

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.2
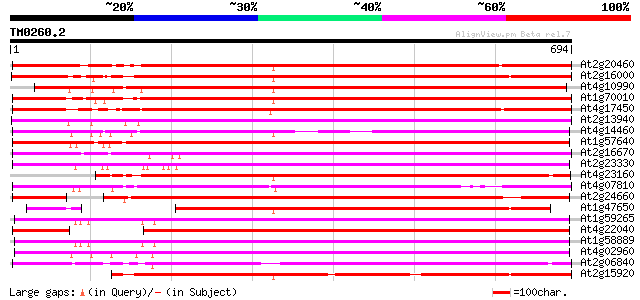
(694 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g20460 putative retroelement pol polyprotein 645 0.0
At2g16000 putative retroelement pol polyprotein 627 e-180
At4g10990 putative retrotransposon polyprotein 627 e-180
At1g70010 hypothetical protein 607 e-174
At4g17450 retrotransposon like protein 604 e-173
At2g13940 putative retroelement pol polyprotein 590 e-168
At4g14460 retrovirus-related like polyprotein 583 e-166
At1g57640 563 e-161
At2g16670 putative retroelement pol polyprotein 556 e-158
At2g23330 putative retroelement pol polyprotein 555 e-158
At4g23160 putative protein 546 e-155
At4g07810 putative polyprotein 520 e-147
At2g24660 putative retroelement pol polyprotein 514 e-146
At1g47650 hypothetical protein 514 e-146
At1g59265 polyprotein, putative 503 e-142
At4g22040 LTR retrotransposon like protein 501 e-142
At1g58889 polyprotein, putative 501 e-142
At4g02960 putative polyprotein of LTR transposon 501 e-142
At2g06840 putative retroelement pol polyprotein 498 e-141
At2g15920 putative retroelement pol polyprotein 498 e-141
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 645 bits (1663), Expect = 0.0
Identities = 334/697 (47%), Positives = 456/697 (64%), Gaps = 33/697 (4%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+S LKTFG LCY+ST ++ R K PR++ CVF+GY G KG+ L D++S V +SR+V F
Sbjct: 786 YSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEF 845
Query: 64 HETEFPFKTMEHTLDPCASVSLPSFMGSEALDYTPLPQGQTPDPQPQVPEVLPVPQTATR 123
HE FP + + + + V P PL G + P++ P Q + R
Sbjct: 846 HEELFPLASSQQSATTASDVFTPM---------DPLSSGNSITSHLPSPQISPSTQISKR 896
Query: 124 HSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTN 183
T+ P+H DY +N HP+SS LSY+ +SP H + N
Sbjct: 897 RITK---FPAHLQDYHCYF------VNKDDS-------HPISSSLSYSQISPSHMLYINN 940
Query: 184 VSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQ 243
+S P +Y +A KEW A+D+E+ A+ R +TW I LPP KK +GCKWV+ VK
Sbjct: 941 ISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHA 1000
Query: 244 DGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAF 303
DGS++R+KAR+VAKGYTQ EGLDY +TFSPVAK T++LLL ++AS+ W+L+QLD+ NAF
Sbjct: 1001 DGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAF 1060
Query: 304 LHGTLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGY 358
L+G L+E IYM+LP G + + PN VC L+KS+YGLKQASRQW+ +L +LG+
Sbjct: 1061 LNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGF 1120
Query: 359 EQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFF 418
E+ DHTL ++ + LL+YVDD+++ + + Q++ L + F++++LG LK+F
Sbjct: 1121 EKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYF 1180
Query: 419 LGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYR 478
LGLEVAR+++GI L+QRKYALELLT A +L CKP PM + LS+ G LL+D YR
Sbjct: 1181 LGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYR 1240
Query: 479 RLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATAS 538
RLVG+L+YLT TRPD+ + V +L Q+ ++P H A +VL+Y+KG GQGLF+SA
Sbjct: 1241 RLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDD 1300
Query: 539 LKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVC 598
L L+ Y+D+DW CPD+RRS G+++F+GSSLISWR+KKQ TVSRSS+EAEYRALA C
Sbjct: 1301 LTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASC 1360
Query: 599 EVQWLSYLFQFLHLRVPFPVP-LFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAG 657
E+ WLS L L LRV VP L+ D+ +A++IA NP FHERTKHIE+DCH VR KL G
Sbjct: 1361 EMAWLSTL--LLALRVHSGVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNG 1418
Query: 658 LIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
+ LL V T+DQ+ADI TKPL P F + K+ + N
Sbjct: 1419 QLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSIQN 1455
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 627 bits (1618), Expect = e-180
Identities = 329/703 (46%), Positives = 446/703 (62%), Gaps = 39/703 (5%)
Query: 3 VFSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVL 62
V+ L+TFG LCY+ST + R K PR++ C+F+GY G KG+ L D++S V +SR+V
Sbjct: 774 VYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQ 833
Query: 63 FHETEFPFKTMEHTLDPCASVSLPSFMGSEALDYTPLPQG------QTPDPQPQVPEVLP 116
FHE FP +P + SL F P+ G +P P LP
Sbjct: 834 FHEEVFPLAK-----NPGSESSLKLFT-----PMVPVSSGIISDTTHSPSSLPSQISDLP 883
Query: 117 VPQTATRHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPK 176
PQ + S R R+ P+H +DY + K+P+SS +SY+ +SP
Sbjct: 884 -PQIS---SQRVRKPPAHLNDYHC-------------NTMQSDHKYPISSTISYSKISPS 926
Query: 177 HYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWV 236
H + N++ PT Y +A KEW +A+D E+ A+ + NTW I LP KK +GCKWV
Sbjct: 927 HMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWV 986
Query: 237 YRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQ 296
+ +K DG+++RYKARLVAKGYTQ EGLDY DTFSPVAK TT++LLL ++AS+ WFL Q
Sbjct: 987 FTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQ 1046
Query: 297 LDVDNAFLHGTLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQASRQWYNTLVA 351
LDV NAFL+G L+EEI+M++P G + + N V L++S+YGLKQASRQW+ +
Sbjct: 1047 LDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSS 1106
Query: 352 ALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKD 411
+L SLG+++ DHTL LK +L+YVDD+++ S + L +F+++D
Sbjct: 1107 SLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRD 1166
Query: 412 LGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLL 471
LGDLK+FLGLEVAR+ GI + QRKYALELL G+L CKPV PM L + + G L+
Sbjct: 1167 LGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLI 1226
Query: 472 DDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGL 531
+D YRR+VG+L+YLT TRPD+ + V +L Q+ ++P H AA RVL+Y+KG GQGL
Sbjct: 1227 EDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGL 1286
Query: 532 FFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYR 591
F+SA++ L L+ ++DSDWA C D+RRS +++F+G SLISWR+KKQ TVSRSS+EAEYR
Sbjct: 1287 FYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYR 1346
Query: 592 ALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVR 651
ALA CE+ WL L L P P+ L+ D+ +AI+IA NP FHERTKHI+LDCH VR
Sbjct: 1347 ALALATCEMVWLFTLLVSLQASPPVPI-LYSDSTAAIYIATNPVFHERTKHIKLDCHTVR 1405
Query: 652 TKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
+L G + LL V T DQ+ADI TKPL P F K+ + N
Sbjct: 1406 ERLDNGELKLLHVRTEDQVADILTKPLFPYQFEHLKSKMSILN 1448
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 627 bits (1617), Expect = e-180
Identities = 323/704 (45%), Positives = 450/704 (63%), Gaps = 49/704 (6%)
Query: 31 QKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLFHETEFPFKT---MEHTLDPCASVSLPS 87
Q V Y G KG+ + D++S + ++R+V+FHET+FPFKT ++ ++D + LP
Sbjct: 328 QNSVVERYPSGYKGYKVLDLESHSISITRNVVFHETKFPFKTSKFLKESVDMFPNSILPL 387
Query: 88 FMGSEALDYTPLPQ--------GQTPDPQPQVPEVLPVPQTATRHST------------- 126
++ PL T + + P+P T +T
Sbjct: 388 PAPLHFVESMPLDDDLRADDNNASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIA 447
Query: 127 ---RSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIK--------------HPLSSVLS 169
R+ + P++ +Y +S+ L+S S +ST I+ +P+S+ +S
Sbjct: 448 RPKRNAKAPAYLSEYHC---NSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAIS 504
Query: 170 YTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKK 229
Y L+P +++ + EP +TQA + ++W A ++EL AL +N TW++ L K
Sbjct: 505 YDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKN 564
Query: 230 PIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAAS 289
+GCKWV+ +K+ DGSI+RYKARLVA+G+TQ EG+DY++TFSPVAK +++LLL LAA+
Sbjct: 565 VVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAA 624
Query: 290 QGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQASRQ 344
GW L Q+DV NAFLHG LDEEIYM LP G + P VC L KSLYGLKQASRQ
Sbjct: 625 TGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQ 684
Query: 345 WYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLH 404
WY L + + Q P D+T+ +K + TSI +L+YVDD+++ N ++ +K L
Sbjct: 685 WYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLR 744
Query: 405 SQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLS 464
S+F+IKDLG +FFLGLE+ARS++GI + QRKYA LL D GL GCKP PMD LHL+
Sbjct: 745 SEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLT 804
Query: 465 QTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVK 524
+ GTLL + TSYR LVGRLLYL TRPD+ + V LSQ+L++PTD+H QAA +VLRY+K
Sbjct: 805 KEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLK 864
Query: 525 GAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRS 584
G PGQGL +SA++ L L +SD+DW C D+RRS+ G+ I+LG+SLI+W++KKQ+ VSRS
Sbjct: 865 GNPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRS 924
Query: 585 SSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIE 644
S+E+EYR+LA CE+ WL L + LH+ + P LFCDN+SA+H+A NP FHERTKHIE
Sbjct: 925 STESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIE 984
Query: 645 LDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVL 688
+DCH VR +++AG + L V T +QLADI TKPL P P F L
Sbjct: 985 IDCHTVRDQIKAGKLKTLHVPTGNQLADILTKPLHPVQSPIFSL 1028
>At1g70010 hypothetical protein
Length = 1315
Score = 607 bits (1564), Expect = e-174
Identities = 320/708 (45%), Positives = 446/708 (62%), Gaps = 38/708 (5%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+ +K FG LCYAST ++R K PRA+ C FIGY G KG+ L D+++ +++SRHV+F
Sbjct: 624 YDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVF 683
Query: 64 HETEFPFKTMEHTLDPCASVSLPSFMGSEALDYTPLPQGQT-----PDPQPQVPEVLP-- 116
HE FPF L S +F L+ TP Q Q+ P E+LP
Sbjct: 684 HEELFPF------LGSDLSQEEQNFFPD--LNPTPPMQRQSSDHVNPSDSSSSVEILPSA 735
Query: 117 -----VPQTATRHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYT 171
VP+ + + S R + P++ DY S VSST H + LSY
Sbjct: 736 NPTNNVPEPSVQTSHRKAKKPAYLQDYYC-----------HSVVSST--PHEIRKFLSYD 782
Query: 172 NLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPI 231
++ + F + T EP+ YT+A + + WRDAM E + L +TW + LP K+ I
Sbjct: 783 RINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCI 842
Query: 232 GCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQG 291
GC+W++++K+ DGS++RYKARLVA+GYTQ EG+DY +TFSPVAK +++LLL +AA
Sbjct: 843 GCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFK 902
Query: 292 WFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQASRQWY 346
L QLD+ NAFL+G LDEEIYMRLP G +S + PN VC L+KSLYGLKQASRQWY
Sbjct: 903 LSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWY 962
Query: 347 NTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQ 406
+ L LG+ Q DHT LK + +L+Y+DD+++ N+ + +KS + S
Sbjct: 963 LKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSF 1022
Query: 407 FRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQT 466
F+++DLG+LK+FLGLE+ RS KGI ++QRKYAL+LL + G LGCKP PMD ++ +
Sbjct: 1023 FKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHD 1082
Query: 467 GGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGA 526
G + YRRL+GRL+YL TRPD+ + V +L+Q+ +P H QA ++L+Y+KG
Sbjct: 1083 SGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGT 1142
Query: 527 PGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSS 586
GQGLF+SAT+ L+L+ Y+++D+ C D+RRS +GY +FLG SLI W+++KQ VS+SS+
Sbjct: 1143 IGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSA 1202
Query: 587 EAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELD 646
EAEYR+L+ E+ WL+ + L + + P LFCDN++AIHIA N FHERTKHIE D
Sbjct: 1203 EAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESD 1262
Query: 647 CHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
CH VR +L GL L ++T Q+AD FTKPL PS F + K+GL N
Sbjct: 1263 CHSVRERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGLLN 1310
>At4g17450 retrotransposon like protein
Length = 1433
Score = 604 bits (1558), Expect = e-173
Identities = 315/698 (45%), Positives = 441/698 (63%), Gaps = 39/698 (5%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+ LKTFG LCY+ST + R K +PRA+ CVF+GY G KG+ L DI++ V +SRHV+F
Sbjct: 761 YESLKTFGCLCYSSTSPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIF 820
Query: 64 HETEFPFKTMEHTLDPCASVSLPSFMGSEALDYTPLPQGQTPDPQPQVPEVLPVPQTATR 123
HE FPF + S + + D+ PL Q P + LP+ QT+
Sbjct: 821 HEDIFPF--------------ISSTIKDDIKDFFPLLQF------PARTDDLPLEQTSI- 859
Query: 124 HSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTN 183
+ +H H ++ ++ + SK KH L Y N + +AF N
Sbjct: 860 -------IDTHPHQ-DVSSSKALVPFDPLSKRQKKPPKH-LQDFHCYNNTTEPFHAFINN 910
Query: 184 VSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQ 243
+++ V P Y++A K W DAM +E+ A+ R NTW +V LPP KK IGCKWV+ +KH
Sbjct: 911 ITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNA 970
Query: 244 DGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAF 303
DGSI+RYKARLVAKGYTQ EGLDY +TFSPVAK T++R++L LAA W +HQLD+ NAF
Sbjct: 971 DGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDISNAF 1030
Query: 304 LHGTLDEEIYMRLPPGVS-----SPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGY 358
L+G LDEEIYM++PPG + + P+ +C L KS+YGLKQASRQWY L LK +G+
Sbjct: 1031 LNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLKGMGF 1090
Query: 359 EQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFF 418
++ DHTL +K + + +L+YVDD+++ NS + + + L S F+++DLG K+F
Sbjct: 1091 QKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYF 1150
Query: 419 LGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYR 478
LG+E+ARS KGI + QRKY LELL+ G LG KP P+D ++ L++ G L D TSYR
Sbjct: 1151 LGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYR 1210
Query: 479 RLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATAS 538
+LVG+L+YL TRPD+ Y V L Q+ +PT +H A +VLRY+KG GQGLF+SA
Sbjct: 1211 KLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDK 1270
Query: 539 LKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVC 598
L+ Y+DSD+ C D+RR + Y +F+G L+SW++KKQ TVS S++EAE+RA++
Sbjct: 1271 FDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTK 1330
Query: 599 EVQWLSYLFQFLHLRVPF--PVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQA 656
E+ WLS LF +VPF P L+CDN +A+HI N FHERTK +ELDC+ R +++
Sbjct: 1331 EMIWLSRLFD--DFKVPFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYKTREAVES 1388
Query: 657 GLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
G + + V T +Q+AD TK + P+ F + K+G+ N
Sbjct: 1389 GFLKTMFVETGEQVADPLTKAIHPAQFHKLIGKMGVCN 1426
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 590 bits (1520), Expect = e-168
Identities = 327/752 (43%), Positives = 446/752 (58%), Gaps = 60/752 (7%)
Query: 3 VFSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVL 62
V+S L+ FGS CY + R++ K R++ C+F+GY G KG+ ++DI+ E L+SR V+
Sbjct: 745 VYSQLRVFGSACYVHRVTRDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVI 804
Query: 63 FHETEFPFK-------------------------------------------TMEHTLDP 79
F E FP+ T E LD
Sbjct: 805 FREEVFPYAGVNSSTLASTSLPTVSEDDDWAIPPLEVRGSIDSVETERVVCTTDEVVLDT 864
Query: 80 CASVS-LPS--FMGSEALDYTPL---PQGQ--TPDPQPQVPEVLPVPQTAT--RHSTRSR 129
S S +P+ F+ + +PL P G TP VP P+P + R S R+
Sbjct: 865 SVSDSEIPNQEFVPDDTPPSSPLSVSPSGSPNTPTTPIVVPVASPIPVSPPKQRKSKRAT 924
Query: 130 RVPSHFHDYQLA----LPSSIAALNSSSKVSST---GIKHPLSSVLSYTNLSPKHYAFTT 182
P +DY L PSSI AL + SST PL+ +S S H A+
Sbjct: 925 HPPPKLNDYVLYNAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLA 984
Query: 183 NVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHK 242
++ VEP + +A Q K W DAM E++AL N TW IVDLPP K IG +WV++ K+
Sbjct: 985 AITDNVEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYN 1044
Query: 243 QDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNA 302
DG+++RYKARLV +G QVEG DY +TF+PV + TT+R LL A+ W ++Q+DV NA
Sbjct: 1045 SDGTVERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNA 1104
Query: 303 FLHGTLDEEIYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCP 362
FLHG L+EE+YM+LPPG P++VC L+KSLYGLKQA R W+ L +L G+ Q
Sbjct: 1105 FLHGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSY 1164
Query: 363 HDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLE 422
D++L + +L+YVDD+L+ GN +Q K +L F +KDLG LK+FLG+E
Sbjct: 1165 EDYSLFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIE 1224
Query: 423 VARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVG 482
V+R +GI L+QRKYAL+++ D+G LG +P TP++ HL+ G LL DP YRRLVG
Sbjct: 1225 VSRGPEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVG 1284
Query: 483 RLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQ 542
RLLYL TRP+L+Y V L+Q++ +P + H AA RV+RY+KG+PGQG+ +A L L+
Sbjct: 1285 RLLYLLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLE 1344
Query: 543 AYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQW 602
Y DSDW CP TRRSI+ Y + LG S ISW+TKKQ TVS SS+EAEYRA++ + E++W
Sbjct: 1345 VYCDSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKW 1404
Query: 603 LSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLL 662
L L + L + P L+CD+++AIHIA NP FHERTKHIE DCH VR ++ G+I
Sbjct: 1405 LRKLLKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQ 1464
Query: 663 PVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
V T +QLAD+FTK L + F + KLG+ N
Sbjct: 1465 HVRTTEQLADVFTKALGRNQFLYLMSKLGVQN 1496
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 583 bits (1503), Expect = e-166
Identities = 317/740 (42%), Positives = 439/740 (58%), Gaps = 107/740 (14%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+S LK FG LC+ ST RTK PRA+ CVF+GY G KG+ + D++S V +SR+V+F
Sbjct: 794 YSLLKNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVF 853
Query: 64 HETEFPFKTME---HTLDPCASVSLPSFMGSEALDYTPL----------PQGQTPDPQPQ 110
E FPFKT E +D + LP ++ PL +T D
Sbjct: 854 KEHVFPFKTSELLNKAVDMFPNSILPLPAPLHFVETMPLIDEDSLIPTTTDSRTADNHAS 913
Query: 111 -----VPEVLPVPQTATR------------HSTRSRRVPSHFHDYQLALPSSIAAL---- 149
+P ++P P + T S R+ R PS+ +Y +L SI+ L
Sbjct: 914 SSSSALPSIIP-PSSNTETQDIDSNAVPITRSKRTTRAPSYLSEYHCSLVPSISTLPPTD 972
Query: 150 ------------NSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAA 197
+SS +T +P+S+V+SY +P ++ ++ EP T++QA
Sbjct: 973 SSIPIHPLPEIFTASSPKKTT--PYPISTVVSYDKYTPLCQSYIFAYNTETEPKTFSQAM 1030
Query: 198 QCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAK 257
+ ++W +EL+A+ N TW + LPP K +GCKWV+ +K+ DG+++RYKARLVA+
Sbjct: 1031 KSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGTVERYKARLVAQ 1090
Query: 258 GYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLP 317
G+TQ EG+D+LDTFSPVAK T+ +++L LAA GW L Q+DV +AFLHG LDEEI+M LP
Sbjct: 1091 GFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHGDLDEEIFMSLP 1150
Query: 318 PGVSSPR-----PNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRT 372
G + P PN VC L KS+YGLKQASRQWY VAAL
Sbjct: 1151 QGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRFVAAL------------------- 1191
Query: 373 STSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICL 432
+Y+DD+++ N+ E++ +K+ L S+F+IKDLG +FFLGL
Sbjct: 1192 --------VYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLGL----------- 1232
Query: 433 TQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRP 492
LGCKP PMD TLHL + GT L +PT+YR+L+GRLLYLT TRP
Sbjct: 1233 ---------------LGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLIGRLLYLTITRP 1277
Query: 493 DLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGC 552
D+ Y V QLSQ++++P+D+H QAA +VLRY+K PGQGL +SA + L +SD+DWA C
Sbjct: 1278 DITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEICLNGFSDADWAAC 1337
Query: 553 PDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHL 612
DTRRSI+G+ I+LG+SLISW++KKQ SRSS+E+EYR++A CE+ WL L + LH+
Sbjct: 1338 KDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEIIWLQQLLKDLHI 1397
Query: 613 RVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLAD 672
+ P LFCDN+SA+H + NP FHERTKHIE+DCH VR +++AG + L V T +Q AD
Sbjct: 1398 PLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLKALHVPTENQHAD 1457
Query: 673 IFTKPLPPSLFPTFVLKLGL 692
I TK L P F + ++ L
Sbjct: 1458 ILTKALHPGPFHHLLRQMSL 1477
>At1g57640
Length = 1444
Score = 563 bits (1452), Expect = e-161
Identities = 304/736 (41%), Positives = 445/736 (60%), Gaps = 54/736 (7%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+S L+ FGSLCYA N K R+++CVF+GY HG KG+ LFD++ ++ +SR V+F
Sbjct: 709 YSHLRVFGSLCYAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIF 768
Query: 64 HETEFPFKTM------EHTLDPCA-------SVSLPSFMGSEALDYTPLPQGQTPDPQPQ 110
ETEFP+ M E L C ++ + +G + T P T P
Sbjct: 769 QETEFPYSKMSCNEEDERVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATG---PI 825
Query: 111 VPEV-----------------------------LPVPQTAT-----RHSTRSRRVPSHFH 136
+PE+ LP+ T R S+R + P
Sbjct: 826 IPEINQESSSPSEFVSLSSLDPFLASSTVQTADLPLSSTTPAPIQLRRSSRQTQKPMKLK 885
Query: 137 DYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTNVSSTVEPTTYTQA 196
++ ++ ++ S S +S+ +P+ + + H AF V++ +EPTTY +A
Sbjct: 886 NFV----TNTVSVESISPEASSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEA 941
Query: 197 AQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVA 256
K WR+AM E+E+L N T+ IV+LPP K+ +G KWVY++K++ DG+I+RYKARLV
Sbjct: 942 MVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVV 1001
Query: 257 KGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRL 316
G Q EG+DY +TF+PVAK +T+RL L +AA++ W +HQ+DV NAFLHG L EE+YM+L
Sbjct: 1002 LGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKL 1061
Query: 317 PPGVSSPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSI 376
P G P++VC L KSLYGLKQA R W++ L +ALK G+ Q D++L
Sbjct: 1062 PQGFQCDDPSKVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIF 1121
Query: 377 TALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRK 436
+L+YVDD++++G+ + + KS+L S F +KDLG LK+FLG+EV+R+A+G L+QRK
Sbjct: 1122 VHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRK 1181
Query: 437 YALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNY 496
Y L+++++ GLLG +P P++ LS + LL D + YRRLVGRL+YL TRP+L+Y
Sbjct: 1182 YVLDIISEMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSY 1241
Query: 497 VVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTR 556
V L+Q++ +P H AA RV+RY+K PGQG+ S+T++L++ + DSD+A CP TR
Sbjct: 1242 SVHTLAQFMQNPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTR 1301
Query: 557 RSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPF 616
RS+ GY + LG + ISW+TKKQ TVSRSS+EAEYRA+A + E+ WL + L +
Sbjct: 1302 RSLTGYFVQLGDTPISWKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQ 1361
Query: 617 PVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTK 676
+ +F D++SAI ++ NP HERTKH+E+DCH +R + G+I V + QLADI TK
Sbjct: 1362 AMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTK 1421
Query: 677 PLPPSLFPTFVLKLGL 692
L F+ KLG+
Sbjct: 1422 ALGEKEVRYFLRKLGI 1437
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 556 bits (1434), Expect = e-158
Identities = 312/724 (43%), Positives = 428/724 (59%), Gaps = 37/724 (5%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
F L+ FGSLCYA NR K R+++CVF+GY HG KG+ LFD++ E +SR V+F
Sbjct: 609 FDHLRVFGSLCYAHNRNRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVF 668
Query: 64 HETEFPFKTM--EHTLDPCASVSLPSFMGSEALDYTPLPQGQTPDPQPQVPEVLPVPQTA 121
E EFPF+ ++ ++ + + L + GQ P P + P+ +A
Sbjct: 669 SELEFPFRISHEQNVIEEEEEALWAPIV--DGLIEEEVHLGQNAGPTPPICVSSPISPSA 726
Query: 122 TRHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKH-PLSSVLSYT--------- 171
T S+RS S D ++ + + ++SS S T ++ PLS T
Sbjct: 727 T--SSRSEHSTSSPLDTEVVPTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAV 784
Query: 172 ----NLSPKHYAFTTNVSSTVEPTTYTQAAQCK--------EWRDAMDK---------EL 210
S ++ A + V TT Q + K + RD + +
Sbjct: 785 PPPRRQSTRNKAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYVAI 844
Query: 211 EALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDT 270
+A N+TW I DLPP K+ IG +WVY+VKH DGS++RYKARLVA G Q EG DY +T
Sbjct: 845 DAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGET 904
Query: 271 FSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPRPNQVCL 330
F+PVAK T+RL L +A + W +HQ+DV NAFLHG L EE+YM+LPPG + PN+VC
Sbjct: 905 FAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFEASHPNKVCR 964
Query: 331 LQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTG 390
L+K+LYGLKQA R W+ L ALK G++Q D++L + +L+YVDD+++TG
Sbjct: 965 LRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITG 1024
Query: 391 NSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGC 450
NS Q K +L S F +KDLG LK+FLG+EVARS GI + QRKYAL+++++ GLLG
Sbjct: 1025 NSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIISETGLLGV 1084
Query: 451 KPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTD 510
KP P++ L + LL DP YRRLVGRL+YL TR DL + V L++++ P +
Sbjct: 1085 KPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILARFMQEPRE 1144
Query: 511 LHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSL 570
H AA RV+RY+K PGQG+F + ++ + DSDWAG P +RRS+ GY + G S
Sbjct: 1145 DHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFGDSP 1204
Query: 571 ISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHI 630
ISW+TKKQ TVS+SS+EAEYRA++ + E+ WL L L + P+ + CD++SAI+I
Sbjct: 1205 ISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSAIYI 1264
Query: 631 AQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKL 690
A NP FHERTKHIE+D H VR + G+I V T QLADIFTKPL F F +KL
Sbjct: 1265 ATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCFSAFRIKL 1324
Query: 691 GLYN 694
G+ N
Sbjct: 1325 GIRN 1328
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 555 bits (1430), Expect = e-158
Identities = 311/771 (40%), Positives = 444/771 (57%), Gaps = 82/771 (10%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+ L++FG LCYA RN+ K R++KCVFIGY HG K + ++D+++ ++ SR V F
Sbjct: 719 YDMLRSFGCLCYAHIRPRNKDKFTSRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRF 778
Query: 64 HETEFPFKTMEHTLDPC---------------ASVSLPSFMGSEALDYTPLPQGQTPDPQ 108
HE +P+ T + P S + S + +P G P
Sbjct: 779 HEDIYPYATATQSNVPLPPPTPPMVNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPP 838
Query: 109 PQVPE-----VLPVPQ---TATRHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGI 160
+ + PVP+ + S+ SR + D + + + + SS SS G+
Sbjct: 839 RSIDQSPSTSTNPVPEEIGSIVPSSSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGL 898
Query: 161 KHPL--------SSVL---SYTNLSPKHYAFTTNVSSTVE------PTTYT--------- 194
L SVL TN + K + N+ S + PT+ +
Sbjct: 899 PELLGKGCREKKKSVLLKDFVTNTTSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTL 958
Query: 195 ------------------------QAAQCKEWRDA---------MDKELEALSRNNTWLI 221
+ + K ++DA M KE++AL N+TW I
Sbjct: 959 YPLSDFLTNSGYSANHIAFMAAILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDI 1018
Query: 222 VDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLR 281
DLP KK I KWVY++K+ DG+++R+KARLV G Q EG+D+ +TF+PVAK TT+R
Sbjct: 1019 TDLPHGKKAISSKWVYKLKYNSDGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVR 1078
Query: 282 LLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPRPNQVCLLQKSLYGLKQA 341
+LA+AA++ W +HQ+DV NAFLHG L+EE+YMRLPPG P++VC L+KSLYGLKQA
Sbjct: 1079 TILAVAAAKDWEVHQMDVHNAFLHGDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQA 1138
Query: 342 SRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKS 401
R W++ L AL+++G+ Q D++L + +I +L+YVDD+++ GN+ + I KS
Sbjct: 1139 PRCWFSKLSTALRNIGFTQSYEDYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKS 1198
Query: 402 FLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTL 461
LH F +KDLG LK+FLGLEV+R G CL+QRKYAL+++ + GLLGCKP P+
Sbjct: 1199 QLHKCFHMKDLGKLKYFLGLEVSRGPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNH 1258
Query: 462 HLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLR 521
L+ G + +P YRRLVGR +YLT TRPDL+Y V LSQ++ +P H +AA R++R
Sbjct: 1259 KLASITGPVFTNPEQYRRLVGRFIYLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVR 1318
Query: 522 YVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTV 581
Y+KG+P QG+F + +SL + AY DSD+ CP TRRS++ Y ++LG S ISW+TKKQ TV
Sbjct: 1319 YLKGSPAQGIFLRSDSSLIINAYCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTV 1378
Query: 582 SRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTK 641
S SS+EAEYRA+A + E++WL L + L + P+ L CD+++AIHIA NP FHERTK
Sbjct: 1379 SYSSAEAEYRAMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTK 1438
Query: 642 HIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
HIE DCH VR + LI + T DQ+AD+ TK LP F + LG+
Sbjct: 1439 HIESDCHKVRDAVLDKLITTEHIYTEDQVADLLTKSLPRPTFERLLSTLGV 1489
>At4g23160 putative protein
Length = 1240
Score = 546 bits (1408), Expect = e-155
Identities = 284/594 (47%), Positives = 386/594 (64%), Gaps = 25/594 (4%)
Query: 107 PQPQVPEVLPVPQTATRHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSS 166
P + +P P T H R R P++ DY S+A+L H +S
Sbjct: 18 PSANIQNDVPEPSVHTSH--RRTRKPAYLQDYYC---HSVASLTI----------HDISQ 62
Query: 167 VLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPP 226
LSY +SP +++F ++ EP+TY +A + W AMD E+ A+ +TW I LPP
Sbjct: 63 FLSYEKVSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGAMDDEIGAMETTHTWEICTLPP 122
Query: 227 TKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLAL 286
KKPIGCKWVY++K+ DG+I+RYKARLVAKGYTQ EG+D+++TFSPV K T+++L+LA+
Sbjct: 123 NKKPIGCKWVYKIKYNSDGTIERYKARLVAKGYTQQEGIDFIETFSPVCKLTSVKLILAI 182
Query: 287 AASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQA 341
+A + LHQLD+ NAFL+G LDEEIYM+LPPG ++ + PN VC L+KS+YGLKQA
Sbjct: 183 SAIYNFTLHQLDISNAFLNGDLDEEIYMKLPPGYAARQGDSLPPNAVCYLKKSIYGLKQA 242
Query: 342 SRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKS 401
SRQW+ L G+ Q DHT LK T+T +L+YVDD+++ N+ + +KS
Sbjct: 243 SRQWFLKFSVTLIGFGFVQSHSDHTYFLKITATLFLCVLVYVDDIIICSNNDAAVDELKS 302
Query: 402 FLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTL 461
L S F+++DLG LK+FLGLE+ARSA GI + QRKYAL+LL + GLLGCKP PMD ++
Sbjct: 303 QLKSCFKLRDLGPLKYFLGLEIARSAAGINICQRKYALDLLDETGLLGCKPSSVPMDPSV 362
Query: 462 HLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLR 521
S G D +YRRL+GRL+YL TR D+++ V +LSQ+ +P HQQA ++L
Sbjct: 363 TFSAHSGGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQFSEAPRLAHQQAVMKILH 422
Query: 522 YVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTV 581
Y+KG GQGLF+S+ A ++LQ +SD+ + C DTRRS NGY +FLG+SLISW++KKQ V
Sbjct: 423 YIKGTVGQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSKKQQVV 482
Query: 582 SRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTK 641
S+SS+EAEYRAL+ E+ WL+ F+ L L + P LFCDN +AIHIA N FHERTK
Sbjct: 483 SKSSAEAEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIATNAVFHERTK 542
Query: 642 HIELDCHVVRTK--LQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLY 693
HIE DCH VR + QA L + D FT+ L P L T + + ++
Sbjct: 543 HIESDCHSVRERSVYQATLSYSFQAYDEQ---DGFTEYLSPILRGTIMYIVSMF 593
>At4g07810 putative polyprotein
Length = 1366
Score = 520 bits (1338), Expect = e-147
Identities = 298/723 (41%), Positives = 419/723 (57%), Gaps = 76/723 (10%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
++ L+ F LCYAST R K RA CVFIGY G KG+ + D++S V ++R+V+F
Sbjct: 683 YNSLRVFCCLCYASTHQHERHKFTERATSCVFIGYESGFKGYKILDLESNTVSVTRNVVF 742
Query: 64 HETEFPFKTMEHTL------DPCASVS---------LPSFMGSEALDYTPLPQGQTPDPQ 108
HET FPF T D +S + + L+ P+ + T
Sbjct: 743 HETIFPFIDKHSTQNVSFFDDSVLPISEKQKENRFQIYDYFNVLNLEVCPVIEPTTVPAH 802
Query: 109 PQVPEVLPVPQTATRHS-----------TRSRRVPSHFHDYQLALPSSIAALNSSSKVSS 157
+ P+ T T + R PS+ Y S++ SSS
Sbjct: 803 THTRSLAPLSTTVTNDQFGNDMDNTLMPRKETRAPSYLSQYHC---SNVLKEPSSSL--- 856
Query: 158 TGIKHPLSSVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNN 217
G H LSS LSY LS ++ F + + EPTT+ +AA ++W DAM+ EL+AL +
Sbjct: 857 HGTAHSLSSHLSYDKLSNEYRLFCFAIIAEKEPTTFKEAALLQKWLDAMNVELDALVSTS 916
Query: 218 TWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKT 277
T I L K+ IGCKWV+++K+K DG+I+RYKARLVA GYTQ EG+DY+DTFSP+AK
Sbjct: 917 TREICSLHDGKRAIGCKWVFKIKYKSDGTIERYKARLVANGYTQQEGVDYIDTFSPIAKL 976
Query: 278 TTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPRPNQ------VCLL 331
T++RL+LALAA W + Q+DV NAFLHG +EEIYM+LP G + PR + VC L
Sbjct: 977 TSVRLILALAAIHNWSISQMDVTNAFLHGDFEEEIYMQLPQGYT-PRKGELLPKRPVCRL 1035
Query: 332 QKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGN 391
KSLYGLKQASRQW++ L G+ Q D TL ++ + ALL+YVDD++L N
Sbjct: 1036 VKSLYGLKQASRQWFHKFSGVLIQNGFMQSLFDPTLFVRVREDTFLALLVYVDDIMLVSN 1095
Query: 392 SFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCK 451
+ VK L +F++KDLG ++FLGLE+ARS +GI ++QRKYALELL + G LGCK
Sbjct: 1096 KDSAVIEVKQILAKEFKLKDLGQKRYFLGLEIARSKEGISISQRKYALELLEEFGFLGCK 1155
Query: 452 PVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDL 511
PV TPM+ L LSQ G LL D + YR+L+GRL+YLT TRPD+ + V +L+QY+++P +
Sbjct: 1156 PVPTPMELNLKLSQEDGALLLDASHYRKLIGRLVYLTVTRPDICFAVNKLNQYMSAPREP 1215
Query: 512 HQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLI 571
H AA R+LRY+K PGQG+F+ A+++L +A++D+DW+ CP++ SI S++
Sbjct: 1216 HLMAARRILRYLKNDPGQGVFYPASSTLTFRAFADADWSNCPESSISI---------SIV 1266
Query: 572 SWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIA 631
W + S+EA +L L +P + ++ D++SA+HIA
Sbjct: 1267 FW--------LKLSTEA--------------------WLVLSLPDTIFVYYDDESALHIA 1298
Query: 632 QNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLG 691
+N FHE TK+ D HVVR K+ G I L V T + D+ TKPL F + K+G
Sbjct: 1299 KNSVFHESTKNFLHDIHVVREKVAVGFIKTLHVDTEHNIVDLLTKPLTALRFNYLLSKMG 1358
Query: 692 LYN 694
L++
Sbjct: 1359 LHH 1361
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 514 bits (1324), Expect = e-146
Identities = 278/587 (47%), Positives = 366/587 (61%), Gaps = 34/587 (5%)
Query: 117 VPQTATRHSTRSRRVPSHFHDYQL-----------ALPSSIAALNSSSKVSSTGIKHPLS 165
V T+ R R R DY L ALP S + SSS V T +PLS
Sbjct: 557 VEATSPRQRKRQIRQSVRLQDYVLYNATVSPINPHALPDSSS--QSSSMVQGTSSLYPLS 614
Query: 166 SVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLP 225
+S S H AF +++ EP + +A + K W DAM KE++AL N TW IVDLP
Sbjct: 615 DYVSDDCFSAGHKAFLAAITANDEPKHFKEAVRIKVWNDAMFKEVDALEINKTWDIVDLP 674
Query: 226 PTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLA 285
P K IG +WVY+ K+ DGSI+RYKARLV +G QVEG DY +TF+PV K TT+R LL
Sbjct: 675 PGKVAIGSQWVYKTKYNADGSIERYKARLVVQGNKQVEGEDYNETFAPVVKMTTVRTLLR 734
Query: 286 LAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQW 345
L A+ W ++Q+DV+NAFLHG LDEE+YM+LPPG P++VC L+KSLYGLKQA R W
Sbjct: 735 LVAANQWEVYQMDVNNAFLHGDLDEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCW 794
Query: 346 YNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHS 405
+ L AL G+ Q D++ + +L+YVDD+L+ GN +Q K +L
Sbjct: 795 FKKLSDALLRFGFVQGHEDYSFFSYTRNGIELRVLVYVDDLLICGNDGYMLQKFKEYLGR 854
Query: 406 QFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQ 465
F +KDLG LK+FLG+EV+R ++GI L+QRKYAL+++TD+G LGC+P TP++ HL+
Sbjct: 855 CFSMKDLGKLKYFLGIEVSRGSEGIFLSQRKYALDIITDSGNLGCRPALTPLEQNHHLAT 914
Query: 466 TGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKG 525
G LL D YRRLVGRLLYL TRP+L+Y V LSQ++ +P + H AA R++R++KG
Sbjct: 915 DDGPLLADAKPYRRLVGRLLYLLHTRPELSYSVHVLSQFMQTPREAHLAAAMRIVRFLKG 974
Query: 526 APGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSS 585
+PGQG+ +A L+L Y DSD+ CP TRRS++ Y + LG S ISW+TKKQ TVS SS
Sbjct: 975 SPGQGILLNADTELQLDVYCDSDFQTCPKTRRSLSAYVVLLGGSPISWKTKKQDTVSHSS 1034
Query: 586 SEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIEL 645
+EAEYRA++ + E++WL L + L NP FHERTKHIE
Sbjct: 1035 AEAEYRAMSVALREIKWLRKLLKEL---------------------ANPVFHERTKHIES 1073
Query: 646 DCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
DCH VR ++ G+I V T +QLADIFTK L S F + KLG+
Sbjct: 1074 DCHSVRDAVRDGIITTHHVRTNEQLADIFTKALGRSQFLYLMSKLGI 1120
Score = 65.1 bits (157), Expect = 1e-10
Identities = 28/67 (41%), Positives = 43/67 (63%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+ L+ FGS CY +R++ K R++ CVFIGY KG+ +FD++ +E L+SR V+F
Sbjct: 307 YEHLRVFGSACYVHRASRDKDKFGERSRLCVFIGYPFAQKGWKVFDMEKKEFLVSRDVVF 366
Query: 64 HETEFPF 70
E FP+
Sbjct: 367 REDVFPY 373
>At1g47650 hypothetical protein
Length = 1409
Score = 514 bits (1324), Expect = e-146
Identities = 255/469 (54%), Positives = 332/469 (70%), Gaps = 6/469 (1%)
Query: 206 MDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGL 265
+ E+ A+ +TW I LP KK +GCKWV+ +K DGS++RYKARLVAKGYTQ EGL
Sbjct: 747 VSSEIGAMETTHTWEITTLPAGKKAVGCKWVFSLKFLADGSLERYKARLVAKGYTQKEGL 806
Query: 266 DYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPR- 324
DY DTFSPVAK TT++LLL ++AS+ WFL QLDV NAFL+G L+EEIYM+LP G ++ +
Sbjct: 807 DYTDTFSPVAKMTTIKLLLKISASKKWFLKQLDVSNAFLNGELEEEIYMKLPEGYAARKG 866
Query: 325 ----PNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALL 380
PN VC L++S+YGLKQASRQW+ A+L LG+++ DHTL +K +L
Sbjct: 867 ITLPPNSVCRLKRSIYGLKQASRQWFKKFSASLLDLGFKKTHGDHTLFIKEYDGEFVIVL 926
Query: 381 LYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALE 440
+YVDD+ + S + L +F+++DLGDLK+FLGLE+AR+ GI + QRKYALE
Sbjct: 927 VYVDDIAIASTSEGAAIQLTQDLQQRFKLRDLGDLKYFLGLEIARTEAGISICQRKYALE 986
Query: 441 LLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQ 500
LL G++ CKPV PM + + +T G LLDD YRR+VG+L+YLT TR D+ + V +
Sbjct: 987 LLASTGMVNCKPVSVPMIPNVKMMKTDGDLLDDREQYRRIVGKLMYLTITRLDITFAVNK 1046
Query: 501 LSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSIN 560
L Q+ ++P H QAA RVL+Y+KG+ GQGLF+SA+A L L+ ++DSDWA CPD+RRS
Sbjct: 1047 LCQFSSAPRTSHLQAAYRVLQYIKGSVGQGLFYSASADLTLKGFADSDWASCPDSRRSTT 1106
Query: 561 GYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPL 620
G+S+F+G SLISWR+KKQ VSRSS+EAEYRALA + CE+ WL L L PV L
Sbjct: 1107 GFSMFVGDSLISWRSKKQHVVSRSSAEAEYRALALVTCELVWLHTLLMSLKASYLVPV-L 1165
Query: 621 FCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQ 669
+ D+ +AI+IA NP FHERTKHIELDCH VR KL G + LL V T DQ
Sbjct: 1166 YSDSTTAIYIATNPVFHERTKHIELDCHTVREKLDNGELKLLHVRTEDQ 1214
Score = 58.5 bits (140), Expect = 1e-08
Identities = 29/68 (42%), Positives = 41/68 (59%), Gaps = 5/68 (7%)
Query: 21 RNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLFHETEFPFKTMEHTLDPC 80
+ R K PR++ CVF+GY G KG+ L D++S V +SR+V+FHE FP +DP
Sbjct: 679 KKRHKFQPRSKACVFLGYPAGYKGYKLMDLESHTVFISRNVVFHEEVFPL-----AVDPK 733
Query: 81 ASVSLPSF 88
+ SL F
Sbjct: 734 SESSLKLF 741
>At1g59265 polyprotein, putative
Length = 1466
Score = 503 bits (1294), Expect = e-142
Identities = 292/773 (37%), Positives = 415/773 (52%), Gaps = 87/773 (11%)
Query: 7 LKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLFHET 66
L+ FG CY N+ KLD ++++CVF+GY +L +++ + +SRHV F E
Sbjct: 688 LRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDEN 747
Query: 67 EFPFKTMEHTLDPC---------------------------------------ASVSLP- 86
FPF TL P +S S P
Sbjct: 748 CFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPSAPF 807
Query: 87 --SFMGSEALD------YTPLPQGQTPDPQPQVPEVLPVPQTATRHSTR--SRRVPSHFH 136
S + S LD + P+ P P P HS++ S+ P++
Sbjct: 808 RNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNES 867
Query: 137 DYQLALPSSIAALNSSSKVSSTGIKH-----------------PLSSVLSYTNLSPKH-- 177
QLA S A +SSS S T PL+ +++ N +P +
Sbjct: 868 PSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTH 927
Query: 178 ----------------YAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLI 221
Y+ ++++ EP T QA + + WR+AM E+ A N+TW +
Sbjct: 928 SMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDL 987
Query: 222 VDLPPTKKPI-GCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTL 280
V PP+ I GC+W++ K+ DGS++RYKARLVAKGY Q GLDY +TFSPV K+T++
Sbjct: 988 VPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSI 1047
Query: 281 RLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPG-VSSPRPNQVCLLQKSLYGLK 339
R++L +A + W + QLDV+NAFL GTL +++YM PPG + RPN VC L+K+LYGLK
Sbjct: 1048 RIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLK 1107
Query: 340 QASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAV 399
QA R WY L L ++G+ D +L + + SI +L+YVDD+L+TGN +
Sbjct: 1108 QAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNT 1167
Query: 400 KSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDS 459
L +F +KD +L +FLG+E R G+ L+QR+Y L+LL ++ KPV TPM
Sbjct: 1168 LDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAP 1227
Query: 460 TLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRV 519
+ LS GT L DPT YR +VG L YL TRPD++Y V +LSQ++ PT+ H QA R+
Sbjct: 1228 SPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRI 1287
Query: 520 LRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQT 579
LRY+ G P G+F +L L AYSD+DWAG D S NGY ++LG ISW +KKQ
Sbjct: 1288 LRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQK 1347
Query: 580 TVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHER 639
V RSS+EAEYR++A E+QW+ L L +R+ P ++CDN A ++ NP FH R
Sbjct: 1348 GVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSR 1407
Query: 640 TKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
KHI +D H +R ++Q+G + ++ VST DQLAD TKPL + F F K+G+
Sbjct: 1408 MKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 501 bits (1291), Expect = e-142
Identities = 250/527 (47%), Positives = 358/527 (67%)
Query: 166 SVLSYTNLSPKHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLP 225
S +S + H AF V++ +EPTTY +A K WR+AM E+E+L N T+ IV+LP
Sbjct: 576 SKMSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIVNLP 635
Query: 226 PTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLA 285
P K+ +G KWVY++K++ DG+I+RYKARLV G Q EG+DY +TF+PVAK +T+RL L
Sbjct: 636 PGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRLFLG 695
Query: 286 LAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQW 345
+AA++ W +HQ+DV NAFLHG L EE+YM+LP G P++VC L KSLYGLKQA R W
Sbjct: 696 VAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQAPRCW 755
Query: 346 YNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHS 405
++ L +ALK G+ Q D++L +L+YVDD++++G+ + + KS+L S
Sbjct: 756 FSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDAVAQFKSYLES 815
Query: 406 QFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQ 465
F +KDLG LK+FLG+EV+R+A+G L+QRKY L+++++ GLLG +P P++ LS
Sbjct: 816 CFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQNHKLSL 875
Query: 466 TGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKG 525
+ LL D + YRRLVGRL+YL TRP+L+Y V L+Q++ +P H AA RV+RY+K
Sbjct: 876 STSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNAAIRVVRYLKS 935
Query: 526 APGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSS 585
PGQG+ S+T++L++ + DSD+A CP TRRS+ GY + LG + ISW+TKKQ T+SRSS
Sbjct: 936 NPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTKKQPTISRSS 995
Query: 586 SEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIEL 645
+EAEYRA+A + E+ WL + L + + +F D++SAI ++ NP HERTKH+E+
Sbjct: 996 AEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEV 1055
Query: 646 DCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
DCH +R + G+I V + QLADI TK L F+ KLG+
Sbjct: 1056 DCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGI 1102
Score = 79.7 bits (195), Expect = 5e-15
Identities = 34/70 (48%), Positives = 48/70 (68%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLF 63
+S L+ FGSLCYA N K R+++CVF+GY HG KG+ LFD++ ++ +SR V+F
Sbjct: 509 YSHLRVFGSLCYAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIF 568
Query: 64 HETEFPFKTM 73
ETEFP+ M
Sbjct: 569 QETEFPYSKM 578
>At1g58889 polyprotein, putative
Length = 1466
Score = 501 bits (1290), Expect = e-142
Identities = 291/773 (37%), Positives = 414/773 (52%), Gaps = 87/773 (11%)
Query: 7 LKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLFHET 66
L+ FG CY N+ KLD ++++CVF+GY +L +++ + +SRHV F E
Sbjct: 688 LRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDEN 747
Query: 67 EFPFKTMEHTLDPC---------------------------------------ASVSLP- 86
FPF TL P +S S P
Sbjct: 748 CFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPSAPF 807
Query: 87 --SFMGSEALD------YTPLPQGQTPDPQPQVPEVLPVPQTATRHSTR--SRRVPSHFH 136
S + S LD + P+ P P P HS++ S+ P++
Sbjct: 808 RNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNES 867
Query: 137 DYQLALPSSIAALNSSSKVSSTGIKH-----------------PLSSVLSYTNLSPKH-- 177
QLA S A +SSS S T PL+ +++ N +P +
Sbjct: 868 PSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPLNTH 927
Query: 178 ----------------YAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLI 221
Y+ ++++ EP T QA + + WR+AM E+ A N+TW +
Sbjct: 928 SMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDL 987
Query: 222 VDLPPTKKPI-GCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTL 280
V PP+ I GC+W++ K+ DGS++RYKAR VAKGY Q GLDY +TFSPV K+T++
Sbjct: 988 VPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSPVIKSTSI 1047
Query: 281 RLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPG-VSSPRPNQVCLLQKSLYGLK 339
R++L +A + W + QLDV+NAFL GTL +++YM PPG + RPN VC L+K+LYGLK
Sbjct: 1048 RIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLK 1107
Query: 340 QASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAV 399
QA R WY L L ++G+ D +L + + SI +L+YVDD+L+TGN +
Sbjct: 1108 QAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNT 1167
Query: 400 KSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDS 459
L +F +KD +L +FLG+E R G+ L+QR+Y L+LL ++ KPV TPM
Sbjct: 1168 LDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAP 1227
Query: 460 TLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRV 519
+ LS GT L DPT YR +VG L YL TRPD++Y V +LSQ++ PT+ H QA R+
Sbjct: 1228 SPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRI 1287
Query: 520 LRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQT 579
LRY+ G P G+F +L L AYSD+DWAG D S NGY ++LG ISW +KKQ
Sbjct: 1288 LRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQK 1347
Query: 580 TVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHER 639
V RSS+EAEYR++A E+QW+ L L +R+ P ++CDN A ++ NP FH R
Sbjct: 1348 GVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSR 1407
Query: 640 TKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
KHI +D H +R ++Q+G + ++ VST DQLAD TKPL + F F K+G+
Sbjct: 1408 MKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 501 bits (1289), Expect = e-142
Identities = 296/777 (38%), Positives = 406/777 (52%), Gaps = 91/777 (11%)
Query: 7 LKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFDIKSREVLLSRHVLFHET 66
LK FG CY NR KL+ ++++C F+GY +L I + + SRHV F E
Sbjct: 667 LKVFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDER 726
Query: 67 EFPFKTMEH---TLDPCASVSLPSFMGSEALDYTPL---------PQGQTPDPQPQVPEV 114
FPF T T S S P++ L TPL P T P P
Sbjct: 727 CFPFSTTNFGVSTSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSP 786
Query: 115 LPVPQTATRH---------STRSRRVPSHFHDYQLALPSSIAALNSSSKV---------- 155
L Q ++ + S+ PSH A P NS+S +
Sbjct: 787 LCTTQVSSSNLPSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPS 846
Query: 156 ----------------------SSTGIKHPLSSVLSYTNLSP------------------ 175
ST I P S S T+ P
Sbjct: 847 PNSPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAP 906
Query: 176 ------------------KHYAFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNN 217
+ Y++ T++++ EP T QA + WR AM E+ A N+
Sbjct: 907 VNTHSMATRAKDGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNH 966
Query: 218 TW-LIVDLPPTKKPIGCKWVYRVKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAK 276
TW L+ PP+ +GC+W++ K DGS++RYKARLVAKGY Q GLDY +TFSPV K
Sbjct: 967 TWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIK 1026
Query: 277 TTTLRLLLALAASQGWFLHQLDVDNAFLHGTLDEEIYMRLPPG-VSSPRPNQVCLLQKSL 335
+T++R++L +A + W + QLDV+NAFL GTL +E+YM PPG V RP+ VC L+K++
Sbjct: 1027 STSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAI 1086
Query: 336 YGLKQASRQWYNTLVAALKSLGYEQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEE 395
YGLKQA R WY L L ++G+ D +L + + SI +L+YVDD+L+TGN
Sbjct: 1087 YGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVL 1146
Query: 396 IQAVKSFLHSQFRIKDLGDLKFFLGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRT 455
++ L +F +K+ DL +FLG+E R +G+ L+QR+Y L+LL +L KPV T
Sbjct: 1147 LKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVAT 1206
Query: 456 PMDSTLHLSQTGGTLLDDPTSYRRLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQA 515
PM ++ L+ GT L DPT YR +VG L YL TRPDL+Y V +LSQY+ PTD H A
Sbjct: 1207 PMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNA 1266
Query: 516 ATRVLRYVKGAPGQGLFFSATASLKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRT 575
RVLRY+ G P G+F +L L AYSD+DWAG D S NGY ++LG ISW +
Sbjct: 1267 LKRVLRYLAGTPDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSS 1326
Query: 576 KKQTTVSRSSSEAEYRALAAIVCEVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPT 635
KKQ V RSS+EAEYR++A E+QW+ L L +++ P ++CDN A ++ NP
Sbjct: 1327 KKQKGVVRSSTEAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPV 1386
Query: 636 FHERTKHIELDCHVVRTKLQAGLIHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGL 692
FH R KHI LD H +R ++Q+G + ++ VST DQLAD TKPL F F K+G+
Sbjct: 1387 FHSRMKHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGV 1443
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 498 bits (1283), Expect = e-141
Identities = 287/696 (41%), Positives = 393/696 (56%), Gaps = 55/696 (7%)
Query: 4 FSFLKTFGSLCYASTLNRNRTKLDPRAQKCVFIGYRHGTKGFLLFD-IKSREVLLSRHVL 62
+ L+TFG LCYA R++ K R++KC+FIGY H T D I +
Sbjct: 452 YDMLRTFGCLCYAHIRPRDKDKFASRSRKCIFIGYPHETATPNTHDSIDPTSTSSDENNT 511
Query: 63 FHETEFPFKTMEHTLDPCASVSLPSFMGSEALDYTPLPQGQTPDPQPQVPEVLPVPQTAT 122
E P H+ +S+S P + ++ ++ P +PE+L
Sbjct: 512 PPEPVTPQAEQPHSP---SSISSPHIVHNKGSVHSRHLNEDHDSSSPGLPELLG------ 562
Query: 123 RHSTRSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSP----KHY 178
R + P + DY ++SS SS G LS +N+SP H
Sbjct: 563 -KGHRPKHPPVYLKDYVAH------KVHSSPHTSSPG--------LSDSNVSPTVSANHI 607
Query: 179 AFTTNVSSTVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYR 238
AF + + E + KEW DAM KE+EAL N+TW + DLP KK I KWVY+
Sbjct: 608 AFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALEANHTWDVTDLPHGKKAISSKWVYK 667
Query: 239 VKHKQDGSIDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLD 298
+K DG+++R+KARLV G Q EG+D+ +TF+PVAK TT+RLLLA+AA++ W + Q+D
Sbjct: 668 LKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPVAKMTTVRLLLAVAAAKDWDVFQMD 727
Query: 299 VDNAFLHGTLDEEIYMRLPPGVSSPRPNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGY 358
V NAFLHG L++E SLYGLKQA R W+ L AL+ LG+
Sbjct: 728 VHNAFLHGDLEQE----------------------SLYGLKQAPRCWFAKLSTALRKLGF 765
Query: 359 EQCPHDHTLLLKRTSTSITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFF 418
Q D++L ++ L+YVDD ++ GN+ + I K LH F +KDLG LK+F
Sbjct: 766 TQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLKAIDHFKEHLHKCFHMKDLGKLKYF 825
Query: 419 LGLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYR 478
LGLEV+R A G CL+Q+KYAL+++ +AGLLG KP PM+ L + D+P YR
Sbjct: 826 LGLEVSRGADGFCLSQQKYALDIINEAGLLGYKPSAVPMELHHKLGSISSPVFDNPAQYR 885
Query: 479 RLVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATAS 538
RLV R +YLT TRPDL+Y V LSQ++ +P + H A R++RY+KG+P QG+ + +
Sbjct: 886 RLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWHATLRLVRYLKGSPDQGILLRSDRA 945
Query: 539 LKLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVC 598
L L AY DSD+ CP TRRS++ Y ++LG + ISW+TKKQ TVS SS+EAEYRA+A +
Sbjct: 946 LSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQDTVSSSSAEAEYRAMAYTLK 1005
Query: 599 EVQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGL 658
E++WL L L + P+ LFCD+Q+AIHIA NP FHERTKHIE DCH VR + +
Sbjct: 1006 EIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHERTKHIEKDCHQVRDAVTDKV 1065
Query: 659 IHLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
I +ST D+ TK LP F + LG N
Sbjct: 1066 ISTPHIST----TDLLTKALPRPTFERLLSTLGTCN 1097
>At2g15920 putative retroelement pol polyprotein
Length = 1100
Score = 498 bits (1282), Expect = e-141
Identities = 267/575 (46%), Positives = 363/575 (62%), Gaps = 41/575 (7%)
Query: 127 RSRRVPSHFHDYQLALPSSIAALNSSSKVSSTGIKHPLSSVLSYTNLSPKHYAFTTNVSS 186
R R+ +H DY K + + K+P+SS +SY+ +SP H + N++
Sbjct: 550 RVRKPSTHLSDYVC-------------KNTQSDQKYPISSTISYSQISPSHMCYIDNITK 596
Query: 187 TVEPTTYTQAAQCKEWRDAMDKELEALSRNNTWLIVDLPPTKKPIGCKWVYRVKHKQDGS 246
+ T + +A KEW +A+D E+ A+ N W I LP KK +GCKWV+ +K DGS
Sbjct: 597 ILISTNFPEAQGTKEWCEAVDVEIGAMESTNKWEITTLPKDKKAVGCKWVFTLKILADGS 656
Query: 247 IDRYKARLVAKGYTQVEGLDYLDTFSPVAKTTTLRLLLALAASQGWFLHQLDVDNAFLHG 306
++RYKARLVAKGYTQ EGLDY +TFSPVAK TT++LLL + AS+ WFL QLDV N+FL+G
Sbjct: 657 LERYKARLVAKGYTQKEGLDYTNTFSPVAKMTTIKLLLKIYASKKWFLKQLDVSNSFLNG 716
Query: 307 TLDEEIYMRLPPGVSSPR-----PNQVCLLQKSLYGLKQASRQWYNTLVAALKSLGYEQC 361
L+EEIYM+LP G + + N VC L++S+YGLKQASRQW+ A+L SLG+ +
Sbjct: 717 ELEEEIYMKLPEGSAERKGIILPSNAVCRLKRSIYGLKQASRQWFKKFSASLLSLGFFKI 776
Query: 362 PHDHTLLLKRTSTS--ITALLLYVDDVLLTGNSFEEIQAVKSFLHSQFRIKDLGDLKFFL 419
DHTL L + +L+YVDD+++ + + LH+ F+++DLGDLK+FL
Sbjct: 777 HGDHTLFLIDCGGEYVVVLVLVYVDDIVIASTNED--------LHNLFKLRDLGDLKYFL 828
Query: 420 GLEVARSAKGICLTQRKYALELLTDAGLLGCKPVRTPMDSTLHLSQTGGTLLDDPTSYRR 479
LE++R+ GI L RK A ELL G++ CKPV PM + + L + G LL+D YRR
Sbjct: 829 CLEISRTEVGISLCHRKNAWELLASIGMINCKPVSVPMITNVKLMKADGELLEDREQYRR 888
Query: 480 LVGRLLYLTTTRPDLNYVVQQLSQYLASPTDLHQQAATRVLRYVKGAPGQGLFFSATASL 539
+VG+L+ LT TRPD+ H QAA RVL+Y+KG GQGLF+SA++ L
Sbjct: 889 IVGKLMDLTITRPDITL------------RTTHLQAAHRVLQYIKGIVGQGLFYSASSDL 936
Query: 540 KLQAYSDSDWAGCPDTRRSINGYSIFLGSSLISWRTKKQTTVSRSSSEAEYRALAAIVCE 599
L+ ++DSDW C D+RRS G+++F+G SLIS R+KKQ VSRSS+EAEYRALA CE
Sbjct: 937 TLKGFADSDWVSCADSRRSTTGFTMFVGDSLISLRSKKQRVVSRSSAEAEYRALALATCE 996
Query: 600 VQWLSYLFQFLHLRVPFPVPLFCDNQSAIHIAQNPTFHERTKHIELDCHVVRTKLQAGLI 659
+ L L L P+ LF ++ +AI+IA NP FHERTKH E+DC+ VR K+ G +
Sbjct: 997 LVSLHTLLVSLTAATSVPI-LFSNSTTAIYIAINPVFHERTKHTEIDCNTVREKVDDGEL 1055
Query: 660 HLLPVSTRDQLADIFTKPLPPSLFPTFVLKLGLYN 694
LL V T DQ+ADI TKPL P F K+ + N
Sbjct: 1056 KLLHVRTEDQVADIMTKPLSPHQFEHLKFKMSILN 1090
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,525,608
Number of Sequences: 26719
Number of extensions: 661678
Number of successful extensions: 2166
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 107
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1747
Number of HSP's gapped (non-prelim): 222
length of query: 694
length of database: 11,318,596
effective HSP length: 106
effective length of query: 588
effective length of database: 8,486,382
effective search space: 4989992616
effective search space used: 4989992616
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0260.2


