

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.8
(312 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
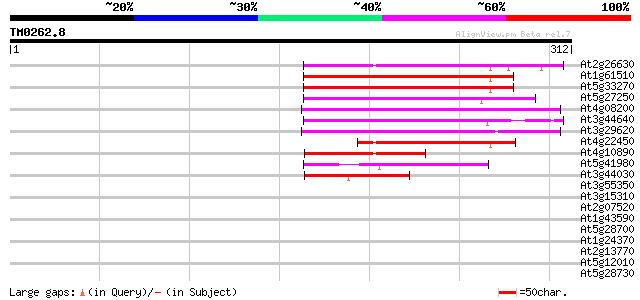
At2g26630 En/Spm-like transposon protein 124 9e-29
At1g61510 hypothetical protein 118 5e-27
At5g33270 putative protein 114 5e-26
At5g27250 putative protein 109 2e-24
At4g08200 putative protein 108 4e-24
At3g44640 putative protein 107 8e-24
At3g29620 hypothetical protein 101 6e-22
At4g22450 hypothetical protein 77 2e-14
At4g10890 putative protein 76 3e-14
At5g41980 unknown protein 71 9e-13
At3g44030 putative protein 60 1e-09
At3g55350 unknown protein 40 0.001
At3g15310 unknown protein 40 0.002
At2g07520 pseudogene 39 0.003
At1g43590 hypothetical protein 36 0.031
At5g28700 putative protein 35 0.052
At1g24370 hypothetical protein 35 0.052
At2g13770 hypothetical protein 34 0.089
At5g12010 putative protein 34 0.12
At5g28730 putative protein 33 0.26
>At2g26630 En/Spm-like transposon protein
Length = 292
Score = 124 bits (310), Expect = 9e-29
Identities = 76/166 (45%), Positives = 96/166 (57%), Gaps = 22/166 (13%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQF-RNGPRITGRVEVFNYYHSSLRNVIERSFG 222
GKYYLVD+GYPT G+LGP+++ RYHL QF R GP +T R E+FN HS LR+VIER+FG
Sbjct: 119 GKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTAR-ELFNRKHSGLRSVIERTFG 177
Query: 223 CCKAKWKILG-SMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF------------DAN 269
KAKW+I+ P Y L KI+TA MALHNFIR + R D +F D
Sbjct: 178 VWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHREDHDFLQWQSIEEYHVDDEE 237
Query: 270 YENEDGG-----GENEAGPSTVTWEEPDFQ--SSLQMEQIREHIKN 308
E E+ G GE E G E ++ ME +R++I N
Sbjct: 238 EEEEEEGEEEEEGEEEEGEGEEEEEHVAYEPTGDRAMEALRDNITN 283
>At1g61510 hypothetical protein
Length = 608
Score = 118 bits (295), Expect = 5e-27
Identities = 61/121 (50%), Positives = 80/121 (65%), Gaps = 4/121 (3%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GKYYLVD+GYPT G+LGP+++ RYHL F G T E+FN HSSLR+VIER+FG
Sbjct: 456 GKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGV 515
Query: 224 CKAKWKILG-SMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF---DANYENEDGGGEN 279
KAKW+IL P Y++K KI+T+ MALHN+IR + + D +F + E G EN
Sbjct: 516 WKAKWRILDRKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHGDEN 575
Query: 280 E 280
+
Sbjct: 576 D 576
>At5g33270 putative protein
Length = 343
Score = 114 bits (286), Expect = 5e-26
Identities = 60/121 (49%), Positives = 79/121 (64%), Gaps = 4/121 (3%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
GKYYLVD+GYPT G+LGP+++ RYHL F G T E+FN HSSLR+VIER+FG
Sbjct: 191 GKYYLVDSGYPTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGV 250
Query: 224 CKAKWKILGSMP-SYDLKTQNKIITACMALHNFIRRNDRSDEEF---DANYENEDGGGEN 279
KAKW+IL Y++K KI+T+ MALHN+IR + + D +F + E G EN
Sbjct: 251 WKAKWRILDRKHLKYEVKKWIKIVTSTMALHNYIRDSQQEDSDFRHWEIVESYEQHGDEN 310
Query: 280 E 280
+
Sbjct: 311 D 311
>At5g27250 putative protein
Length = 348
Score = 109 bits (273), Expect = 2e-24
Identities = 58/135 (42%), Positives = 78/135 (56%), Gaps = 6/135 (4%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRN-GPRITGRVEVFNYYHSSLRNVIERSFG 222
GKYYL D G+P FL P + RYHL FR G T + E+FN H+SLRNVIER FG
Sbjct: 188 GKYYLADCGFPNRRNFLAPLRSTRYHLQDFRGEGRDPTNQNELFNLRHASLRNVIERIFG 247
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDR-----SDEEFDANYENEDGGG 277
K+++ I S P + KTQ +I+ +C ALHNF+R+ R SDEE + + +N +
Sbjct: 248 IFKSRFLIFKSAPPFSFKTQAEIVLSCAALHNFLRQKCRSDEFSSDEEDETDVDNANQNS 307
Query: 278 ENEAGPSTVTWEEPD 292
E G V +E +
Sbjct: 308 EENGGEENVETQEQE 322
>At4g08200 putative protein
Length = 202
Score = 108 bits (270), Expect = 4e-24
Identities = 57/144 (39%), Positives = 83/144 (57%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
+ KYYLVD+GY G+L Y++++YH F+N E FN HSSLR V ER+F
Sbjct: 50 ISKYYLVDSGYGLHRGYLISYRQSQYHPSHFQNQAPPNNYKEKFNRLHSSLRLVTERTFR 109
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENEDGGGENEAG 282
K KWKI+ + YD++T K++ MALHNF+R+++ D +F+AN+E +D +
Sbjct: 110 VWKGKWKIMHNRARYDVRTTKKLVVETMALHNFVRKSNILDPDFEANWEQDDNHQPSLKE 169
Query: 283 PSTVTWEEPDFQSSLQMEQIREHI 306
V + F S ME IR+ I
Sbjct: 170 EVEVQDDGQIFDSRQYMEGIRDDI 193
>At3g44640 putative protein
Length = 377
Score = 107 bits (267), Expect = 8e-24
Identities = 64/149 (42%), Positives = 83/149 (54%), Gaps = 12/149 (8%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRN-GPRITGRVEVFNYYHSSLRNVIERSFG 222
GK+YLVD GY FL P+++ RYHL FR G + E+FN H+SLRNVIER FG
Sbjct: 221 GKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIFG 280
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDE---EFDANYENEDGGGEN 279
K+++ I S P Y KTQ +++ AC LHNF+ + RSDE E N EN D EN
Sbjct: 281 IFKSRFLIFKSAPPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEENSDDNEEN 340
Query: 280 EAGPSTVTWEEPDFQSSLQMEQIREHIKN 308
+EE L+ +Q RE+ N
Sbjct: 341 -------NYEENGDVGILESQQ-REYANN 361
>At3g29620 hypothetical protein
Length = 222
Score = 101 bits (251), Expect = 6e-22
Identities = 57/144 (39%), Positives = 79/144 (54%), Gaps = 1/144 (0%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
+GKYYLVD+GY G+LGP+++ YH QF+N E FN+ HS LR VIER+FG
Sbjct: 71 IGKYYLVDSGYALRRGYLGPFRQTWYHHNQFQNQAPPNNHKEKFNWRHSLLRCVIERTFG 130
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDANYENEDGGGENEAG 282
K KW+I+ Y++ T KI+ A MALHNF+ + D +FD ++ +D
Sbjct: 131 VWKGKWRIMQDRAWYNIVTTRKIMVATMALHNFVWKLGIPDLDFDIDW-MQDNDHHPTLD 189
Query: 283 PSTVTWEEPDFQSSLQMEQIREHI 306
T E+ S ME R+ I
Sbjct: 190 DEDETVEQDMTGSRQHMEGFRDEI 213
>At4g22450 hypothetical protein
Length = 457
Score = 76.6 bits (187), Expect = 2e-14
Identities = 44/92 (47%), Positives = 58/92 (62%), Gaps = 5/92 (5%)
Query: 194 RNGPRITGRVEVFNYYHSSLRNVIERSFGCCKAKWKILG-SMPSYDLKTQNKIITACMAL 252
R GP R E+FN HSSLR+VIER+FG KAKW+IL P Y++K KI+T+ MAL
Sbjct: 336 RGGPPTNSR-ELFNRRHSSLRSVIERTFGVWKAKWRILDRKHPKYEVKKWIKIVTSTMAL 394
Query: 253 HNFIRRNDRSDEEF---DANYENEDGGGENEA 281
HN+IR + + D +F + E G EN+A
Sbjct: 395 HNYIRDSQQEDSDFRHWEIVESYEQHGDENDA 426
>At4g10890 putative protein
Length = 380
Score = 75.9 bits (185), Expect = 3e-14
Identities = 38/68 (55%), Positives = 51/68 (74%), Gaps = 2/68 (2%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQF-RNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
KYYLV++ YPT G+LGP+++ YHL QF R GP +T + E+FN H LR+VI+R+FG
Sbjct: 94 KYYLVNSVYPTTTGYLGPHRRILYHLGQFGRGGPPVTVQ-ELFNRKHLDLRSVIDRTFGV 152
Query: 224 CKAKWKIL 231
KAKW+IL
Sbjct: 153 WKAKWRIL 160
>At5g41980 unknown protein
Length = 374
Score = 70.9 bits (172), Expect = 9e-13
Identities = 41/106 (38%), Positives = 53/106 (49%), Gaps = 14/106 (13%)
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVE---VFNYYHSSLRNVIERS 220
GKYY+VD YP GF+ PY +G R E +FN H L I R+
Sbjct: 217 GKYYIVDNKYPNLPGFIAPY-----------HGVSTNSREEAKEMFNERHKLLHRAIHRT 265
Query: 221 FGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEF 266
FG K ++ IL S P Y L+TQ K++ A ALHN++R D F
Sbjct: 266 FGALKERFPILLSAPPYPLQTQVKLVIAACALHNYVRLEKPDDLVF 311
>At3g44030 putative protein
Length = 775
Score = 60.5 bits (145), Expect = 1e-09
Identities = 31/63 (49%), Positives = 40/63 (63%), Gaps = 5/63 (7%)
Query: 165 KYYLVDAGYPTFMGFLGPYKKNR-----YHLPQFRNGPRITGRVEVFNYYHSSLRNVIER 219
KYYLVD+ YP GFL Y+ +R YH+ QF GP + E+FN H+SLR+VIER
Sbjct: 712 KYYLVDSCYPNKQGFLALYRSSRNRVVRYHMSQFYPGPPPRNKHELFNQCHTSLRSVIER 771
Query: 220 SFG 222
+ G
Sbjct: 772 TVG 774
>At3g55350 unknown protein
Length = 406
Score = 40.4 bits (93), Expect = 0.001
Identities = 31/106 (29%), Positives = 43/106 (40%), Gaps = 14/106 (13%)
Query: 163 LGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFG 222
L +Y + D+G+P L PY+ LPQ FN HS + +
Sbjct: 272 LREYIVGDSGFPLLPWLLTPYQGKPTSLPQTE-----------FNKRHSEATKAAQMALS 320
Query: 223 CCKAKWKIL-GSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFD 267
K +W+I+ G M D +II C LHN I D D+ D
Sbjct: 321 KLKDRWRIINGVMWMPDRNRLPRIIFVCCLLHNII--IDMEDQTLD 364
>At3g15310 unknown protein
Length = 415
Score = 40.0 bits (92), Expect = 0.002
Identities = 28/97 (28%), Positives = 48/97 (48%), Gaps = 13/97 (13%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP + F+ + LPQ R + +F + + R +ER+FG +
Sbjct: 278 YYLTDGIYPKWATFIQSIR-----LPQNR-------KATLFATHQEADRKDVERAFGVLQ 325
Query: 226 AKWKILGSMPS-YDLKTQNKIITACMALHNFIRRNDR 261
A++ I+ + +D + I+ AC+ LHN I ++R
Sbjct: 326 ARFHIIKNPALVWDKEKIGNIMKACIILHNMIVEDER 362
>At2g07520 pseudogene
Length = 222
Score = 39.3 bits (90), Expect = 0.003
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 17/96 (17%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP++ F+ + LPQ ++F R IER+FG
Sbjct: 95 YYLADGIYPSYPTFVKSIR-----LPQSEPD-------KLFVQLQEGCRKDIERAFGVLH 142
Query: 226 AKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDR 261
A++KI+ ++ + I+ +C+ LHN I N+R
Sbjct: 143 ARFKIIWNIADLAI-----IMRSCIILHNMIVENER 173
>At1g43590 hypothetical protein
Length = 168
Score = 35.8 bits (81), Expect = 0.031
Identities = 38/150 (25%), Positives = 64/150 (42%), Gaps = 28/150 (18%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP + F+ LPQ + +F + R +ER+FG +
Sbjct: 14 YYLTDEIYPKWATFI-----QSISLPQDE-------KASLFATNQEACRKDVERAFGVLQ 61
Query: 226 AKWKILGSMPS--YDLKTQNKIITACMALHNFIRRNDRSD-EEFDANYENEDGGGENEAG 282
A++ I+ P+ +D I+ AC+ LHN I ++R ++D + + A
Sbjct: 62 ARFAIV-KHPALIWDKIKIGNIMRACIILHNMIVEDERDGYTQYDVS---------DFAH 111
Query: 283 PSTVTWEEPDFQSSLQMEQIREHIKNMFPT 312
P + + + DF S M ++ NM T
Sbjct: 112 PESASSSQVDFTYSTDMP---SNLGNMMAT 138
>At5g28700 putative protein
Length = 292
Score = 35.0 bits (79), Expect = 0.052
Identities = 26/97 (26%), Positives = 45/97 (45%), Gaps = 13/97 (13%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP + F+ Q + P+ + +F + R +ER+FG +
Sbjct: 166 YYLTDGIYPKWATFI-----------QSISNPQ-GDKASLFVTTQKACRKDVERAFGVFQ 213
Query: 226 AKWKILGSMPSYDLKTQ-NKIITACMALHNFIRRNDR 261
A++ I+ + K + I+ AC+ LHN I ++R
Sbjct: 214 ARFSIVKHPALFHDKVKIGNIMRACIILHNMIVEDER 250
>At1g24370 hypothetical protein
Length = 413
Score = 35.0 bits (79), Expect = 0.052
Identities = 25/97 (25%), Positives = 48/97 (48%), Gaps = 13/97 (13%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP + + H P+ GP+ ++F + R +ER+FG +
Sbjct: 161 YYLADGIYPKWSTLV-----QTIHDPR---GPK----KKLFAMKQEACRKDVERAFGVLQ 208
Query: 226 AKWKIL-GSMPSYDLKTQNKIITACMALHNFIRRNDR 261
+++ I+ G ++ + I+T+C+ +HN I ++R
Sbjct: 209 SRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDER 245
>At2g13770 hypothetical protein
Length = 244
Score = 34.3 bits (77), Expect = 0.089
Identities = 25/97 (25%), Positives = 47/97 (47%), Gaps = 13/97 (13%)
Query: 166 YYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
YYL D YP + + H P+ GP+ ++F + R +ER+FG +
Sbjct: 126 YYLADGIYPKWSTLV-----QTIHDPR---GPK----KKLFAMKQEACRKDVERAFGVLQ 173
Query: 226 AKWKIL-GSMPSYDLKTQNKIITACMALHNFIRRNDR 261
++ I+ G ++ + I+T+C+ +HN I ++R
Sbjct: 174 LRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDER 210
>At5g12010 putative protein
Length = 502
Score = 33.9 bits (76), Expect = 0.12
Identities = 22/93 (23%), Positives = 34/93 (35%), Gaps = 11/93 (11%)
Query: 172 GYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCKAKWKIL 231
G+P L PY + +T FN S ++ V + +FG K +W L
Sbjct: 376 GHPLLDWVLVPYTQQN-----------LTWTQHAFNEKMSEVQGVAKEAFGRLKGRWACL 424
Query: 232 GSMPSYDLKTQNKIITACMALHNFIRRNDRSDE 264
L+ ++ AC LHN + E
Sbjct: 425 QKRTEVKLQDLPTVLGACCVLHNICEMREEKME 457
>At5g28730 putative protein
Length = 296
Score = 32.7 bits (73), Expect = 0.26
Identities = 12/21 (57%), Positives = 16/21 (76%)
Query: 165 KYYLVDAGYPTFMGFLGPYKK 185
KYYLVD+GY G+L PY++
Sbjct: 186 KYYLVDSGYANKRGYLAPYRR 206
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.152 0.496
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,640,001
Number of Sequences: 26719
Number of extensions: 271880
Number of successful extensions: 1059
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1022
Number of HSP's gapped (non-prelim): 31
length of query: 312
length of database: 11,318,596
effective HSP length: 99
effective length of query: 213
effective length of database: 8,673,415
effective search space: 1847437395
effective search space used: 1847437395
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0262.8


