

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.11
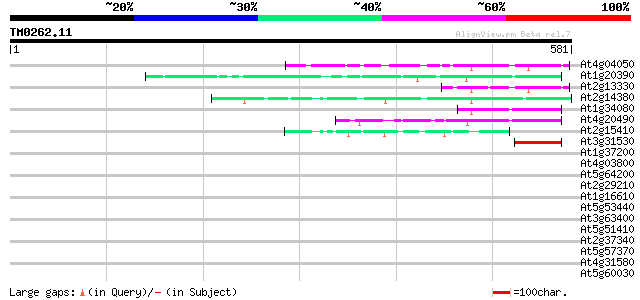
(581 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04050 putative transposon protein 85 1e-16
At1g20390 hypothetical protein 72 1e-12
At2g13330 F14O4.9 69 9e-12
At2g14380 putative retroelement pol polyprotein 64 2e-10
At1g34080 putative protein 56 6e-08
At4g20490 putative protein 53 4e-07
At2g15410 putative retroelement pol polyprotein 46 7e-05
At3g31530 hypothetical protein 45 8e-05
At1g37200 hypothetical protein 42 0.001
At4g03800 40 0.005
At5g64200 unknown protein 39 0.010
At2g29210 proline-rich protein like 39 0.010
At1g16610 arginine/serine-rich protein 37 0.030
At5g53440 putative protein 35 0.15
At3g63400 cyclophylin (ROC22) 34 0.20
At5g51410 arginine-aspartate-rich RNA binding protein-like 34 0.26
At2g37340 unknown protein 34 0.26
At5g57370 unknown protein 33 0.33
At4g31580 splicing factor 9G8-like SR protein / SRZ-22 33 0.57
At5g60030 KED - like protein 32 0.74
>At4g04050 putative transposon protein
Length = 681
Score = 84.7 bits (208), Expect = 1e-16
Identities = 89/310 (28%), Positives = 140/310 (44%), Gaps = 47/310 (15%)
Query: 286 KLDSKRFCEFHDSPDNNTDECLNLKDKVK*LIRAGRLSKYVALSYGDLPLPRSPTSRRSP 345
+LD ++C++H +T+EC VK LI AG +K + + P P ++P
Sbjct: 379 ELDLGKYCKYHKKRGYSTEECR----AVKKLIAAGGKTKKGSNPKVETPPPDEQEEEQTP 434
Query: 346 TPPRQRARTPPRHRTDDRYCRAP--QNGRSPDRRRSFDRGRIPDRHGRDEVIRHHRMNQI 403
++R RTP T AP +N R P R F R E + R++ I
Sbjct: 435 KQ-KKRERTPEGITTPPP---APHKKNMRFPLRLLKFCRSAT-------EHLSPKRIDFI 483
Query: 404 SVGSIAGGFAAGGHTSNSRKSSI*VIMSAMGRPRPRHSEPSRQKVTIWFSDDDYDTYTGK 463
GS + ++ RK+ + + + P+ Q +T W S+
Sbjct: 484 MGGSQLCNDSINSIKTHQRKAD------SYTKGKSPMMGPNHQ-ITFWESETT--DLNKP 534
Query: 464 EDDPIVIEALIGN----------GSSADVMFYDAYKKLHLSVKDLLLYDHDLVGFTGDRV 513
DD +VI +GN GSS DV+FYDA+K++ +L L GF GD
Sbjct: 535 HDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSELQGRKTPLTGFAGDTT 594
Query: 514 LPLG*FDTCLSLEDPNICRTVK----LIVVECQTKYNAILGRPSLNSYRAIISTHHLMLK 569
LG +++ P I R V+ +VV + +NAILGRP L++ +A+ ST+H +K
Sbjct: 595 FSLG------TIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKAVPSTYHQCIK 648
Query: 570 YP*AGKAVAV 579
+P + K +AV
Sbjct: 649 FP-SDKGIAV 657
>At1g20390 hypothetical protein
Length = 1791
Score = 71.6 bits (174), Expect = 1e-12
Identities = 96/446 (21%), Positives = 180/446 (39%), Gaps = 45/446 (10%)
Query: 141 KESLKTFLNRFNKEAGDIHYLLPQVRLVLVR*AL-HPGPFLTSLDGKKAKTVEEFQARSE 199
KESL++F++RF +I + + +V +R A+ + F + T+E+ R+
Sbjct: 267 KESLRSFVDRFKLVVTNIT-VPDEAAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRAS 325
Query: 200 KYINMEDAAALRASNQADQQKAPEWAKDPGDTTRIRESRRKNQDEKKPKRKRYENYAPLN 259
++I +E+ + A + K P D I+ D +P++ N +
Sbjct: 326 RFIELEEEKLILARKH-NSTKTPACK----DAVVIKVGP---DDSNEPRQHLDRNPSAGR 377
Query: 260 SLSQILRERASTDLRDRPPPMLTKGDKLDSKRFCEFHDSPDNNTDECLNLKDKVK*LIRA 319
+ L + D + + +CE+H S ++T+ C L+ + ++
Sbjct: 378 KPTSFLVSTETPDAKPWNKYIRDADSPAAGPMYCEYHKSRAHSTENCRFLQGLLMAKYKS 437
Query: 320 GRLSKYVALSYGDLPLPRSPTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRSPDRRRS 379
G ++ + R P + ++ ++R T R +D+ + P +
Sbjct: 438 GGIT---------IECDRPPINNKN----QRRNETTARQYLNDQ-TKPPTPAEQGIITSA 483
Query: 380 FDRGRIPDRHGRDEVIRHHRMNQISVGSIAGGFAAGGHTSNS----RKSSI*VIMSAMGR 435
D R+G+ + Q+ V I GG + S RK + V+
Sbjct: 484 DDPAAKRQRNGKAIAAEPVVVRQVHV--IMGGLQNCSDSVRSIKQYRKKAEMVVAWPSST 541
Query: 436 PRPRHSEPSRQKVTIWFSDDDYDTYTGKEDDPIVIE----------ALIGNGSSADVMFY 485
R+ Q I F+D D + +DP+V+E LI GSS D++F
Sbjct: 542 STTRNPN---QSAPISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFK 598
Query: 486 DAYKKLHLSVKDLLLYDHDLVGFTGDRVLPLG*FDTCLSLEDPNICRTVKLIVVECQTKY 545
D ++++ + + L GF GD V+ +G L + + VK +V+ Y
Sbjct: 599 DVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIK--LPIFVGGLIAWVKFVVIGKPAVY 656
Query: 546 NAILGRPSLNSYRAIISTHHLMLKYP 571
N ILG P ++ +AI ST+H +K+P
Sbjct: 657 NVILGTPWIHQMQAIPSTYHQCVKFP 682
>At2g13330 F14O4.9
Length = 889
Score = 68.6 bits (166), Expect = 9e-12
Identities = 54/148 (36%), Positives = 78/148 (52%), Gaps = 27/148 (18%)
Query: 448 VTIWFSD-DDYDTYTGKEDDPIVIEALIGN----------GSSADVMFYDAYKKL-HLSV 495
+T W S+ D D DD +VI +GN GSS DV+FYDA+K+ HL
Sbjct: 47 ITFWESEITDLDK---PHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDS 103
Query: 496 KDLLLYDHDLVGFTGDRVLPLG*FDTCLSLEDPNICRTVK----LIVVECQTKYNAILGR 551
K L L GF GD +G +++ P I R V+ +VV+ + +NAILGR
Sbjct: 104 K-LQGRKTPLTGFAGDTTFSIG------TIQLPTIARGVRQLTNFLVVDKKAPFNAILGR 156
Query: 552 PSLNSYRAIISTHHLMLKYP*AGKAVAV 579
P L+ +A+ ST+H +K+P + K +AV
Sbjct: 157 PWLHVMKAVPSTYHQCIKFP-SYKGIAV 183
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 63.9 bits (154), Expect = 2e-10
Identities = 95/402 (23%), Positives = 158/402 (38%), Gaps = 59/402 (14%)
Query: 210 LRASNQADQQKAPEWAKDPGDTTRIRESRRKN-----------QDEKKPKRKRYE-NYAP 257
LR++ QA QK + + +I E +K + + K K K YE N P
Sbjct: 97 LRSTIQAMSQKVHQATSSTPELEQILEKNQKTPFAVHISNVSVRHDNKIKLKSYEGNTDP 156
Query: 258 LNSLSQILRERASTDLRDRPPPMLTKGDKLDSKRFCEFHDSPDNNTDECLNLKDKVK*LI 317
LS +T P ++LD ++C+FH ++T C +L+ +
Sbjct: 157 QQFLSSF---SVATQRDHFHPSEAEVENRLD--QYCDFHKRSGHSTAACRHLQSIL---- 207
Query: 318 RAGRLSKYVALSYGDLPLPRSPTSRRSPTPPRQRARTPPR--HRTDDRYCRAPQNGRSPD 375
L+KY GD+ + + T + R HR P GR +
Sbjct: 208 ----LNKY---KKGDIEVQHRQYKSHNNTYAARGGRDGNNVFHRL------GPHTGRQQE 254
Query: 376 RRRSFDRGRIPD----RHGRDEVIRHHR--MNQISVGSIAGGFAAGGHTSNSRKSSI*VI 429
+ + R PD + R+ +H+ + + V I GG A + S K I
Sbjct: 255 APPANEEERHPDMEPPKKNRENDQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYI--- 311
Query: 430 MSAMGRPRPRHSEPSRQKVTIWFSDDDYDTYTGKEDDPIVIEALIGN----------GSS 479
G S +++ + F+ +D +DP+VIE IG GSS
Sbjct: 312 --KSGAATLWSSPATKEMTPLTFTSEDLFGVDLPHNDPLVIELHIGESEVTRILIDTGSS 369
Query: 480 ADVMFYDAYKKLHLSVKDLLLYDHDLVGFTGDRVLPLG*FDTCLSLEDPNICRTVKLIVV 539
+V+F D +K+ + + + L GF G+ ++ G + L K +VV
Sbjct: 370 VNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWH--KFVVV 427
Query: 540 ECQTKYNAILGRPSLNSYRAIISTHHLMLKYP*AGKAVAVRG 581
+ T YN ILG P ++ +AI S++H +K P + +RG
Sbjct: 428 DKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIGIETIRG 469
>At1g34080 putative protein
Length = 811
Score = 55.8 bits (133), Expect = 6e-08
Identities = 36/118 (30%), Positives = 58/118 (48%), Gaps = 12/118 (10%)
Query: 464 EDDPIVIEALIGN----------GSSADVMFYDAYKKLHLSVKDLLLYDHDLVGFTGDRV 513
+DD +VI +GN GSS D++F D ++ +S KD+ LV FT +
Sbjct: 473 DDDALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETS 532
Query: 514 LPLG*FDTCLSLEDPNICRTVKLIVVECQTKYNAILGRPSLNSYRAIISTHHLMLKYP 571
+ LG L + + + ++ V + YN ILG P L +A+ ST+H +K+P
Sbjct: 533 MSLG--TITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKFP 588
>At4g20490 putative protein
Length = 853
Score = 53.1 bits (126), Expect = 4e-07
Identities = 65/251 (25%), Positives = 105/251 (40%), Gaps = 39/251 (15%)
Query: 338 SPTSRRSPTPPRQRARTPPRHRT---DDRY--CRAPQNGRS--PDRRRSFDRGRIPDRHG 390
S S+ PT Q HR DDR R P+N P R R R PD
Sbjct: 364 SKQSQAGPTHEEQHVD----HRLLLEDDRVEEARVPENELPGPPKRWRGQQPQRAPD--- 416
Query: 391 RDEVIRHHRMNQISVGSIAGGFAAGGHTSNSRKSSI*VIMSAMGRPRPRHSEPSRQKVTI 450
H +IS+ I G + + ++ K + S P +EP Q+ I
Sbjct: 417 -------HPRGRISM--IMAGLSDDEDSVSALKKRARQVCSVRVAPE---AEPLSQEPII 464
Query: 451 WFSDDDYDTYTGKEDDPIVIEA----------LIGNGSSADVMFYDAYKKLHLSVKDLLL 500
F+ +D +D +VI+ LI GSS ++MF K+ +S ++
Sbjct: 465 -FTSEDASGIQHPHNDALVIKLVMEDFDVERILIDTGSSVNLMFLKTLFKMGISESKIMP 523
Query: 501 YDHDLVGFTGDRVLPLG*FDTCLSLEDPNICRTVKLIVVECQTKYNAILGRPSLNSYRAI 560
+ G+ G+ + +G + L ++ I + K +V++ + YNAILG P + S +AI
Sbjct: 524 KIRPMTGYDGEAKMSIG--EIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAI 581
Query: 561 ISTHHLMLKYP 571
ST+ ++ P
Sbjct: 582 PSTNLHTIRQP 592
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 45.8 bits (107), Expect = 7e-05
Identities = 64/248 (25%), Positives = 93/248 (36%), Gaps = 49/248 (19%)
Query: 285 DKLDSKRFCEFHDSPDNNTDECLNLKDKVK*LIRAGRLSKYVALSYGDLPLPRSPTSRRS 344
D +++ EFH P ++TDEC L+ + L+K+ GDL + P RR+
Sbjct: 458 DAKGDQKYYEFHKIPGHSTDECRQLQTLL--------LTKF---KKGDLDI--EPDRRRT 504
Query: 345 PTPPR----QRARTPPRHRTDDRYCRAPQNGRSPDRRRSFDRGRIP-------DRHGRDE 393
T R +R R DD+ R + R +R DR P R DE
Sbjct: 505 GTNDRDNTHRRVENTDRDNQDDKR-RQDETDRRAERIHDDDRRAEPHKRNHDAPRQFEDE 563
Query: 394 VIRHHRMNQISVGSIAGGFAAGGHTSNSRKSSI*VIMSAMGRPRPRHSEPSRQKV----T 449
R N I GG A + S KS G + E + V
Sbjct: 564 PASKRRRNM-----IMGGLTACRDSVRSIKS-----YRRQGEKQRAWIEKNTSAVECNEP 613
Query: 450 IWFSDDDYDTYTGKEDDPIVIEALIGNGSSADVMFYDAYKKLHLSVKDLLLYDHDLVGFT 509
I FS+ D G +DP+V+E IG + +L + V+ L GF
Sbjct: 614 ITFSEADI-ALPGSHNDPLVVELKIGE---------NVLLQLEIDVRSFQDKVQPLTGFD 663
Query: 510 GDRVLPLG 517
GD ++ +G
Sbjct: 664 GDTIMTVG 671
>At3g31530 hypothetical protein
Length = 831
Score = 45.4 bits (106), Expect = 8e-05
Identities = 20/49 (40%), Positives = 30/49 (60%)
Query: 523 LSLEDPNICRTVKLIVVECQTKYNAILGRPSLNSYRAIISTHHLMLKYP 571
L + + + V + + T YNAI+G P LN +RA+ ST+HL LK+P
Sbjct: 8 LPVRAKGVTKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFP 56
>At1g37200 hypothetical protein
Length = 1564
Score = 41.6 bits (96), Expect = 0.001
Identities = 64/301 (21%), Positives = 121/301 (39%), Gaps = 59/301 (19%)
Query: 286 KLDSKRFCEFHDSPDNNTDECLNLKDKVK*LIRAGRLSKYVALSYGDLPLPRSPTSRRSP 345
+LD ++C++H+ ++T+EC VK LI AG +K G P +P P
Sbjct: 317 ELDLGKYCKYHNKRGHSTEECR----AVKKLIAAGGKTKK-----GSNPKVETP-----P 362
Query: 346 TPPRQRARTPPRHRTDDRYCRAPQNGRSPDRRRSFDRGRIPDRHGRDEVIRHHRMNQISV 405
++ +TP + + + R + G SP P R R +++ ++ +
Sbjct: 363 PDEQEEEQTPKQKKRE----RTLEGGDSPP----------PARRERIDLV----FTELDL 404
Query: 406 GSIAGGFAAGGHTSNSRKSS-I*VIMSAMGRPRPRHSEPSRQKVTIWFSD------DDYD 458
G G ++ +K+ + +S R H P R + S +
Sbjct: 405 GGKTGRSVTTPPSAPHKKNMRFPLRLSKFCRSATEHLSPKRIDFIMGGSQLCNDSINSIK 464
Query: 459 TYTGKEDDPIVIEALIGNGSSADVMFYDAYKKLHLSVKDLLLYDHDLVGFTGDRVLPLG* 518
T+ K D ++L+ ++ L D ++ D+ + R++
Sbjct: 465 THQRKADSYTKGKSLMMGPDHKITLWESETTDLDKPHDDAIVIRIDVGNYKLSRIM---- 520
Query: 519 FDTCLSLEDPNICRTVKLIVVECQTKYNAILGRPSLNSYRAIISTHHLMLKYP*AGKAVA 578
DT S++ + +NAILGRP L++ +A+ ST+H +K+P + K +A
Sbjct: 521 IDTGSSVD---------------KAPFNAILGRPWLHAMKAVPSTYHQCIKFP-SEKGIA 564
Query: 579 V 579
V
Sbjct: 565 V 565
>At4g03800
Length = 637
Score = 39.7 bits (91), Expect = 0.005
Identities = 38/159 (23%), Positives = 64/159 (39%), Gaps = 33/159 (20%)
Query: 430 MSAMGRPRPR-------HSEPSRQKVTIWFSDDDYDTYTGKEDDPIVI----------EA 472
+SA+ + RP E + Q ++ F +++ DD ++I
Sbjct: 158 VSAIKKHRPNVLLKSSLSEEVNYQCSSVSFDEEETRHIERPHDDALIITLDVANFKISRI 217
Query: 473 LIGNGSSADVMFYDAYKKLHLSVKDLLLYDHDLVGFTGDRVLPLG*FDTCLSLEDPNICR 532
L+ GSS D++F LV FT + + LG L + + +
Sbjct: 218 LVDTGSSVDLIFLGP--------------PSPLVAFTSESAMSLGTIK--LPVLAGKMSK 261
Query: 533 TVKLIVVECQTKYNAILGRPSLNSYRAIISTHHLMLKYP 571
V +V + YN ILG P + +A+ ST+H LK+P
Sbjct: 262 IVDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLKFP 300
>At5g64200 unknown protein
Length = 303
Score = 38.5 bits (88), Expect = 0.010
Identities = 35/123 (28%), Positives = 50/123 (40%), Gaps = 21/123 (17%)
Query: 339 PTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRSPDRR----------------RSFDR 382
P SRRS + +R+R+P R R+ R R+P+ RSP RR RS+DR
Sbjct: 111 PKSRRSRSRSPRRSRSPRRSRSPPRR-RSPRRSRSPRRRSRDDYREKDYRKRSRSRSYDR 169
Query: 383 GRIPDRHGRDEVIRHHRMNQISVGSIAGGFAAGGHTSNSRKSSI*VIMSAMGRPRPRHSE 442
+ RD H R + S G N +S + ++ R PR S
Sbjct: 170 RERHEEKDRD----HRRRTRSRSASPDEKRRVRGRYDNESRSHSRSLSASPARRSPRSSS 225
Query: 443 PSR 445
P +
Sbjct: 226 PQK 228
>At2g29210 proline-rich protein like
Length = 878
Score = 38.5 bits (88), Expect = 0.010
Identities = 21/45 (46%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query: 337 RSPTSRRSPTPPRQRAR--TPPRHRTDDRYCRAPQNGRSPDRRRS 379
RSP RRSPTP +R+R + D R R+P + RSP R RS
Sbjct: 578 RSPVRRRSPTPVNRRSRRSSSASRSPDRRRRRSPSSSRSPSRSRS 622
Score = 33.9 bits (76), Expect = 0.26
Identities = 21/49 (42%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Query: 337 RSPT----SRRSPTPPRQRARTPP---RHRTDDRYCRAPQNGRSPDRRR 378
RSPT RRSP+PP +R R+PP R R+ R ++ P RRR
Sbjct: 341 RSPTPPARQRRSPSPPARRHRSPPPARRRRSPSPPARRRRSPSPPARRR 389
Score = 31.2 bits (69), Expect = 1.7
Identities = 26/82 (31%), Positives = 38/82 (45%), Gaps = 14/82 (17%)
Query: 342 RRSPTPPRQRARTPPRHRT----DDRYCRAPQNGRSPD--RRRSFDRGRIPDRHGRDEVI 395
+ SP+ Q+AR+P R R+ + R R+ RSPD RRRS R P R V+
Sbjct: 567 KSSPSSRHQKARSPVRRRSPTPVNRRSRRSSSASRSPDRRRRRSPSSSRSPSRSRSPPVL 626
Query: 396 --------RHHRMNQISVGSIA 409
R H+ + S G ++
Sbjct: 627 HRSPSPRGRKHQRERRSPGRLS 648
>At1g16610 arginine/serine-rich protein
Length = 414
Score = 37.0 bits (84), Expect = 0.030
Identities = 26/68 (38%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query: 338 SPTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRSPDRRRSFDR-GRIPDRHGRDEVIR 396
SP RRSP P R+R+ P R R+ R + RSP RR R I R GR R
Sbjct: 301 SPVRRRSPLPLRRRSPPPRRLRSPPRRSPIRRRSRSPIRRPGRSRSSSISPRKGRGPAGR 360
Query: 397 HHRMNQIS 404
R + S
Sbjct: 361 RGRSSSYS 368
Score = 35.0 bits (79), Expect = 0.11
Identities = 23/54 (42%), Positives = 27/54 (49%), Gaps = 10/54 (18%)
Query: 336 PRSPTSRRSPTPPRQRARTPPRHRTDD----RYCRAPQNGR------SPDRRRS 379
P SP RR TPPR+R +P R R+ R R+P G SP RRRS
Sbjct: 254 PGSPIRRRGDTPPRRRPASPSRGRSPSSPPPRRYRSPPRGSPRRIRGSPVRRRS 307
Score = 34.3 bits (77), Expect = 0.20
Identities = 23/53 (43%), Positives = 26/53 (48%), Gaps = 10/53 (18%)
Query: 336 PRSPTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRSPD-----RRRSFDRG 383
P SP RR +P R+R TPPR R +P GRSP R RS RG
Sbjct: 246 PDSPHRRRPGSPIRRRGDTPPRRRP-----ASPSRGRSPSSPPPRRYRSPPRG 293
Score = 33.1 bits (74), Expect = 0.44
Identities = 37/109 (33%), Positives = 47/109 (42%), Gaps = 15/109 (13%)
Query: 331 GDLP---LPRSPTSRRSP-TPPRQRARTPPRHRTDDRYCRAPQNGRS--PDRRRSFDRGR 384
GD P P SP+ RSP +PP +R R+PPR + R +P RS P RRRS R
Sbjct: 262 GDTPPRRRPASPSRGRSPSSPPPRRYRSPPR-GSPRRIRGSPVRRRSPLPLRRRSPPPRR 320
Query: 385 IPD--------RHGRDEVIRHHRMNQISVGSIAGGFAAGGHTSNSRKSS 425
+ R R + R R S+ G AG +S SS
Sbjct: 321 LRSPPRRSPIRRRSRSPIRRPGRSRSSSISPRKGRGPAGRRGRSSSYSS 369
>At5g53440 putative protein
Length = 1181
Score = 34.7 bits (78), Expect = 0.15
Identities = 54/232 (23%), Positives = 86/232 (36%), Gaps = 28/232 (12%)
Query: 182 SLDGKKAKTVEEFQARSEKYINMEDAAALRASNQADQQKAPEWAKDPGDTTRIRESRRKN 241
S D K K + + +S+K+ + +D +A + + KA + A+ PG + R+
Sbjct: 139 SKDVDKEKDRKYKEGKSDKFYDGDDHHKSKAGSDKTESKAQDHARSPGTENYTEKRSRRK 198
Query: 242 QDEKKPKRKRYENYAPLNSLSQILRERASTDLRDRPPPMLTKGDKLDSKRFCEFHDS--- 298
+D+ K ++N S + +R T D KG+K K + +
Sbjct: 199 RDDHGTGDKHHDN-------SDDVGDRVLTSGDDYIKDGKHKGEKSRDKYREDKEEEDIK 251
Query: 299 -------PDNNTDECLNLKDKVK*LIRAGRLSKYVALSYGDLPLPRSPTSRRSPTPPRQR 351
D T E L +K+ + + SK+ +G P S R+R
Sbjct: 252 QKGDKQRDDRPTKEHLRSDEKLT-RDESKKKSKFQDNDHG-----HEPDSELDGYHERER 305
Query: 352 ----ARTPPRHRTDDRYCRAPQNGRSPDRRRSFDRGRIPDRHGRD-EVIRHH 398
R R+ D R DR R DR R DR RD E R+H
Sbjct: 306 NRDYDRESDRNERDRERTRDRDRDYERDRDRDRDRDRERDRDRRDYEHDRYH 357
>At3g63400 cyclophylin (ROC22)
Length = 567
Score = 34.3 bits (77), Expect = 0.20
Identities = 25/60 (41%), Positives = 33/60 (54%), Gaps = 4/60 (6%)
Query: 329 SYGDLPLPRSPTSRR-SPTPPRQRARTPPRHRTDDRYCRAPQN-GRSPD--RRRSFDRGR 384
++ D R P++R S PR R R+PPR R+ RY R ++ RSPD RRR D R
Sbjct: 447 NFQDRNRDRYPSNRSYSERSPRGRFRSPPRRRSPPRYNRRRRSTSRSPDGYRRRLRDGSR 506
Score = 31.2 bits (69), Expect = 1.7
Identities = 18/58 (31%), Positives = 33/58 (56%), Gaps = 6/58 (10%)
Query: 200 KYINMEDAAALRASNQADQQKAPEWAKD----PGDTT--RIRESRRKNQDEKKPKRKR 251
K I+ + + +RA + A+++K + PGD + +E+R+K +EK+ KRKR
Sbjct: 168 KIIDCGETSQIRAHDAAEREKGKSKKSNKNFSPGDVSDREAKETRKKESNEKRIKRKR 225
>At5g51410 arginine-aspartate-rich RNA binding protein-like
Length = 334
Score = 33.9 bits (76), Expect = 0.26
Identities = 22/55 (40%), Positives = 24/55 (43%)
Query: 337 RSPTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRSPDRRRSFDRGRIPDRHGR 391
RS +S R R R R RHR R R P + RS R DR R HGR
Sbjct: 266 RSGSSDRERYRDRDRNRDGDRHRDRGRDYRKPYDRRSRSGREDRDRSRSRSPHGR 320
>At2g37340 unknown protein
Length = 249
Score = 33.9 bits (76), Expect = 0.26
Identities = 46/163 (28%), Positives = 62/163 (37%), Gaps = 31/163 (19%)
Query: 308 NLKDKVK*LIRAGRLSKYVALSYGDLPLPRSPTSRRSPTPPRQRARTPPRHRTD--DRYC 365
N K+ K L R+G S+ S RSP RRSP+ R+R+ R R+ R
Sbjct: 94 NCKNSPKKLRRSGSYSRSPVRS-------RSPRRRRSPSRSLSRSRSYSRSRSPVRRRER 146
Query: 366 RAPQNGRSPDRR------RSFDRGRIPDRHGRDEVI----RHHRMNQISVGSIAGGFAAG 415
+ RSP R R+ DR + D G ++I Q VGS G G
Sbjct: 147 SVEERSRSPKRMDDSLSPRARDRSPVLDDEGSPKIIDGSPPPSPKLQKEVGSDRDG---G 203
Query: 416 GHTSNSRKSSI*VIMSAMGRPRPRHSEPSRQKVTIWFSDDDYD 458
N R S + ++ A G P DDDY+
Sbjct: 204 SPQDNGRNSVVSPVVGAGGDSSKEDRSP---------VDDDYE 237
>At5g57370 unknown protein
Length = 219
Score = 33.5 bits (75), Expect = 0.33
Identities = 40/141 (28%), Positives = 52/141 (36%), Gaps = 40/141 (28%)
Query: 349 RQRARTPPRHRTDDRYCRAPQNGRSPDRRRSFDRGRIPDRHGRDEVIRHHRMNQISVGSI 408
+ R+RTP H R+ R+P+ RS R RS DR R DR RHHR S
Sbjct: 52 KSRSRTPDHHARA-RHVRSPERYRS--RSRSIDRDRDRDRQ------RHHRRRSPS---- 98
Query: 409 AGGFAAGGHTSNSRKSSI*VIMSAMGRPRPR------HSEPSRQKVTIWFSDDDYDTYTG 462
A R RPR E +R++V D++ T T
Sbjct: 99 ---------------------PDAPSRKRPRQGSVDDEKERNRKRVVTDSVDENASTITK 137
Query: 463 KEDDPIVIEALIGNGSSADVM 483
K+D A G G + M
Sbjct: 138 KKDKQPSDAADNGGGEDEEGM 158
>At4g31580 splicing factor 9G8-like SR protein / SRZ-22
Length = 200
Score = 32.7 bits (73), Expect = 0.57
Identities = 24/66 (36%), Positives = 30/66 (45%), Gaps = 5/66 (7%)
Query: 318 RAGRLSKYVALSYGDLPLPRSPTSRRSPTPP----RQRARTPPRHRTDDRYCRAPQNGRS 373
R R Y SY P RRSP+PP R +R+PP +R + A NG
Sbjct: 134 RYRRSPSYGRRSYSPRARSPPPPRRRSPSPPPARGRSYSRSPPPYRAREEVPYANGNGLK 193
Query: 374 PDRRRS 379
+RRRS
Sbjct: 194 -ERRRS 198
Score = 29.3 bits (64), Expect = 6.3
Identities = 19/47 (40%), Positives = 24/47 (50%), Gaps = 7/47 (14%)
Query: 337 RSPTSRRSPTPPRQRARTPPRHRTDDRYCRAPQNGRS----PDRRRS 379
R T RR + R+RTPPR+R Y R + R+ P RRRS
Sbjct: 117 RGGTGRRRS---KSRSRTPPRYRRSPSYGRRSYSPRARSPPPPRRRS 160
>At5g60030 KED - like protein
Length = 292
Score = 32.3 bits (72), Expect = 0.74
Identities = 33/155 (21%), Positives = 61/155 (39%), Gaps = 22/155 (14%)
Query: 182 SLDGKKAKTV-EEFQARSEKYINMEDAAALRASNQADQQKAPEWAKDPG----------- 229
S D K + V +EF R +Y N E + + +K + +KD
Sbjct: 79 SSDAKSERNVIDEFDGRKIRYRNSEAVSVESVYGRERDEKKMKKSKDADVVDEKVNEKLE 138
Query: 230 DTTRIRESRRKNQDEKKPKRKRYENYAPLNSLSQILRERASTDLRDRPPPMLTKG----- 284
R E R + +++KK K + E+ ++ E+ S D ++R K
Sbjct: 139 AEQRSEERRERKKEKKKKKNNKDEDVVDEKVKEKLEDEQKSADRKERKKKKSKKNNDEDV 198
Query: 285 ----DKL-DSKRFCEFHDSPDNNTDECLNLKDKVK 314
+KL D ++ E + N ++ ++ K+K K
Sbjct: 199 VDEKEKLEDEQKSAEIKEKKKNKDEDVVDEKEKEK 233
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.142 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,054,296
Number of Sequences: 26719
Number of extensions: 661381
Number of successful extensions: 2728
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 2571
Number of HSP's gapped (non-prelim): 146
length of query: 581
length of database: 11,318,596
effective HSP length: 105
effective length of query: 476
effective length of database: 8,513,101
effective search space: 4052236076
effective search space used: 4052236076
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0262.11


