

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.10
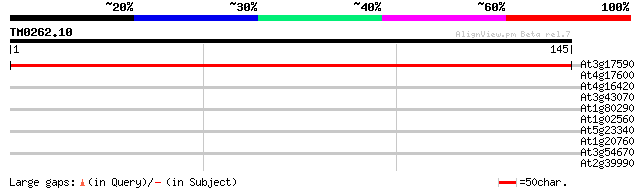
(145 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g17590 unknown protein 235 5e-63
At4g17600 Lil3 protein (Lil3:1) 30 0.58
At4g16420 transcriptional adaptor like protein 29 0.76
At3g43070 putative protein 28 1.3
At1g80290 unknown protein 28 1.3
At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1) 28 1.3
At5g23340 unknown protein 27 3.8
At1g20760 unknown protein 27 3.8
At3g54670 structural maintenance of chromosomes (SMC) - like pro... 26 6.5
At2g39990 26S proteasome regulatory subunit 26 6.5
>At3g17590 unknown protein
Length = 240
Score = 235 bits (600), Expect = 5e-63
Identities = 112/145 (77%), Positives = 127/145 (87%)
Query: 1 MKTPASVFYRNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTV 60
MK S ++ PVKFRMPTAENLVPIRLDI+ + +RYKDAFTWNPSDPD+EV +FA+RTV
Sbjct: 1 MKGLVSTGWKGPVKFRMPTAENLVPIRLDIQFEGQRYKDAFTWNPSDPDNEVVIFAKRTV 60
Query: 61 KDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDL 120
KDLKLP FVTQIAQSI+SQL++FR+YEGQDMY GEKIIPIKLDL VNH L+KDQFLWDL
Sbjct: 61 KDLKLPYAFVTQIAQSIQSQLSDFRAYEGQDMYTGEKIIPIKLDLRVNHTLIKDQFLWDL 120
Query: 121 NNFESDLEELARIFCKDMGIEDPEV 145
NNFESD EE AR CKD+G+EDPEV
Sbjct: 121 NNFESDPEEFARTLCKDLGVEDPEV 145
>At4g17600 Lil3 protein (Lil3:1)
Length = 262
Score = 29.6 bits (65), Expect = 0.58
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Query: 46 SDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDL 105
+ P V V A V+ + PP T + E+ A+ + EG++M E ++ +
Sbjct: 47 TSPTAAVSVEAPEPVEVIVKEPPQSTPAVKKEETATAKNVAVEGEEMKTTESVVKFQDAR 106
Query: 106 CVNHMLVKDQFLWDLNNFESD 126
+N WDL FE D
Sbjct: 107 WING-------TWDLKQFEKD 120
>At4g16420 transcriptional adaptor like protein
Length = 487
Score = 29.3 bits (64), Expect = 0.76
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query: 57 RRTVKDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVK 113
+R ++KL PP Q+ Q + ++ + + D Y+ KI P K+D V MLVK
Sbjct: 426 KRLCSEVKLVPPVYLQMQQVMSHEIFKGNVTKKSDAYSLFKIDPTKVDR-VYDMLVK 481
>At3g43070 putative protein
Length = 426
Score = 28.5 bits (62), Expect = 1.3
Identities = 19/82 (23%), Positives = 35/82 (42%), Gaps = 7/82 (8%)
Query: 15 FRMP--TAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQ 72
F MP A +V + + +D + W S PD + VF+ T+ ++L P F+
Sbjct: 60 FHMPDHVAAKIVSLVFEDGVDMLK-----AWIQSGPDGKAAVFSSETLSSVRLAPFFIHM 114
Query: 73 IAQSIESQLAEFRSYEGQDMYA 94
+S + + ++ YA
Sbjct: 115 AEESSSYHMFYTKCLTAENPYA 136
>At1g80290 unknown protein
Length = 329
Score = 28.5 bits (62), Expect = 1.3
Identities = 15/36 (41%), Positives = 21/36 (57%)
Query: 29 DIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLK 64
D+EID + + AF+ S+PD VG F R DL+
Sbjct: 141 DVEIDQRSLEFAFSVWKSNPDRLVGTFVRSHGFDLQ 176
>At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1)
Length = 298
Score = 28.5 bits (62), Expect = 1.3
Identities = 21/83 (25%), Positives = 35/83 (41%), Gaps = 19/83 (22%)
Query: 3 TPASVFYRNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKD 62
TP +V+ N +P+ + + IR D+++ + Y A+
Sbjct: 60 TPKAVYSGNLWTPEIPSPQGVWSIRDDLQVPSSPYFPAYAQGQG---------------- 103
Query: 63 LKLPPPFVTQIAQSIESQLAEFR 85
PPP V + QSI SQL ++R
Sbjct: 104 ---PPPMVQERFQSIISQLFQYR 123
>At5g23340 unknown protein
Length = 405
Score = 26.9 bits (58), Expect = 3.8
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query: 105 LCVNHMLVKDQFLWDLNNFESDLE-ELARIFCK 136
+CVN L D+ W L+ +SD + E+ + CK
Sbjct: 4 VCVNEALTDDELRWVLSRLDSDKDKEVFGLVCK 36
>At1g20760 unknown protein
Length = 1019
Score = 26.9 bits (58), Expect = 3.8
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 8/75 (10%)
Query: 70 VTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEE 129
V +I + + A FR EG+ M + I+ ++ + +L + +SDLEE
Sbjct: 604 VAEIGSKLTIEEARFREIEGRKMELSQAIVNMEQGGSADGLLQV-----RADRIQSDLEE 658
Query: 130 LARIF---CKDMGIE 141
L + CK G+E
Sbjct: 659 LMKALTERCKKHGLE 673
>At3g54670 structural maintenance of chromosomes (SMC) - like
protein
Length = 1265
Score = 26.2 bits (56), Expect = 6.5
Identities = 9/19 (47%), Positives = 15/19 (78%)
Query: 111 LVKDQFLWDLNNFESDLEE 129
L +++FLW L N E+D+E+
Sbjct: 229 LKRERFLWQLYNIENDIEK 247
>At2g39990 26S proteasome regulatory subunit
Length = 293
Score = 26.2 bits (56), Expect = 6.5
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 33 DAKRYKDAFTWNPSDPDHEVGVFARRTVKDL-KLPPP-FVTQIAQSIESQL 81
D +Y D+ PD+ +G F V L KLPP F + S++ QL
Sbjct: 217 DVYKYVDSVVGGQIAPDNNIGRFIADAVASLPKLPPQVFDNLVNDSLQDQL 267
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,484,129
Number of Sequences: 26719
Number of extensions: 140485
Number of successful extensions: 293
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 286
Number of HSP's gapped (non-prelim): 10
length of query: 145
length of database: 11,318,596
effective HSP length: 90
effective length of query: 55
effective length of database: 8,913,886
effective search space: 490263730
effective search space used: 490263730
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0262.10


