

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0263.4
(195 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
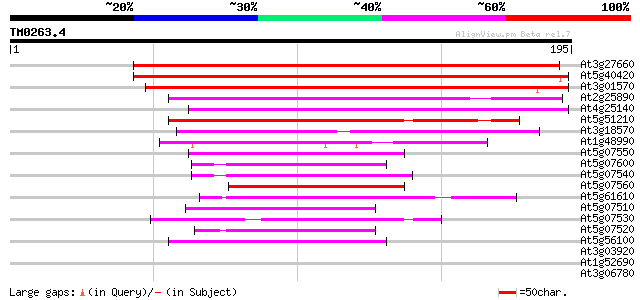
Score E
Sequences producing significant alignments: (bits) Value
At3g27660 oleosin isoform 182 1e-46
At5g40420 oleosin 180 5e-46
At3g01570 putative oleosin 169 1e-42
At2g25890 putative oleosin protein 104 3e-23
At4g25140 oleosin, 18.5K 91 3e-19
At5g51210 oleosin-like 91 4e-19
At3g18570 unknown protein 74 6e-14
At1g48990 oleosin like protein gb|AAC42242.1 62 2e-10
At5g07550 glycine-rich protein PUTG1 59 1e-09
At5g07600 unknown protein 56 1e-08
At5g07540 glycine-rich protein atGRP-6 56 1e-08
At5g07560 oleosin-like protein 55 2e-08
At5g61610 putative protein 54 5e-08
At5g07510 glycine-rich protein 52 3e-07
At5g07530 glycine-rich protein atGRP-7 51 3e-07
At5g07520 glycine-rich protein atGRP 50 8e-07
At5g56100 unknown protein (At5g56100) 41 5e-04
At3g03920 putative GAR1 protein 32 0.28
At1g52690 unknown protein 31 0.36
At3g06780 unknown protein 31 0.48
>At3g27660 oleosin isoform
Length = 191
Score = 182 bits (462), Expect = 1e-46
Identities = 93/148 (62%), Positives = 108/148 (72%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GG G S + RGPS +QI+A+ AGVPIGGTLL LAG++L S+IGL V PL +LFS
Sbjct: 32 GYGGYGAGSDYKSRGPSTNQILALIAGVPIGGTLLTLAGLTLAGSVIGLLVSIPLFLLFS 91
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLAV GIL SG FGLTGL S SW++NY+R T VPEQLDYAK RMAD
Sbjct: 92 PVIVPAALTIGLAVTGILASGLFGLTGLSSVSWVLNYLRGTSDTVPEQLDYAKRRMADAV 151
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPITT 191
Y G K KE+G +Q KAHEA+ T T
Sbjct: 152 GYAGMKGKEMGQYVQDKAHEARETEFMT 179
>At5g40420 oleosin
Length = 199
Score = 180 bits (456), Expect = 5e-46
Identities = 89/152 (58%), Positives = 112/152 (73%), Gaps = 1/152 (0%)
Query: 44 GGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFS 103
G GGGG+ S+ PE GPS++Q++++ GVP+ G+LL LAG+ L S+IGL V PL +LFS
Sbjct: 34 GYGGGGYKSMMPESGPSSTQVLSLLIGVPVVGSLLALAGLLLAGSVIGLMVALPLFLLFS 93
Query: 104 PVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
PVIVPA +TIGLA+ G L SG FGLTGL S SW+MNY+R TR VPEQL+YAK RMAD
Sbjct: 94 PVIVPAALTIGLAMTGFLASGMFGLTGLSSISWVMNYLRGTRRTVPEQLEYAKRRMADAV 153
Query: 164 DYVGQKTKEVGHDIQTKAHEAKRTPIT-THDT 194
Y GQK KE+G +Q KA + K+ I+ HDT
Sbjct: 154 GYAGQKGKEMGQHVQNKAQDVKQYDISKPHDT 185
>At3g01570 putative oleosin
Length = 183
Score = 169 bits (427), Expect = 1e-42
Identities = 83/150 (55%), Positives = 109/150 (72%), Gaps = 3/150 (2%)
Query: 48 GGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIV 107
GG L+P+ GPS++Q++AV GVPIGGTLL +AG++L S+IGL + PL ++FSPVIV
Sbjct: 21 GGIKVLYPQSGPSSTQVLAVFVGVPIGGTLLTIAGLTLAGSVIGLMLAFPLFLIFSPVIV 80
Query: 108 PAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVG 167
PA IGLA+ G L SGA GLTGL S SW++NYIR +PE+L+ AK+R+ADMA+YVG
Sbjct: 81 PAAFVIGLAMTGFLASGAIGLTGLSSMSWVLNYIRRAGQHIPEELEEAKHRLADMAEYVG 140
Query: 168 QKTKEVGHDIQTKAH---EAKRTPITTHDT 194
Q+TK+ G I+ KAH EAK + DT
Sbjct: 141 QRTKDAGQTIEDKAHDVREAKTFDVRDRDT 170
>At2g25890 putative oleosin protein
Length = 174
Score = 104 bits (259), Expect = 3e-23
Identities = 53/137 (38%), Positives = 82/137 (59%), Gaps = 7/137 (5%)
Query: 56 ERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGL 115
E PS QI+ IG +LL+L+G++L ++IGL V TPL++LFSPV+VPAV+TIGL
Sbjct: 45 ESSPSTRQIVRFVTAATIGLSLLVLSGLTLTGTVIGLIVATPLMVLFSPVLVPAVITIGL 104
Query: 116 AVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGH 175
G L SG G+ + +W+ Y+ + +++DYA+ R+A +K KE+GH
Sbjct: 105 LTMGFLFSGGCGVAAATALTWIYKYVTGKHPMGADKVDYARMRIA-------EKAKELGH 157
Query: 176 DIQTKAHEAKRTPITTH 192
++ + +T TTH
Sbjct: 158 YTHSQPQQTHQTTTTTH 174
>At4g25140 oleosin, 18.5K
Length = 173
Score = 91.3 bits (225), Expect = 3e-19
Identities = 45/132 (34%), Positives = 74/132 (55%)
Query: 63 QIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILT 122
QI A V GG+LL+L+ ++L+ ++I L V TPLL++FSP++VPA++T+ L + G L+
Sbjct: 42 QIAKAATAVTAGGSLLVLSSLTLVGTVIALTVATPLLVIFSPILVPALITVALLITGFLS 101
Query: 123 SGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTKAH 182
SG FG+ + FSW+ Y ++LD A+ ++ A + + + G H
Sbjct: 102 SGGFGIAAITVFSWIYKYATGEHPQGSDKLDSARMKLGSKAQDLKDRAQYYGQQHTGGEH 161
Query: 183 EAKRTPITTHDT 194
+ RT H T
Sbjct: 162 DRDRTRGGQHTT 173
>At5g51210 oleosin-like
Length = 141
Score = 90.9 bits (224), Expect = 4e-19
Identities = 48/122 (39%), Positives = 74/122 (60%), Gaps = 7/122 (5%)
Query: 56 ERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGL 115
E P A Q++ A V GG+LL+L+G++L ++I L V TPLL++FSPV+VPAV+T+ L
Sbjct: 19 EAHPKARQMVKAATAVTAGGSLLVLSGLTLAGTVIALTVATPLLVIFSPVLVPAVVTVAL 78
Query: 116 AVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGH 175
+ G L SG FG+ + +FSWL R G ++++ A+ ++ Q TK H
Sbjct: 79 IITGFLASGGFGIAAITAFSWL---YRHMTGSGSDKIENARMKVGSRV----QDTKYGQH 131
Query: 176 DI 177
+I
Sbjct: 132 NI 133
>At3g18570 unknown protein
Length = 166
Score = 73.6 bits (179), Expect = 6e-14
Identities = 41/126 (32%), Positives = 66/126 (51%), Gaps = 4/126 (3%)
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P++SQ+ A G LL L G+S+ A+++G VF PL+I+ SP+ +P + +G
Sbjct: 41 PTSSQLFGFLALFISTGILLFLLGVSVTAAVLGFIVFLPLIIISSPIWIPVFVVVG---- 96
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQ 178
G LT F + + SW Y R + Q+DYA++R+ D A +V +E G
Sbjct: 97 GFLTVSGFLVGTVALVSWTYRYFRGMHPVGSNQMDYARSRIYDTASHVKDYAREYGGYFH 156
Query: 179 TKAHEA 184
+A +A
Sbjct: 157 GRAKDA 162
>At1g48990 oleosin like protein gb|AAC42242.1
Length = 169
Score = 62.0 bits (149), Expect = 2e-10
Identities = 42/118 (35%), Positives = 66/118 (55%), Gaps = 11/118 (9%)
Query: 53 LFPERGPSAS-QIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVP--A 109
L + P+ S Q+ A GG LLLL GI++ A ++G F PL+I+ SP+ +P
Sbjct: 40 LLQSQSPNHSGQLFGFLAFFISGGILLLLTGITVTAFVLGFIAFLPLIIISSPIWIPLFL 99
Query: 110 VMTIGLAVAG-ILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYV 166
++T L++AG IL +GA SWL Y + + +Q+DYA++R+ D A +V
Sbjct: 100 IVTGFLSLAGLILATGAV-------VSWLYRYFKGMHPLRSDQVDYARSRIHDTAAHV 150
>At5g07550 glycine-rich protein PUTG1
Length = 106
Score = 59.3 bits (142), Expect = 1e-09
Identities = 29/75 (38%), Positives = 41/75 (54%)
Query: 63 QIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILT 122
+II + LL AGI+L S++ + TPL ++FSPV+VPA + L +G
Sbjct: 3 EIIQAVFSAGVALALLTFAGITLGGSVVACIISTPLFVIFSPVLVPATIATTLLASGFTA 62
Query: 123 SGAFGLTGLMSFSWL 137
SG+FG T SWL
Sbjct: 63 SGSFGATAFTILSWL 77
>At5g07600 unknown protein
Length = 230
Score = 55.8 bits (133), Expect = 1e-08
Identities = 27/68 (39%), Positives = 40/68 (58%), Gaps = 4/68 (5%)
Query: 64 IIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTS 123
+IAV A + + +G++L + +GL V TPL I+FSP++VPA + I L G T
Sbjct: 15 VIAVVASI----VFFVFSGLTLAGTAVGLTVTTPLFIIFSPILVPATIAITLLTTGFTTG 70
Query: 124 GAFGLTGL 131
GA G T +
Sbjct: 71 GALGATAI 78
>At5g07540 glycine-rich protein atGRP-6
Length = 244
Score = 55.8 bits (133), Expect = 1e-08
Identities = 28/77 (36%), Positives = 42/77 (54%), Gaps = 4/77 (5%)
Query: 64 IIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTS 123
+IA A V L+ AGI+ AS++GL + TPL ++FSP++VPA + V G +
Sbjct: 15 VIAAVASV----VFLVFAGITFGASIVGLTIATPLFVIFSPILVPATIATTFLVGGATAA 70
Query: 124 GAFGLTGLMSFSWLMNY 140
A G+T WL +
Sbjct: 71 VALGVTAFALILWLFKH 87
>At5g07560 oleosin-like protein
Length = 153
Score = 55.1 bits (131), Expect = 2e-08
Identities = 26/61 (42%), Positives = 38/61 (61%)
Query: 77 LLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSW 136
LLLLAGISL + + L PL ++FSPV+VPA + + +G++ G G++GL W
Sbjct: 44 LLLLAGISLTGTAVALFASMPLFLVFSPVLVPAGIATTILASGLMAGGTSGVSGLTILMW 103
Query: 137 L 137
L
Sbjct: 104 L 104
>At5g61610 putative protein
Length = 225
Score = 53.9 bits (128), Expect = 5e-08
Identities = 29/110 (26%), Positives = 55/110 (49%), Gaps = 6/110 (5%)
Query: 67 VAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSGAF 126
+ AG+ + + ++L G++ + + L V P+L++ SP++VPAV+T G L SG+
Sbjct: 12 IYAGIAVV-SFMILTGLTFAGTAVALTVMIPVLVVLSPILVPAVITSSFLATGFLASGSL 70
Query: 127 GLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHD 176
G G+ WL +R + +A+ D + + + K+ G D
Sbjct: 71 GALGIALLIWLYKKEEYSRATL-----HARGGRPDGPNKLAESGKQSGGD 115
>At5g07510 glycine-rich protein
Length = 193
Score = 51.6 bits (122), Expect = 3e-07
Identities = 23/66 (34%), Positives = 36/66 (53%)
Query: 62 SQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGIL 121
S++ V + L+LAG++L S L + TPL I+FSP++VPA + + G+
Sbjct: 10 SEVFQVVIAAVVSIVFLVLAGLTLAGSATALTITTPLFIIFSPILVPATIATAVITTGLT 69
Query: 122 TSGAFG 127
GA G
Sbjct: 70 AGGALG 75
>At5g07530 glycine-rich protein atGRP-7
Length = 543
Score = 51.2 bits (121), Expect = 3e-07
Identities = 34/101 (33%), Positives = 49/101 (47%), Gaps = 8/101 (7%)
Query: 50 FSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPA 109
FS L P I +VA+ + +G + LAG S L V TP+ I+FSPV+VPA
Sbjct: 56 FSFLMPLLEVIKIIIASVASVIFVGFACVTLAG-----SAAALVVSTPVFIIFSPVLVPA 110
Query: 110 VMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPE 150
+ + G G+FG T L WL ++ G+ P+
Sbjct: 111 TIATVVLATGFTAGGSFGATALGLIMWL---VKRRMGVKPK 148
>At5g07520 glycine-rich protein atGRP
Length = 228
Score = 50.1 bits (118), Expect = 8e-07
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query: 65 IAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTSG 124
+ +A V I L+ AG++L+ S L + TPL I+FSP++VPA + + G T G
Sbjct: 14 VVIATVVSI--VFLVFAGLTLVGSATALTITTPLFIIFSPILVPATIATAVITTGFTTGG 71
Query: 125 AFG 127
A G
Sbjct: 72 ALG 74
>At5g56100 unknown protein (At5g56100)
Length = 150
Score = 40.8 bits (94), Expect = 5e-04
Identities = 21/76 (27%), Positives = 39/76 (50%)
Query: 56 ERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGL 115
ER +S + A A V + G L L S +A++ + +PLL++F+P + V +
Sbjct: 18 ERTTGSSVVAATVAAVAVVGPLFGLMSFSFVATVTLFLIASPLLLIFAPAFMVTVAVLVS 77
Query: 116 AVAGILTSGAFGLTGL 131
A+ G+ + A + G+
Sbjct: 78 AMVGVGVAAAMWMMGI 93
>At3g03920 putative GAR1 protein
Length = 202
Score = 31.6 bits (70), Expect = 0.28
Identities = 18/44 (40%), Positives = 24/44 (53%), Gaps = 2/44 (4%)
Query: 28 RYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQIIAVAAGV 71
R+ GG F R+GGGGG F F + GP S+++ VA V
Sbjct: 24 RFGGGGGRFGGGGGRFGGGGGRFGG-FRDEGP-PSEVVEVATFV 65
>At1g52690 unknown protein
Length = 169
Score = 31.2 bits (69), Expect = 0.36
Identities = 16/45 (35%), Positives = 20/45 (43%)
Query: 143 ETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTKAHEAKRT 187
ETRG E+ A M D KT+E Q KAHE ++
Sbjct: 13 ETRGKAQEKTGEAMGTMGDKTQAAKDKTQETAQSAQQKAHETAQS 57
>At3g06780 unknown protein
Length = 201
Score = 30.8 bits (68), Expect = 0.48
Identities = 13/30 (43%), Positives = 18/30 (59%)
Query: 21 GGANPPQRYEAGGINFNNQPQRYGGGGGGF 50
GG++ P + GG + + YGGGGGGF
Sbjct: 111 GGSDGPFGFGGGGNDGGGKGWNYGGGGGGF 140
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,353,215
Number of Sequences: 26719
Number of extensions: 188739
Number of successful extensions: 860
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 783
Number of HSP's gapped (non-prelim): 81
length of query: 195
length of database: 11,318,596
effective HSP length: 94
effective length of query: 101
effective length of database: 8,807,010
effective search space: 889508010
effective search space used: 889508010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0263.4


