVTASTYKINPNTELVLGIAEKWCPDTKTFVFPWGETTIT 125
Query: 185 LLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIKD-----------HYVENGE 233
L DV + G V+G + + I + K KD ++++G+
Sbjct: 126 LEDVMLLLGFSVLGSPVFVTLDSSGEIIREKLEKEWKKVKKDKGNATQRTWKERFMDSGD 185
Query: 234 VSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQMINEA 293
+ EHVAFL+ WLS +VF ++ I +LP+A L RL + + +
Sbjct: 186 --ELEHVAFLVLWLSYFVFPSRYYHIYGAILPIAVHLSSDNDEVDGRLTIAQMVGPSKKK 243
Query: 294 IADIRD 299
+D+++
Sbjct: 244 YSDVKN 249
>At1g50820 hypothetical protein
Length = 528
Score = 57.8 bits (138), Expect = 1e-08
Identities = 50/197 (25%), Positives = 84/197 (42%), Gaps = 18/197 (9%)
Query: 137 WKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTLLDVAAITGLWV 196
W+ GIF+ + S I + ++L W T +F P+G + TL DV + G V
Sbjct: 75 WRKAGIFEAVIASTYKIPKDTDLVLGLAEKWCPDTKTFIFPWGEATITLEDVMVLLGFSV 134
Query: 197 VGDDYHSSAAPTNPIAIPTDNIAFSKFIKD------------HYVENGEVSDAEHVAFLL 244
+G ++ + + + K D ++ +G+ + EHV FL+
Sbjct: 135 LGLPVFATVDSSGKEIMAKLEKEWKKIKNDKVCLVTKLAWMERFMNSGD--ELEHVGFLV 192
Query: 245 YWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARLVLGNLYQMINEAIADIR----DP 300
WLS + F + I +LP+A L G +A+A VL +LY ++ IR
Sbjct: 193 LWLSYFAFPSHLFHISEAILPVAVHLSSGTKMALAPAVLAHLYADLSLLQGHIRVFSESL 252
Query: 301 KVASLNAAGPFWLLQLW 317
L+ F L+Q+W
Sbjct: 253 IKVQLDLNALFKLVQVW 269
>At2g25010 unknown protein
Length = 509
Score = 48.1 bits (113), Expect = 1e-05
Identities = 52/228 (22%), Positives = 93/228 (39%), Gaps = 40/228 (17%)
Query: 126 LDRVEASKFGHWKVTGIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTL 185
++ V+ + FG+++ G P++ N +++ + + W R TN+FH+P G ++ TL
Sbjct: 52 INLVDKAGFGYFRKIG----------PMSLNNSLISALVERWRRETNTFHLPLGEMTITL 101
Query: 186 LDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSK-----------------FIKDHY 228
+VA + GL + GD S + +A+ K ++K +
Sbjct: 102 DEVALVLGLEIDGDPIVGSKV-GDEVAMDMCGRLLGKLPSAANKEVNCSRVKLNWLKRTF 160
Query: 229 VENGEVSDAEHV-----AFLLYWLSAYVFC-TKSLRIPAKLLPLANLLHEGRIIAMARLV 282
E E + + V A+LLY + + +F T ++ K LPL + A
Sbjct: 161 SECPEDASFDVVKCHTRAYLLYLIGSTIFATTDGDKVSVKYLPLFEDFDQAGRYAWGAAA 220
Query: 283 LGNLYQMINEAIADIRDPKVASLNAAGPFWLLQLWMNAVFEPSLPAKN 330
L LY+ + A + N G LLQ W + P K+
Sbjct: 221 LACLYRALGNASLK------SQSNICGCLTLLQCWSYFHLDIGRPEKS 262
>At1g17930 unknown protein
Length = 478
Score = 41.2 bits (95), Expect = 0.001
Identities = 51/206 (24%), Positives = 85/206 (40%), Gaps = 37/206 (17%)
Query: 119 EGAYLRWLDRVEASKFGHWKVT------------GIFDLIQLSRSPITYNPAMLLSSLFF 166
E LR +R S HWK+T G F L+ I+ N +++ + +
Sbjct: 20 ERGVLRCQERT--SLLHHWKLTKEQIALVEKAGFGWFRLV----GSISLNNSLISALVER 73
Query: 167 WERSTNSFHVPFGTLSPTLLDVAAITGLWVVG---------DDYHSSAAPTNPIAIP--- 214
W R TN+FH P G ++ TL +V+ I GL V G D+ S +P
Sbjct: 74 WRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEKDEDPSQVCLRLLGKLPKGE 133
Query: 215 -TDNIAFSKFIKDHYVENGEVSDAEHV-----AFLLYWLSAYVFCTKS-LRIPAKLLPLA 267
+ N +K++K+ + E + + + + A+L+Y + + +F T +I L L
Sbjct: 134 LSGNRVTAKWLKESFAECPKGATMKEIEYHTRAYLIYIVGSTIFATTDPSKISVDYLILF 193
Query: 268 NLLHEGRIIAMARLVLGNLYQMINEA 293
+ A L LY+ I A
Sbjct: 194 EDFEKAGEYAWGAAALAFLYRQIGNA 219
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 37.0 bits (84), Expect = 0.027
Identities = 44/196 (22%), Positives = 84/196 (42%), Gaps = 28/196 (14%)
Query: 141 GIFDLIQLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTLLDVAAITGLWVVGDD 200
G++ + +++ + Y A++ + + W T++FH+P G ++ TL DV + GL V G
Sbjct: 68 GLYGVYKVAFIQLDY--ALITALVERWRPETHTFHLPAGEITVTLQDVNILLGLRVDGPA 125
Query: 201 YHSSAA-------------PTNPIAIPTDNIAFSKFIKDHY----VENGEVSDAEHV-AF 242
S P + +++ + ++++++ + EV+ H AF
Sbjct: 126 VTGSTKYNWADLCEDLLGHRPGPKDLHGSHVSLA-WLRENFRNLPADPDEVTLKCHTRAF 184
Query: 243 LLYWLSAYVFCTKSLR-IPAKLLPLANLLHEGRIIAMARLVLGNLYQMINEAIADIRDPK 301
+L +S +++ KS + LPL E ++ L LY+ + R K
Sbjct: 185 VLALMSGFLYGDKSKHDVALTFLPLLRDFDEVAKLSWGSATLALLYREL------CRASK 238
Query: 302 VASLNAAGPFWLLQLW 317
GP LLQLW
Sbjct: 239 RTVSTICGPLVLLQLW 254
>At1g61840 hypothetical protein
Length = 814
Score = 30.0 bits (66), Expect = 3.3
Identities = 21/84 (25%), Positives = 33/84 (39%), Gaps = 5/84 (5%)
Query: 368 FFELKRFRSSFAPYHKPSYGPRWLRNSYPNLPGSENLSQHQVELWQTILSPRVLTINFAS 427
F EL+ F YH P Y P S P S+ + H + L P + +S
Sbjct: 203 FHELENNGKLFFVYHHPKYQPIPQTKS----PSSDKTANHDLPFQPLFLCPHLRVTKNSS 258
Query: 428 NDFTL-CGYNPQLVSRQFRLSQDL 450
+ T Y+P+ + + +DL
Sbjct: 259 DSLTYPISYSPEYIVSTTKSDEDL 282
>At4g37770 1-aminocyclopropane-1-carboxylate synthase - like
protein
Length = 469
Score = 29.6 bits (65), Expect = 4.4
Identities = 17/60 (28%), Positives = 30/60 (49%), Gaps = 5/60 (8%)
Query: 185 LLDVAAITGLWVVGDDYHSSAAPTNPIAIPTDNIAFSKFIKDHYVENGEVSDAEHVAFLL 244
LLD + + ++ D+ +S TNP I+ + +KD +EN +V D H+ + L
Sbjct: 216 LLDFISRKKIHLISDEIYSGTVFTNP-----GFISVMEVLKDRKLENTDVFDRVHIVYSL 270
>At3g08960 hypothetical protein
Length = 754
Score = 29.6 bits (65), Expect = 4.4
Identities = 25/99 (25%), Positives = 43/99 (43%), Gaps = 10/99 (10%)
Query: 397 NLPGSENLSQHQVELWQTILSPRVLTINFASNDFTLCGYNPQLVSRQFRLSQDLPNTLFD 456
N P S NL + + LW+T LS + + L Y +++ R F Q + ++ D
Sbjct: 635 NSPDSLNLLEDSMALWETTLSYAPMMV---PQLLALFPYMVEIIERSFDHLQ-VAVSIMD 690
Query: 457 KSLILYPGTI------TKRSVFDTTIGYYNEEELLGLSP 489
+IL G + + D +G N++ LL + P
Sbjct: 691 SYIILDGGEFLNMHASSVAKILDLIVGNVNDKGLLSILP 729
>At3g32280 hypothetical protein
Length = 474
Score = 29.3 bits (64), Expect = 5.7
Identities = 16/46 (34%), Positives = 24/46 (51%)
Query: 236 DAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRIIAMARL 281
+ EHVAFL+ WL +VF + + + P+A L G A+ L
Sbjct: 10 ELEHVAFLVLWLRYFVFPSGFHYLYVTMFPIAIHLSSGTKTALVVL 55
>At5g04490 unknown protein
Length = 304
Score = 28.9 bits (63), Expect = 7.4
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query: 147 QLSRSPITYNPAMLLSSLFFWERSTNSFHVPFGTLSPTLLDVAAITGLW-VVGDDYHSSA 205
+L + P+ Y A+L S++FFW S P G +S L + G+ ++G + S+
Sbjct: 167 ELLKGPLFYVLALLFSAVFFWRES------PIGMIS--LAMMCGGDGIADIMGRKFGSTK 218
Query: 206 APTNP 210
P NP
Sbjct: 219 IPYNP 223
>At1g72550 putative phenylalanyl-tRNA synthetase beta-subunit; PheHB
Length = 598
Score = 28.9 bits (63), Expect = 7.4
Identities = 19/54 (35%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Query: 60 PLKKAKKGRPNSLPEGYEHFHPSIRGDELILAFQPSYRFPFLKDPKRTFRSAPP 113
PLK+ K R + L E Y+ ++ + + + S FP L D KRT S PP
Sbjct: 186 PLKQTKSFRADELIEFYKS---DMKLKKFLHIIENSPVFPVLYDSKRTVLSLPP 236
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,051,765
Number of Sequences: 26719
Number of extensions: 582070
Number of successful extensions: 1215
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1179
Number of HSP's gapped (non-prelim): 26
length of query: 532
length of database: 11,318,596
effective HSP length: 104
effective length of query: 428
effective length of database: 8,539,820
effective search space: 3655042960
effective search space used: 3655042960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0265.3


