

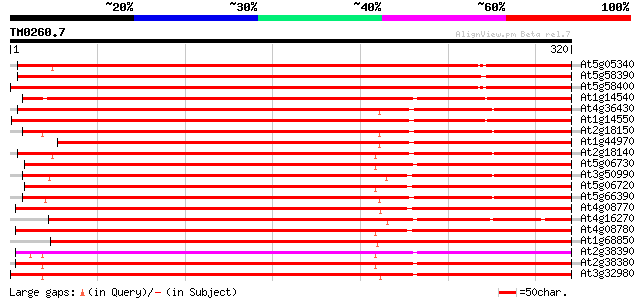
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.7
(320 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g05340 peroxidase 347 6e-96
At5g58390 peroxidase 335 1e-92
At5g58400 peroxidase 327 5e-90
At1g14540 peroxidase, putative 300 7e-82
At4g36430 peroxidase like protein 296 1e-80
At1g14550 peroxidase, putative 296 1e-80
At2g18150 putative peroxidase 295 3e-80
At1g44970 peroxidase like protein 290 9e-79
At2g18140 putative peroxidase 285 2e-77
At5g06730 peroxidase 281 4e-76
At3g50990 peroxidase-like protein 278 4e-75
At5g06720 peroxidase (emb|CAA68212.1) 276 1e-74
At5g66390 peroxidase (emb|CAA66964.1) 274 4e-74
At4g08770 peroxidase C2 precursor like protein 273 9e-74
At4g16270 peroxidase like protein 268 3e-72
At4g08780 peroxidase C2 precursor like protein 265 3e-71
At1g68850 peroxidase ATP23a 264 6e-71
At2g38390 peroxidase 260 6e-70
At2g38380 peroxidase like protein 260 8e-70
At3g32980 peroxidase, putative 259 1e-69
>At5g05340 peroxidase
Length = 324
Score = 347 bits (889), Expect = 6e-96
Identities = 177/320 (55%), Positives = 227/320 (70%), Gaps = 6/320 (1%)
Query: 5 LSCLIFLITICLVGKTSAV----LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
+S L+ ++T+ L G + V L+T FY+ +CP L T+++AV++AV+ E RMGAS+LR
Sbjct: 7 ISILVLVVTLLLQGDNNYVVEAQLTTNFYSTSCPNLLSTVQTAVKSAVNSEARMGASILR 66
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
L FHDCFV GCD S+LLDDT SFTGE+N+ PN NS RGF VID+IK +E CPGVVSCA
Sbjct: 67 LFFHDCFVNGCDGSILLDDTSSFTGEQNAAPNRNSARGFNVIDNIKSAVEKACPGVVSCA 126
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFN 180
DIL +AARDSV ALGG W++++GRRD+ TAS A+NS++PAP LS LIS++ G +
Sbjct: 127 DILAIAARDSVVALGGPNWNVKVGRRDARTASQAAANSNIPAPTSSLSQLISSFSAVGLS 186
Query: 181 ATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDST 240
+MV LSGAHTIG SRC +R RIYNETNI+ +FA + Q C G D N +P D T
Sbjct: 187 TRDMVALSGAHTIGQSRCTNFRARIYNETNINAAFATTRQRTCPRASGSGDGNLAPLDVT 246
Query: 241 TQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMS 300
T FDN +++NL+ Q+GL+HSDQ L+ NG G+TDS V YS N F DF AM+KM
Sbjct: 247 TAASFDNNYFKNLMTQRGLLHSDQVLF-NG-GSTDSIVRGYSNNPSSFNSDFTAAMIKMG 304
Query: 301 LLSPLTGSDGQIRKNCRVVN 320
+SPLTGS G+IRK C N
Sbjct: 305 DISPLTGSSGEIRKVCGRTN 324
>At5g58390 peroxidase
Length = 316
Score = 335 bits (860), Expect = 1e-92
Identities = 170/317 (53%), Positives = 221/317 (69%), Gaps = 3/317 (0%)
Query: 5 LSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFH 64
L ++ ++ + L ++ A L+ FY E+CP +R V+ AV++EPRMGASLLRL FH
Sbjct: 2 LKVVLLMMIMMLASQSEAQLNRDFYKESCPSLFLVVRRVVKRAVAREPRMGASLLRLFFH 61
Query: 65 DCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILT 124
DCFV GCD S+LLDDT SF GEK S P+ NS+RGFEVID IK ++E MCPG+VSCADIL
Sbjct: 62 DCFVNGCDGSLLLDDTPSFLGEKTSGPSNNSVRGFEVIDKIKFKVEKMCPGIVSCADILA 121
Query: 125 VAARDSVFALGGQRWDLQLGRRDSTTASLDASNSD-LPAPFLDLSGLISAYGKKGFNATE 183
+ ARDSV LGG W ++LGRRDSTTA+ A+NS +P P LS LI+ + +G + +
Sbjct: 122 ITARDSVLLLGGPGWSVKLGRRDSTTANFAAANSGVIPPPITTLSNLINRFKAQGLSTRD 181
Query: 184 MVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQF 243
MV LSGAHTIG ++C+ +R+RIYN +NID SFA S + NC G DN + D +
Sbjct: 182 MVALSGAHTIGRAQCVTFRNRIYNASNIDTSFAISKRRNCPATSGSGDNKKANLDVRSPD 241
Query: 244 KFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLS 303
+FD+ FY+ LL++KGL+ SDQ L+ N G TDS V +YS N F DFA AM+KM +S
Sbjct: 242 RFDHGFYKQLLSKKGLLTSDQVLFNN--GPTDSLVIAYSHNLNAFYRDFARAMIKMGDIS 299
Query: 304 PLTGSDGQIRKNCRVVN 320
PLTGS+GQIR+NCR N
Sbjct: 300 PLTGSNGQIRQNCRRPN 316
>At5g58400 peroxidase
Length = 325
Score = 327 bits (838), Expect = 5e-90
Identities = 170/321 (52%), Positives = 220/321 (67%), Gaps = 3/321 (0%)
Query: 1 SRGSLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
SR + ++ L + L + A L T FY+++CP L T+R V+ V+KE R+ ASLLR
Sbjct: 7 SRQRAAFVVLLFIVMLGSQAQAQLRTDFYSDSCPSLLPTVRRVVQREVAKERRIAASLLR 66
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
L FHDCFV GCDAS+LLDDT SF GEK + PN NS+RG+EVID IK ++E +CPGVVSCA
Sbjct: 67 LFFHDCFVNGCDASILLDDTRSFLGEKTAGPNNNSVRGYEVIDAIKSRVERLCPGVVSCA 126
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSD-LPAPFLDLSGLISAYGKKGF 179
DIL + ARDSV +GG+ W ++LGRRDS TAS +NS LP P L LI+ + G
Sbjct: 127 DILAITARDSVLLMGGRGWSVKLGRRDSITASFSTANSGVLPPPTSTLDNLINLFRANGL 186
Query: 180 NATEMVTLSGAHTIGLSRCIFYRDRIYNETNIDPSFAASMQVNCSFDGGDTDNNASPFDS 239
+ +MV LSGAHTIG +RC+ +R RIYN TNID SFA S + +C G DNNA+ D
Sbjct: 187 SPRDMVALSGAHTIGQARCVTFRSRIYNSTNIDLSFALSRRRSCPAATGSGDNNAAILDL 246
Query: 240 TTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKM 299
T KFD +++ L+N +GL+ SDQ L+ NG G+TDS V SYS++ F DF AM+KM
Sbjct: 247 RTPEKFDGSYFMQLVNHRGLLTSDQVLF-NG-GSTDSIVVSYSRSVQAFYRDFVAAMIKM 304
Query: 300 SLLSPLTGSDGQIRKNCRVVN 320
+SPLTGS+GQIR++CR N
Sbjct: 305 GDISPLTGSNGQIRRSCRRPN 325
>At1g14540 peroxidase, putative
Length = 315
Score = 300 bits (768), Expect = 7e-82
Identities = 154/314 (49%), Positives = 214/314 (68%), Gaps = 6/314 (1%)
Query: 8 LIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCF 67
L+ L+++C + A LS FY++TC AL TIRS++ A+S+E RM ASL+RLHFHDCF
Sbjct: 7 LVLLLSLCCFSQ--AQLSPTFYDQTCQNALSTIRSSIRTAISRERRMAASLIRLHFHDCF 64
Query: 68 VQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAA 127
V GCDASV+L T + E++S N S RGFEVID K +ES+CPGVVSCADI+ VAA
Sbjct: 65 VNGCDASVMLVATPTMESERDSLANFQSARGFEVIDQAKSAVESVCPGVVSCADIIAVAA 124
Query: 128 RDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTL 187
RD+ +GG R+D+++GRRDST A ++ DLP L+ L + +KG N ++V L
Sbjct: 125 RDASEYVGGPRYDVKVGRRDSTNAFRAIADRDLPNFRASLNDLSELFLRKGLNTRDLVAL 184
Query: 188 SGAHTIGLSRCIFYRDRIY-NETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFD 246
SGAHT+G ++C+ ++ R+Y N ++ID F+++ + C +GGDT +P D T FD
Sbjct: 185 SGAHTLGQAQCLTFKGRLYDNSSDIDAGFSSTRKRRCPVNGGDT--TLAPLDQVTPNSFD 242
Query: 247 NAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLT 306
N +Y+NL+ +KGL+ SDQ L+ G+ +TDS VT YS+N RF DF+ AM+KM + LT
Sbjct: 243 NNYYRNLMQKKGLLESDQVLFGTGA-STDSIVTEYSRNPSRFASDFSAAMIKMGDIQTLT 301
Query: 307 GSDGQIRKNCRVVN 320
GSDGQIR+ C VN
Sbjct: 302 GSDGQIRRICSAVN 315
>At4g36430 peroxidase like protein
Length = 331
Score = 296 bits (758), Expect = 1e-80
Identities = 151/324 (46%), Positives = 211/324 (64%), Gaps = 11/324 (3%)
Query: 5 LSCLIFLITICLVGKT-SAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
L LI + +CL K+ L +Y +CP+ +RS V AV++E RM ASLLRLHF
Sbjct: 10 LLSLICFVPLCLCDKSYGGKLFPGYYAHSCPQVNEIVRSVVAKAVARETRMAASLLRLHF 69
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFVQGCD S+LLD + EKNS PN+ S RGF+V+D IK +LE CPG VSCAD+L
Sbjct: 70 HDCFVQGCDGSLLLDSSGRVATEKNSNPNSKSARGFDVVDQIKAELEKQCPGTVSCADVL 129
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATE 183
T+AARDS GG W + LGRRDS +ASL SN+++PAP ++S + ++G + T+
Sbjct: 130 TLAARDSSVLTGGPSWVVPLGRRDSRSASLSQSNNNIPAPNNTFQTILSKFNRQGLDITD 189
Query: 184 MVTLSGAHTIGLSRCIFYRDRIYNET-------NIDPSFAASMQVNCSFDGGDTDNNASP 236
+V LSG+HTIG SRC +R R+YN++ ++ SFAA+++ C GG D S
Sbjct: 190 LVALSGSHTIGFSRCTSFRQRLYNQSGNGSPDMTLEQSFAANLRQRCPKSGG--DQILSV 247
Query: 237 FDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAM 296
D + FDN++++NL+ KGL++SDQ L+ + + + V Y+++ G F E FA +M
Sbjct: 248 LDIISAASFDNSYFKNLIENKGLLNSDQVLFSSNEKSRE-LVKKYAEDQGEFFEQFAESM 306
Query: 297 VKMSLLSPLTGSDGQIRKNCRVVN 320
+KM +SPLTGS G+IRKNCR +N
Sbjct: 307 IKMGNISPLTGSSGEIRKNCRKIN 330
>At1g14550 peroxidase, putative
Length = 321
Score = 296 bits (757), Expect = 1e-80
Identities = 154/322 (47%), Positives = 216/322 (66%), Gaps = 6/322 (1%)
Query: 2 RGSLSCLIFLITICLVGKT-SAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
R SL ++ +++I L A LS FY+++C AL IRS+V A+++E RM ASL+R
Sbjct: 3 RFSLRFVLMMVSIILTSSICQAQLSPTFYDQSCRNALSKIRSSVRTAIARERRMAASLIR 62
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
+HFHDCFV GCDAS+LL+ T + E+++ PN S+RGFEVID K ++E +CPG+VSCA
Sbjct: 63 MHFHDCFVHGCDASILLEGTSTIESERDALPNFKSVRGFEVIDKAKSEVEKVCPGIVSCA 122
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNS-DLPAPFLDLSGLISAYGKKGF 179
DI+ VAARD+ +GG +W +++GRRDST A +NS +LP L L + KKG
Sbjct: 123 DIIAVAARDASEYVGGPKWAVKVGRRDSTAAFKALANSGELPGFKDTLDQLSGLFSKKGL 182
Query: 180 NATEMVTLSGAHTIGLSRCIFYRDRIY-NETNIDPSFAASMQVNCSFDGGDTDNNASPFD 238
N ++V LSGAHTIG S+C +RDR+Y N ++ID FA++ + C GG D N + D
Sbjct: 183 NTRDLVALSGAHTIGQSQCFLFRDRLYENSSDIDAGFASTRKRRCPTVGG--DGNLAALD 240
Query: 239 STTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVK 298
T FDN +Y+NL+ +KGL+ +DQ L+ +G+ +TD V+ YSKN +F DFA AM+K
Sbjct: 241 LVTPNSFDNNYYKNLMQKKGLLVTDQVLFGSGA-STDGIVSEYSKNRSKFAADFATAMIK 299
Query: 299 MSLLSPLTGSDGQIRKNCRVVN 320
M + PLTGS+G+IRK C VN
Sbjct: 300 MGNIEPLTGSNGEIRKICSFVN 321
>At2g18150 putative peroxidase
Length = 338
Score = 295 bits (754), Expect = 3e-80
Identities = 149/327 (45%), Positives = 213/327 (64%), Gaps = 17/327 (5%)
Query: 8 LIFLITICLV-------GKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
LI+ +T+C+ G L FY +CP+A +RS V AV++E RM ASL+R
Sbjct: 13 LIYALTLCICDDDESNYGGDKGNLFPGFYRSSCPRAEEIVRSVVAKAVARETRMAASLMR 72
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
LHFHDCFVQGCD S+LLD + S EKNS PN+ S RGFEV+D+IK LE+ CP VSCA
Sbjct: 73 LHFHDCFVQGCDGSLLLDTSGSIVTEKNSNPNSRSARGFEVVDEIKAALENECPNTVSCA 132
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFN 180
D LT+AARDS GG W + LGRRDST+ASL SN+++PAP + +++ + +G +
Sbjct: 133 DALTLAARDSSVLTGGPSWMVPLGRRDSTSASLSGSNNNIPAPNNTFNTIVTRFNNQGLD 192
Query: 181 ATEMVTLSGAHTIGLSRCIFYRDRIYNET-------NIDPSFAASMQVNCSFDGGDTDNN 233
T++V LSG+HTIG SRC +R R+YN++ ++ S+AA+++ C GG D N
Sbjct: 193 LTDVVALSGSHTIGFSRCTSFRQRLYNQSGNGSPDRTLEQSYAANLRQRCPRSGG--DQN 250
Query: 234 ASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFA 293
S D + +FDN++++NL+ GL++SD+ L+ + + + V Y+++ F E FA
Sbjct: 251 LSELDINSAGRFDNSYFKNLIENMGLLNSDEVLFSSNEQSRE-LVKKYAEDQEEFFEQFA 309
Query: 294 NAMVKMSLLSPLTGSDGQIRKNCRVVN 320
+M+KM +SPLTGS G+IRKNCR +N
Sbjct: 310 ESMIKMGNISPLTGSSGEIRKNCRKIN 336
>At1g44970 peroxidase like protein
Length = 346
Score = 290 bits (741), Expect = 9e-79
Identities = 142/300 (47%), Positives = 202/300 (67%), Gaps = 9/300 (3%)
Query: 28 FYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSFTGEK 87
FY +CP+A + + +E A++KEPRM ASLLRLHFHDCFVQGCDAS+LLDD+ + EK
Sbjct: 49 FYQFSCPQADEIVMTVLEKAIAKEPRMAASLLRLHFHDCFVQGCDASILLDDSATIRSEK 108
Query: 88 NSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQLGRRD 147
N+ PN NS+RGF+VID+IK +LE CP VSCADIL +AAR S GG W+L LGRRD
Sbjct: 109 NAGPNKNSVRGFQVIDEIKAKLEQACPQTVSCADILALAARGSTILSGGPSWELPLGRRD 168
Query: 148 STTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRDRIYN 207
S TASL+ +N+++PAP + L++ + +KG N ++V+LSG HTIG++RC ++ R+YN
Sbjct: 169 SRTASLNGANTNIPAPNSTIQNLLTMFQRKGLNEEDLVSLSGGHTIGVARCTTFKQRLYN 228
Query: 208 ET-------NIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQKGLV 260
+ ++ S+ ++ C GG DNN SP D + +FDN +++ LL KGL+
Sbjct: 229 QNGNNQPDETLERSYYYGLRSICPPTGG--DNNISPLDLASPARFDNTYFKLLLWGKGLL 286
Query: 261 HSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
SD+ L G T + V +Y+++ F + FA +MV M + PLTG +G+IRK+C V+N
Sbjct: 287 TSDEVLLTGNVGKTGALVKAYAEDERLFFQQFAKSMVNMGNIQPLTGFNGEIRKSCHVIN 346
>At2g18140 putative peroxidase
Length = 337
Score = 285 bits (729), Expect = 2e-77
Identities = 149/329 (45%), Positives = 201/329 (60%), Gaps = 16/329 (4%)
Query: 5 LSCLIFLITICLVGKTSAV------LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASL 58
L L + +T+C+ S L FY +CP+A +RS V A +E RM ASL
Sbjct: 10 LLSLTYALTLCICDNASNFGGNKRNLFPDFYRSSCPRAEEIVRSVVAKAFERETRMAASL 69
Query: 59 LRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVS 118
+RLHFHDCFVQGCD S+LLD + S EKNS PN+ S RGFEV+D+IK LE+ CP VS
Sbjct: 70 MRLHFHDCFVQGCDGSLLLDTSGSIVTEKNSNPNSRSARGFEVVDEIKAALENECPNTVS 129
Query: 119 CADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKG 178
CAD LT+AARDS GG W + LGRRDS TAS N DLP P + + +G
Sbjct: 130 CADALTLAARDSSVLTGGPSWTVPLGRRDSATASRAKPNKDLPEPDNLFDTIFLRFSNEG 189
Query: 179 FNATEMVTLSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTD 231
N T++V LSG+HTIG SRC +R R+YN +T ++ S+AA ++ C GG D
Sbjct: 190 LNLTDLVALSGSHTIGFSRCTSFRQRLYNQSGSGSPDTTLEKSYAAILRQRCPRSGG--D 247
Query: 232 NNASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLED 291
N S D + +FDN++++NL+ GL++SDQ L+ + + + V Y+++ F E
Sbjct: 248 QNLSELDINSAGRFDNSYFKNLIENMGLLNSDQVLFSSNEQSRE-LVKKYAEDQEEFFEQ 306
Query: 292 FANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
FA +M+KM +SPLTGS G+IRK CR +N
Sbjct: 307 FAESMIKMGKISPLTGSSGEIRKKCRKIN 335
>At5g06730 peroxidase
Length = 358
Score = 281 bits (718), Expect = 4e-76
Identities = 140/319 (43%), Positives = 201/319 (62%), Gaps = 9/319 (2%)
Query: 9 IFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFV 68
+ +I L G +SA L+ FY+ TCP A +RS ++ A+ + R+G SL+RLHFHDCFV
Sbjct: 18 LIVIVSSLFGTSSAQLNATFYSGTCPNASAIVRSTIQQALQSDARIGGSLIRLHFHDCFV 77
Query: 69 QGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAAR 128
GCD S+LLDDT S EKN+ NANS RGF V+D IK LE+ CPG+VSC+DIL +A+
Sbjct: 78 NGCDGSLLLDDTSSIQSEKNAPANANSTRGFNVVDSIKTALENACPGIVSCSDILALASE 137
Query: 129 DSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLS 188
SV GG W + LGRRD TA+L +NS LP+PF L+ + S + G T++V+LS
Sbjct: 138 ASVSLAGGPSWTVLLGRRDGLTANLSGANSSLPSPFEGLNNITSKFVAVGLKTTDVVSLS 197
Query: 189 GAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTT 241
GAHT G +C+ + +R++N + ++ + +S+Q C +G +T + D +T
Sbjct: 198 GAHTFGRGQCVTFNNRLFNFNGTGNPDPTLNSTLLSSLQQLCPQNGSNT--GITNLDLST 255
Query: 242 QFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSL 301
FDN ++ NL + GL+ SDQ+L+ N T V S++ N F E F +M+KM
Sbjct: 256 PDAFDNNYFTNLQSNNGLLQSDQELFSNTGSATVPIVNSFASNQTLFFEAFVQSMIKMGN 315
Query: 302 LSPLTGSDGQIRKNCRVVN 320
+SPLTGS G+IR++C+VVN
Sbjct: 316 ISPLTGSSGEIRQDCKVVN 334
>At3g50990 peroxidase-like protein
Length = 336
Score = 278 bits (710), Expect = 4e-75
Identities = 150/326 (46%), Positives = 202/326 (61%), Gaps = 16/326 (4%)
Query: 8 LIFLITICLVGKTS------AVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRL 61
L+ L +C+ +T A LS FY +CP A ++S V A +PRM AS+LRL
Sbjct: 11 LVALFPLCICYQTHQSTSSVASLSPQFYENSCPNAQAIVQSYVANAYFNDPRMAASILRL 70
Query: 62 HFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCAD 121
HFHDCFV GCDASVLLD + + EK S N +S RGFEVID+IK LE+ CP VSCAD
Sbjct: 71 HFHDCFVNGCDASVLLDSSGTMESEKRSNANRDSARGFEVIDEIKSALENECPETVSCAD 130
Query: 122 ILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNA 181
+L + ARDS+ GG W++ LGRRD+ ASL S ++P+P L +++ + +G +
Sbjct: 131 LLALVARDSIVICGGPSWEVYLGRRDAREASLIGSMENIPSPESTLQTILTMFNFQGLDL 190
Query: 182 TEMVTLSGAHTIGLSRCIFYRDRIYNET-NIDP------SFAASMQVNCSFDGGDTDNNA 234
T++V L G+HTIG SRCI +R R+YN T N DP +A+ +Q C G D N
Sbjct: 191 TDLVALLGSHTIGNSRCIGFRQRLYNHTGNNDPDQTLNQDYASMLQQGCPISG--NDQNL 248
Query: 235 SPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFAN 294
D T KFDN +Y+NL+N +GL+ SD+ L+ T + V Y++N G F E FA
Sbjct: 249 FNLDYVTPTKFDNYYYKNLVNFRGLLSSDEILFTQSIETME-MVKYYAENEGAFFEQFAK 307
Query: 295 AMVKMSLLSPLTGSDGQIRKNCRVVN 320
+MVKM +SPLTG+DG+IR+ CR VN
Sbjct: 308 SMVKMGNISPLTGTDGEIRRICRRVN 333
>At5g06720 peroxidase (emb|CAA68212.1)
Length = 335
Score = 276 bits (706), Expect = 1e-74
Identities = 137/319 (42%), Positives = 203/319 (62%), Gaps = 9/319 (2%)
Query: 9 IFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFV 68
+ +I + G +SA L+ FY+ TCP A +RS ++ A+ + R+GASL+RLHFHDCFV
Sbjct: 17 LIVIVSSIFGTSSAQLNATFYSGTCPNASAIVRSTIQQALQSDTRIGASLIRLHFHDCFV 76
Query: 69 QGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAAR 128
GCDAS+LLDDT S EKN+ PN NS RGF V+D+IK LE+ CPGVVSC+D+L +A+
Sbjct: 77 NGCDASILLDDTGSIQSEKNAGPNVNSARGFNVVDNIKTALENACPGVVSCSDVLALASE 136
Query: 129 DSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLS 188
SV GG W + LGRRDS TA+L +NS +P+P LS + + G N ++V LS
Sbjct: 137 ASVSLAGGPSWTVLLGRRDSLTANLAGANSSIPSPIESLSNITFKFSAVGLNTNDLVALS 196
Query: 189 GAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNASPFDSTT 241
GAHT G +RC + +R++N + ++ + +++Q C +G + + + D +T
Sbjct: 197 GAHTFGRARCGVFNNRLFNFSGTGNPDPTLNSTLLSTLQQLCPQNG--SASTITNLDLST 254
Query: 242 QFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSL 301
FDN ++ NL + GL+ SDQ+L+ +T + VTS++ N F + FA +M+ M
Sbjct: 255 PDAFDNNYFANLQSNDGLLQSDQELFSTTGSSTIAIVTSFASNQTLFFQAFAQSMINMGN 314
Query: 302 LSPLTGSDGQIRKNCRVVN 320
+SPLTGS+G+IR +C+ VN
Sbjct: 315 ISPLTGSNGEIRLDCKKVN 333
>At5g66390 peroxidase (emb|CAA66964.1)
Length = 336
Score = 274 bits (701), Expect = 4e-74
Identities = 145/323 (44%), Positives = 200/323 (61%), Gaps = 13/323 (4%)
Query: 8 LIFLITICLVGK---TSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFH 64
LI CL K + L FY+++CPKA ++S V A +PRM ASLLRLHFH
Sbjct: 14 LIAFSPFCLCSKAYGSGGYLFPQFYDQSCPKAQEIVQSIVAKAFEHDPRMPASLLRLHFH 73
Query: 65 DCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILT 124
DCFV+GCDAS+LLD + + EK S PN NS RGFE+I++IK LE CP VSCADIL
Sbjct: 74 DCFVKGCDASILLDSSGTIISEKRSNPNRNSARGFELIEEIKHALEQECPETVSCADILA 133
Query: 125 VAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEM 184
+AARDS GG W++ LGRRD+ ASL SN+D+PAP +++ + ++G + ++
Sbjct: 134 LAARDSTVITGGPSWEVPLGRRDARGASLSGSNNDIPAPNNTFQTILTKFKRQGLDLVDL 193
Query: 185 VTLSGAHTIGLSRCIFYRDRIYNET-------NIDPSFAASMQVNCSFDGGDTDNNASPF 237
V+LSG+HTIG SRC +R R+YN++ + +A ++ C GG D
Sbjct: 194 VSLSGSHTIGNSRCTSFRQRLYNQSGNGKPDMTLSQYYATLLRQRCPRSGG--DQTLFFL 251
Query: 238 DSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMV 297
D T FKFDN +++NL+ KGL+ SD+ L+ + + V Y++N F E FA +MV
Sbjct: 252 DFATPFKFDNHYFKNLIMYKGLLSSDEILFTKNKQSKE-LVELYAENQEAFFEQFAKSMV 310
Query: 298 KMSLLSPLTGSDGQIRKNCRVVN 320
KM +SPLTG+ G+IR+ CR VN
Sbjct: 311 KMGNISPLTGAKGEIRRICRRVN 333
>At4g08770 peroxidase C2 precursor like protein
Length = 346
Score = 273 bits (698), Expect = 9e-74
Identities = 146/326 (44%), Positives = 203/326 (61%), Gaps = 11/326 (3%)
Query: 4 SLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
SL L FL+ + V + A LS FY++TCP+ + + A+ +PR+ AS+LRLHF
Sbjct: 4 SLIKLGFLLLLIQVSLSHAQLSPSFYDKTCPQVFDIATTTIVNALRSDPRIAASILRLHF 63
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFV GCDAS+LLD+T SF EK++F NANS RGF+VID +K +E CP VSCAD+L
Sbjct: 64 HDCFVNGCDASILLDNTTSFRTEKDAFGNANSARGFDVIDKMKAAVEKACPKTVSCADLL 123
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFN-AT 182
+AA++SV GG W + GRRDS +D +N +LPAPF L+ L + G + A+
Sbjct: 124 AIAAQESVVLAGGPSWRVPNGRRDSLRGFMDLANDNLPAPFFTLNQLKDRFKNVGLDRAS 183
Query: 183 EMVTLSGAHTIGLSRCIFYRDRIYNETN-------IDPSFAASMQVNCSFDGGDTDNNAS 235
++V LSG HT G ++C F DR+YN +N +D S+ ++++ C +G +
Sbjct: 184 DLVALSGGHTFGKNQCQFIMDRLYNFSNTGLPDPTLDKSYLSTLRKQCPRNG--NQSVLV 241
Query: 236 PFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVN-GSGTTDSQVTSYSKNAGRFLEDFAN 294
FD T FDN +Y NL KGL+ SDQ+L+ + + T V Y+ G+F + FA
Sbjct: 242 DFDLRTPTLFDNKYYVNLKENKGLIQSDQELFSSPDASDTLPLVREYADGQGKFFDAFAK 301
Query: 295 AMVKMSLLSPLTGSDGQIRKNCRVVN 320
AM++MS LSPLTG G+IR NCRVVN
Sbjct: 302 AMIRMSSLSPLTGKQGEIRLNCRVVN 327
>At4g16270 peroxidase like protein
Length = 362
Score = 268 bits (685), Expect = 3e-72
Identities = 149/305 (48%), Positives = 191/305 (61%), Gaps = 12/305 (3%)
Query: 23 VLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDS 82
VL Y +CP+A + S VE V ++PRM ASLLRLHFHDCFV GCDASVLLDDT+
Sbjct: 63 VLDFGLYRNSCPEAESIVYSWVETTVLEDPRMAASLLRLHFHDCFVNGCDASVLLDDTEG 122
Query: 83 FTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQ 142
GEK + PN NSLRGFEVID IK +ES+CP VSCADIL +AARDSV GG RW+++
Sbjct: 123 LVGEKTAPPNLNSLRGFEVIDSIKSDIESVCPETVSCADILAMAARDSVVVSGGPRWEVE 182
Query: 143 LGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYR 202
+GR+DS TAS A+ + LP+P +S LIS + G + T+MV LSG HT+G +RC +
Sbjct: 183 VGRKDSRTASKQAATNGLPSPNSTVSTLISTFQNLGLSQTDMVALSGGHTLGKARCTSFT 242
Query: 203 DRIYNETNIDPS-------FAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLN 255
R+ P+ F S+Q CS G + D T FDN +Y NLL+
Sbjct: 243 ARLQPLQTGQPANHGDNLEFLESLQQLCSTVGPSV--GITQLDLVTPSTFDNQYYVNLLS 300
Query: 256 QKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDFANAMVKMSLLSPLTGSDGQIRKN 315
+GL+ SDQ L V GT + V +Y+ + F EDF NAMVKM + GS+ +IRKN
Sbjct: 301 GEGLLPSDQALAVQDPGTR-AIVETYATDQSVFFEDFKNAMVKMGGIP--GGSNSEIRKN 357
Query: 316 CRVVN 320
CR++N
Sbjct: 358 CRMIN 362
>At4g08780 peroxidase C2 precursor like protein
Length = 346
Score = 265 bits (676), Expect = 3e-71
Identities = 141/326 (43%), Positives = 199/326 (60%), Gaps = 11/326 (3%)
Query: 4 SLSCLIFLITICLVGKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHF 63
SL L FL+ + V + A LS FY++TCP+ + + + A+ +PR+ AS+LRLHF
Sbjct: 4 SLIKLGFLLLLLQVSLSHAQLSPSFYDKTCPQVFDIVTNTIVNALRSDPRIAASILRLHF 63
Query: 64 HDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADIL 123
HDCFV GCDAS+LLD+T SF EK++F NANS RGF+VID +K +E CP VSCAD+L
Sbjct: 64 HDCFVNGCDASILLDNTTSFRTEKDAFGNANSARGFDVIDKMKAAIEKACPRTVSCADML 123
Query: 124 TVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFN-AT 182
+AA++S+ GG W + GRRDS +D +N +LP P L L + G + ++
Sbjct: 124 AIAAKESIVLAGGPSWMVPNGRRDSLRGFMDLANDNLPGPSSTLKQLKDRFKNVGLDRSS 183
Query: 183 EMVTLSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDNNAS 235
++V LSG HT G S+C F DR+YN + +D S+ A+++ C +G +
Sbjct: 184 DLVALSGGHTFGKSQCQFIMDRLYNFGETGLPDPTLDKSYLATLRKQCPRNG--NQSVLV 241
Query: 236 PFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVN-GSGTTDSQVTSYSKNAGRFLEDFAN 294
FD T FDN +Y NL KGL+ SDQ+L+ + + T V +Y+ G F + F
Sbjct: 242 DFDLRTPTLFDNKYYVNLKENKGLIQSDQELFSSPDAADTLPLVRAYADGQGTFFDAFVK 301
Query: 295 AMVKMSLLSPLTGSDGQIRKNCRVVN 320
A+++MS LSPLTG G+IR NCRVVN
Sbjct: 302 AIIRMSSLSPLTGKQGEIRLNCRVVN 327
>At1g68850 peroxidase ATP23a
Length = 336
Score = 264 bits (674), Expect = 6e-71
Identities = 128/306 (41%), Positives = 198/306 (63%), Gaps = 9/306 (2%)
Query: 24 LSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLRLHFHDCFVQGCDASVLLDDTDSF 83
L+ +Y TCP I+ +E V ++PR A ++RLHFHDCFVQGCD SVLLD+T++
Sbjct: 30 LTLDYYKSTCPTVFDVIKKEMECIVKEDPRNAAIIIRLHFHDCFVQGCDGSVLLDETETL 89
Query: 84 TGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCADILTVAARDSVFALGGQRWDLQL 143
GEK + PN NSL+G++++D IK +ES CPGVVSCAD+LT+ ARD+ +GG WD+ +
Sbjct: 90 QGEKKASPNINSLKGYKIVDRIKNIIESECPGVVSCADLLTIGARDATILVGGPYWDVPV 149
Query: 144 GRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFNATEMVTLSGAHTIGLSRCIFYRD 203
GR+DS TAS + + ++LP P L +I+ + +G + +MV L GAHTIG ++C +R
Sbjct: 150 GRKDSKTASYELATTNLPTPEEGLISIIAKFYSQGLSVEDMVALIGAHTIGKAQCRNFRS 209
Query: 204 RIYNE-------TNIDPSFAASMQVNCSFDGGDTDNNASPFDSTTQFKFDNAFYQNLLNQ 256
RIY + + ++ AS++ C G+ D+N + D+ T FDN+ Y LL
Sbjct: 210 RIYGDFQVTSALNPVSETYLASLREICPASSGEGDSNVTAIDNVTPNLFDNSIYHTLLRG 269
Query: 257 KGLVHSDQQLYVNGSGTTDSQVTS-YSKNAGRFLEDFANAMVKM-SLLSPLTGSDGQIRK 314
+GL++SDQ++Y + G ++ S Y+++ F E F+ +MVKM ++L+ + +DG++R+
Sbjct: 270 EGLLNSDQEMYTSLFGIQTRRIVSKYAEDPVAFFEQFSKSMVKMGNILNSESLADGEVRR 329
Query: 315 NCRVVN 320
NCR VN
Sbjct: 330 NCRFVN 335
>At2g38390 peroxidase
Length = 349
Score = 260 bits (665), Expect = 6e-70
Identities = 145/330 (43%), Positives = 197/330 (58%), Gaps = 15/330 (4%)
Query: 4 SLSCLIF--LITICLV---GKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASL 58
SLSC LI CL+ ++A L FY TCP I + + +PR+ ASL
Sbjct: 6 SLSCSAMGALIVGCLLLQASNSNAQLRPDFYFRTCPPIFNIIGDTIVNELRTDPRIAASL 65
Query: 59 LRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVS 118
LRLHFHDCFV+GCDAS+LLD++ SF EK++ PN NS+RGF+VID +K +E CP VS
Sbjct: 66 LRLHFHDCFVRGCDASILLDNSTSFRTEKDAAPNKNSVRGFDVIDRMKAAIERACPRTVS 125
Query: 119 CADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKG 178
CADI+T+A++ SV GG W + LGRRDS A +N+ LP+PF L+ L +A+ G
Sbjct: 126 CADIITIASQISVLLSGGPWWPVPLGRRDSVEAFFALANTALPSPFSTLTQLKTAFADVG 185
Query: 179 FN-ATEMVTLSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDT 230
N +++V LSG HT G ++C F R+YN + +++P++ ++ C +G T
Sbjct: 186 LNRPSDLVALSGGHTFGKAQCQFVTPRLYNFNGTNRPDPSLNPTYLVELRRLCPQNGNGT 245
Query: 231 DNNASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLE 290
FDS T FD +Y NLLN KGL+ SDQ L+ T V YS N F
Sbjct: 246 --VLVNFDSVTPTTFDRQYYTNLLNGKGLIQSDQVLFSTPGADTIPLVNQYSSNTFVFFG 303
Query: 291 DFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
F +AM++M L PLTG+ G+IR+NCRVVN
Sbjct: 304 AFVDAMIRMGNLKPLTGTQGEIRQNCRVVN 333
>At2g38380 peroxidase like protein
Length = 349
Score = 260 bits (664), Expect = 8e-70
Identities = 145/328 (44%), Positives = 198/328 (60%), Gaps = 13/328 (3%)
Query: 4 SLSCLIFLITICLV---GKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGASLLR 60
S S + LI CL+ ++A L FY TCP I + + + +PR+ ASLLR
Sbjct: 8 SCSAIGALILGCLLLQASNSNAQLRPDFYFGTCPFVFDIIGNIIVDELQTDPRIAASLLR 67
Query: 61 LHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGVVSCA 120
LHFHDCFV+GCDAS+LLD++ SF EK++ PNANS RGF VID +K LE CPG VSCA
Sbjct: 68 LHFHDCFVRGCDASILLDNSTSFRTEKDAAPNANSARGFNVIDRMKVALERACPGRVSCA 127
Query: 121 DILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGKKGFN 180
DILT+A++ SV GG W + LGRRDS A +N+ LP+PF +L+ L +A+ G N
Sbjct: 128 DILTIASQISVLLSGGPWWPVPLGRRDSVEAFFALANTALPSPFFNLTQLKTAFADVGLN 187
Query: 181 AT-EMVTLSGAHTIGLSRCIFYRDRIYN-------ETNIDPSFAASMQVNCSFDGGDTDN 232
T ++V LSG HT G ++C F R+YN + +++P++ ++ C +G T
Sbjct: 188 RTSDLVALSGGHTFGRAQCQFVTPRLYNFNGTNSPDPSLNPTYLVELRRLCPQNGNGT-- 245
Query: 233 NASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGTTDSQVTSYSKNAGRFLEDF 292
FD T FD+ +Y NL N KGL+ SDQ+L+ T V YS + F F
Sbjct: 246 VLVNFDVVTPDAFDSQYYTNLRNGKGLIQSDQELFSTPGADTIPLVNQYSSDMSVFFRAF 305
Query: 293 ANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
+AM++M L PLTG+ G+IR+NCRVVN
Sbjct: 306 IDAMIRMGNLRPLTGTQGEIRQNCRVVN 333
>At3g32980 peroxidase, putative
Length = 352
Score = 259 bits (662), Expect = 1e-69
Identities = 144/333 (43%), Positives = 203/333 (60%), Gaps = 15/333 (4%)
Query: 1 SRGSLSCLIFLITI-CLV---GKTSAVLSTIFYNETCPKALRTIRSAVEAAVSKEPRMGA 56
S SLS L+T+ CL+ +SA L+ FY+ TCP +R + + +PR+ A
Sbjct: 4 SYSSLSTWTTLMTLGCLLLHSSISSAQLTPTFYDNTCPSVFTIVRDTIVNELRSDPRIAA 63
Query: 57 SLLRLHFHDCFVQGCDASVLLDDTDSFTGEKNSFPNANSLRGFEVIDDIKKQLESMCPGV 116
S+LRLHFHDCFV GCDAS+LLD+T SF EK++ PNANS RGF VID +K +E+ CP
Sbjct: 64 SILRLHFHDCFVNGCDASILLDNTTSFRTEKDAAPNANSARGFPVIDRMKAAVETACPRT 123
Query: 117 VSCADILTVAARDSVFALGGQRWDLQLGRRDSTTASLDASNSDLPAPFLDLSGLISAYGK 176
VSCADILT+AA+ +V GG W + LGRRDS A +N++LPAPF L L +++
Sbjct: 124 VSCADILTIAAQQAVNLAGGPSWRVPLGRRDSLQAFFALANTNLPAPFFTLPQLKASFQN 183
Query: 177 KGFN-ATEMVTLSGAHTIGLSRCIFYRDRIYNETN-------IDPSFAASMQVNCSFDGG 228
G + +++V LSG HT G ++C F DR+YN +N ++ ++ +++ C +G
Sbjct: 184 VGLDRPSDLVALSGGHTFGKNQCQFIMDRLYNFSNTGLPDPTLNTTYLQTLRGQCPRNGN 243
Query: 229 DTDNNASPFDSTTQFKFDNAFYQNLLNQKGLVHSDQQLYVNGSGT-TDSQVTSYSKNAGR 287
T FD T FDN +Y NL KGL+ +DQ+L+ + + T T V Y+ +
Sbjct: 244 QT--VLVDFDLRTPTVFDNKYYVNLKELKGLIQTDQELFSSPNATDTIPLVREYADGTQK 301
Query: 288 FLEDFANAMVKMSLLSPLTGSDGQIRKNCRVVN 320
F F AM +M ++PLTG+ GQIR+NCRVVN
Sbjct: 302 FFNAFVEAMNRMGNITPLTGTQGQIRQNCRVVN 334
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,005,540
Number of Sequences: 26719
Number of extensions: 292380
Number of successful extensions: 970
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 80
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 645
Number of HSP's gapped (non-prelim): 104
length of query: 320
length of database: 11,318,596
effective HSP length: 99
effective length of query: 221
effective length of database: 8,673,415
effective search space: 1916824715
effective search space used: 1916824715
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0260.7


