

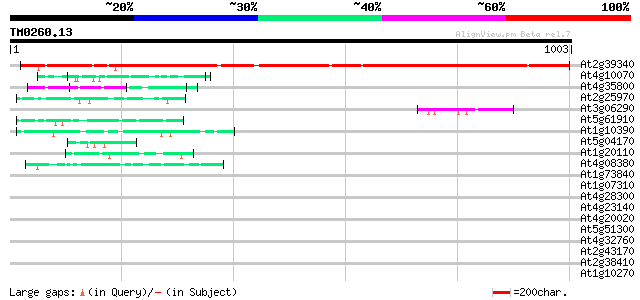
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.13
(1003 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g39340 unknown protein 1018 0.0
At4g10070 putative DNA-directed RNA polymerase 65 2e-10
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 59 2e-08
At2g25970 pseudogene 57 5e-08
At3g06290 hypothetical protein 54 6e-07
At5g61910 unknown protein 47 5e-05
At1g10390 unknown protein 46 9e-05
At5g04170 EF - hand Calcium binding protein - like 45 2e-04
At1g20110 unknown protein 45 3e-04
At4g08380 extensin-like protein 44 5e-04
At1g73840 proline-rich like protein precursor 40 0.005
At1g07310 unknown protein 39 0.011
At4g28300 predicted proline-rich protein 39 0.019
At4g23140 receptor-like protein kinase 5 (RLK5) 39 0.019
At4g20020 DAG-like protein 38 0.025
At5g51300 unknown protein 38 0.033
At4g32760 unknown protein 38 0.033
At2g43170 unknown protein 38 0.033
At2g38410 unknown protein 38 0.033
At1g10270 unknown protein 38 0.033
>At2g39340 unknown protein
Length = 989
Score = 1018 bits (2632), Expect = 0.0
Identities = 555/1013 (54%), Positives = 693/1013 (67%), Gaps = 58/1013 (5%)
Query: 20 ENRH-FDPGQNQPPSYAPTTPGSQAVSWAVQS----STANGGYSNPTYQYDQHPQPPGRS 74
+NR+ D Q Q SY +T GS++ W S + NG YSN Y HPQP G +
Sbjct: 2 QNRYGVDGSQTQKYSYQYST-GSESAPWTGHSVENQAVENGNYSNSNYY---HPQPTGPA 57
Query: 75 IQDGQNVSSVAGN--SSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPN 132
+ Q + + SS GTANV QDY+ YT Y +SS+P+ Y + GY+ YY+ YQQQP+
Sbjct: 58 TGNVQEIPNTVSFTISSTSGTANVAQDYSGYTPYQTSSDPHNYSNTGYSNYYSGYQQQPS 117
Query: 133 QTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQN----- 187
Q+Y QPVGAYQNTGAP QP+SSFQN GSYAG+ +YS TYYNPA DYQT G YQ+
Sbjct: 118 QSYPQPVGAYQNTGAP-QPLSSFQNPGSYAGTPSYSGTYYNPA--DYQTAGGYQSTNYNN 174
Query: 188 -------STGYANQAPPWNNGSYSSYASHPHTNYTPDS---NSSGAATTSVQYQQQQQNP 237
ST Y+NQ P N G+Y+ Y S+P+ NYTPD+ +SS ATT + QQ
Sbjct: 175 QTAGSYPSTNYSNQTPASNQGNYTDYTSNPYQNYTPDAANTHSSTIATTPPVHYQQNYQQ 234
Query: 238 WADYYNQTEVSCAPGTENLSVTSSSTFGG--PIPAATSGYATPNSQPQQSYPPYWRQDSS 295
W +YY+QTEV CAPGTE LS ++S + P+P TS NSQP SY WR ++
Sbjct: 235 WTEYYSQTEVPCAPGTEKLSTPTTSAYSQSFPVPGVTSEMPASNSQPAPSYVQPWRPETD 294
Query: 296 ASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDN-FQDQQKT 354
+S PS QP A S +D YW H A + Q H P Q NYQSPL+ K Y+ FQ Q+
Sbjct: 295 SSHPPSQQPGAAVSTSNDTYWMHQAPSLQAHHPVPPQNNYQSPLETKPLYETPFQGHQRA 354
Query: 355 VSSQGTNLYLP-PPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQP 413
Q N PL P Q+AP +D++RVSK+QIPTNPRIASNL G
Sbjct: 355 TYPQEMNSQSSFHQAPLG------YRQPTQTAPLVDSQRVSKVQIPTNPRIASNLPSGFT 408
Query: 414 KIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCK 473
K++KDS+ + A PAY++VS+PK D ++ PG FPKSLRG+VERA RCK
Sbjct: 409 KMDKDSTAASAAQAPAYVSVSMPKP------KDHTTAMSDPGTFPKSLRGFVERAFARCK 462
Query: 474 DDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPMPDSDVVNKDSV---LSSTHKRS 530
DDK+ ++C+ +++++ KA D TL TR+WD EPL + ++V N +S LSS +S
Sbjct: 463 DDKEKESCEVALRKIVKKAKEDNTLYTRDWDTEPLSTVTTTNVTNSESSSAQLSSLQNKS 522
Query: 531 P-RRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKVVMENKESKEDDLRNTKF 589
P RR KSRWEPL E KP P S + VK+ W ++ K ++ ES + T F
Sbjct: 523 PTRRPKSRWEPLVEGKPFVKPASTFSSAVKFGVWNHQNENNK---KSSESFQKVDAATGF 579
Query: 590 SPFI--QRTSSKAPQRPFKKQRLTDASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPE 647
P Q ++ K+ QRP K+QR + + + +ASS SDK+ LT YYS+AMA + + E
Sbjct: 580 KPTYSGQNSAKKSFQRPVKRQRFSGGAATAIDDEASSDSDKD--LTPYYSSAMALAGSAE 637
Query: 648 EKKRRENRSKRFDLGQGHRTENNHFRRKNAGAGNLYSRRASALVLSKSFEDGVSKAVEDI 707
EKKRR++RSKRF+ QGH N+ + KNA GNL+SRRA+AL LSK F++ S+AVEDI
Sbjct: 638 EKKRRDSRSKRFEKIQGHSRGNDLTKPKNANVGNLHSRRATALRLSKVFDESGSRAVEDI 697
Query: 708 DWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQKNYLYKCDQLKSI 767
DWDALTVKG CQEIEKRYLRLTSAPDPATVRPE+VLEKAL+MVQ+SQKNYL+KCDQLKSI
Sbjct: 698 DWDALTVKGTCQEIEKRYLRLTSAPDPATVRPEDVLEKALIMVQDSQKNYLFKCDQLKSI 757
Query: 768 RQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYN 827
RQDLTVQRI N LT KVYETHAR A+EAGDL EYNQC SQL LYA+G+EG +EFAAY+
Sbjct: 758 RQDLTVQRIHNHLTAKVYETHARLALEAGDLPEYNQCLSQLKTLYAEGVEGCSLEFAAYS 817
Query: 828 LLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLN 887
LL + +HSNNNR+LLSSM+RLS+E KKDEAV+HAL+VRAAVTSGNYV FFRLYK APN+N
Sbjct: 818 LLYITLHSNNNRELLSSMSRLSEEDKKDEAVRHALSVRAAVTSGNYVMFFRLYKTAPNMN 877
Query: 888 TCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKDTEAFEECL 947
+CLMDLYVEKMRYKAV+ M RS RPT+PVSYI VLGF + A+ G+DEK+T+ E+CL
Sbjct: 878 SCLMDLYVEKMRYKAVNFMSRSCRPTIPVSYIVQVLGF--TGAASEGTDEKETDGMEDCL 935
Query: 948 EWLKAHGASIMTDNNKDILVDTKVSSSSLFMPEPEDAVAHGDANLAVNDFLAK 1000
EWLK HGA+I+TD+N D+L+DTK +S+SLFMPEPEDAVAHGD NL VNDF +
Sbjct: 936 EWLKTHGANIITDSNGDMLLDTKATSTSLFMPEPEDAVAHGDRNLDVNDFFTR 988
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 65.1 bits (157), Expect = 2e-10
Identities = 70/256 (27%), Positives = 99/256 (38%), Gaps = 45/256 (17%)
Query: 103 YTSYPSSSNPYGYG----SAGYTGYYNNYQQQPN-QTYSQPVGAYQNTGAPYQPISSFQN 157
Y S P PYG G +GYY QQP + Y G Q P QP ++
Sbjct: 523 YDSNPPMQPPYGGSYPPAGGGQSGYYQ--MQQPGVRPYGMQQGPVQQGYGPPQPAAA--- 577
Query: 158 TGSYAGSANYSSTYYNPAPADYQTTGV--YQNSTGYANQAPPWNNGSYSSYASHPHTNYT 215
+ +G Y PA Y +T + Q GY + P +Y SY+S P ++
Sbjct: 578 --ASSGDVPYQGA--TPAAPSYGSTNMAPQQQQYGYTSSDGPVQQQTYPSYSSAPPSD-- 631
Query: 216 PDSNSSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGY 275
+N + T YQQQ P + Y+QT A +++ +GG + A T GY
Sbjct: 632 AYNNGTQTPATGPAYQQQSVQPASSTYDQTGAQQA---------AAAGYGGQV-APTGGY 681
Query: 276 ATPNSQPQQSYPPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNP--IQP 333
P SQ P Y Q + + A P+ GY A + ++ T P
Sbjct: 682 TYPTSQ-----PAYGSQAAYSQAAPT----------QTGYEQQPATQAAVYATAPGTAPV 726
Query: 334 NYQSPLDLKSSYDNFQ 349
QSP + YD Q
Sbjct: 727 KTQSPQSAYAQYDASQ 742
Score = 55.8 bits (133), Expect = 1e-07
Identities = 83/340 (24%), Positives = 121/340 (35%), Gaps = 70/340 (20%)
Query: 51 STANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSS 110
S+ +GGY+ P Y+ PQ PG Q G + P Y+ + P SS
Sbjct: 372 SSYSGGYNQPAYR----PQGPGGPPQWGSR------------GPHAPHPYDYHPRGPYSS 415
Query: 111 NPYGYGSAGY-----------TGYYNNYQQQPNQTYSQPVGAYQNTGA--------PYQP 151
Y S G+ GY ++ Q+P YS P Y GA P P
Sbjct: 416 QGSYYNSPGFGGYPPQHMPPRGGYGTDWDQRP--PYSGPYNYYGRQGAQSAGPVPPPSGP 473
Query: 152 ISSFQNTG------SYAGSANYSSTYYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSS 205
+ S G SY ++ Y + AP Y TG YQ + G + P +++ +
Sbjct: 474 VPSPAFGGPPLSQVSYGYGQSHGPEYGHAAP--YSQTG-YQQTYGQTYEQPKYDS---NP 527
Query: 206 YASHPHTNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSST-F 264
P+ P + G + Q QQ P+ + P + +S +
Sbjct: 528 PMQPPYGGSYPP--AGGGQSGYYQMQQPGVRPYGMQQGPVQQGYGPPQPAAAASSGDVPY 585
Query: 265 GGPIPAATSGYATPNSQPQQSYPPYWRQDS--SASAVPSFQPAAINSGDHDGYWNHGAQ- 321
G PAA S Y + N PQQ Y D PS+ A + +N+G Q
Sbjct: 586 QGATPAAPS-YGSTNMAPQQQQYGYTSSDGPVQQQTYPSYSSAPPSDA-----YNNGTQT 639
Query: 322 --TSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQG 359
T +Q +QP S+YD QQ + G
Sbjct: 640 PATGPAYQQQSVQP-------ASSTYDQTGAQQAAAAGYG 672
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 58.5 bits (140), Expect = 2e-08
Identities = 74/289 (25%), Positives = 109/289 (37%), Gaps = 35/289 (12%)
Query: 32 PSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVAGNSSNL 91
P Y+PT+PG S ++ ++PTY P PG S S +S
Sbjct: 1550 PGYSPTSPGYSPTSPGYSPTSPGYSPTSPTYS----PSSPGYS-----PTSPAYSPTSPS 1600
Query: 92 GTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQP 151
+ P SY +S Y S Y+ +Y + YS AY T Y P
Sbjct: 1601 YSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSY-SPTSPAYSPTSPAYSPTSPAYSP 1659
Query: 152 IS-SFQNTGSYAGSANYSST--YYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYAS 208
S S+ T S +YS T Y+P Y T S Y+ +P ++ S +
Sbjct: 1660 TSPSYSPT-----SPSYSPTSPSYSPTSPSYSPT-----SPSYSPTSPAYSPTSPGYSPT 1709
Query: 209 HPHTNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPI 268
P +Y+P S S G + S Q + +P ++ +P LS S + P
Sbjct: 1710 SP--SYSPTSPSYGPTSPSYNPQSAKYSP--------SIAYSPSNARLSPASPYSPTSPN 1759
Query: 269 PAATSGYATPNSQPQQSYPPYWRQDS--SASAVPSFQPAAINSGDHDGY 315
+ TS +P S P + S S+ A P + P+A S GY
Sbjct: 1760 YSPTSPSYSPTSPSYSPSSPTYSPSSPYSSGASPDYSPSAGYSPTLPGY 1808
Score = 52.8 bits (125), Expect = 1e-06
Identities = 60/231 (25%), Positives = 86/231 (36%), Gaps = 11/231 (4%)
Query: 107 PSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSAN 166
PSSS Y S GY+ Y + YS Y T Y P S + S A S
Sbjct: 1539 PSSSPGYSPSSPGYSPTSPGYSPT-SPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSP- 1596
Query: 167 YSSTYYNPAPADYQTTGVYQNSTG--YANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAA 224
+S Y+P Y T + T Y+ +P ++ S S + P Y+P S +
Sbjct: 1597 -TSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPA--YSPTSPAYSPT 1653
Query: 225 TTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQ 284
+ + +P + Y+ T S +P + + S TS S P A S + S
Sbjct: 1654 SPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSY--SPTSPAYSPTSPGYSPTSP 1711
Query: 285 SYPPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNY 335
SY P S PS+ P + Y A+ S +P PNY
Sbjct: 1712 SYSP--TSPSYGPTSPSYNPQSAKYSPSIAYSPSNARLSPASPYSPTSPNY 1760
Score = 44.3 bits (103), Expect = 4e-04
Identities = 46/182 (25%), Positives = 75/182 (40%), Gaps = 14/182 (7%)
Query: 32 PSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRS-IQDGQNVSSVAGNSSN 90
P+Y+PT+P S + ++ + ++P+Y P P S + +S A + ++
Sbjct: 1648 PAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYS----PTSPSYSPTSPSYSPTSPAYSPTS 1703
Query: 91 LGTANVPQDYNAYT-SYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPY 149
G + Y+ + SY +S Y SA Y+ + P+ P Y T Y
Sbjct: 1704 PGYSPTSPSYSPTSPSYGPTSPSYNPQSAKYSP---SIAYSPSNARLSPASPYSPTSPNY 1760
Query: 150 QPIS-SFQNTG-SYAGSANYSSTYYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYA 207
P S S+ T SY+ S S TY +P + Y S GY+ P ++ S Y
Sbjct: 1761 SPTSPSYSPTSPSYSPS---SPTYSPSSPYSSGASPDYSPSAGYSPTLPGYSPSSTGQYT 1817
Query: 208 SH 209
H
Sbjct: 1818 PH 1819
Score = 35.8 bits (81), Expect = 0.12
Identities = 38/161 (23%), Positives = 66/161 (40%), Gaps = 38/161 (23%)
Query: 32 PSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVAGNSSNL 91
P+Y+PT+PG S + ++ + G ++P+Y + Q + S+A + SN
Sbjct: 1697 PAYSPTSPGYSPTSPSYSPTSPSYGPTSPSY-----------NPQSAKYSPSIAYSPSN- 1744
Query: 92 GTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQP 151
+ S ++PY S Y+ + +YS +Y + Y P
Sbjct: 1745 -------------ARLSPASPYSPTSPNYS--------PTSPSYSPTSPSYSPSSPTYSP 1783
Query: 152 ISSFQNTGS--YAGSANYSSTYYNPAPADYQTTGVYQNSTG 190
S + + S Y+ SA YS T +P+ +TG Y G
Sbjct: 1784 SSPYSSGASPDYSPSAGYSPTLPGYSPS---STGQYTPHEG 1821
>At2g25970 pseudogene
Length = 632
Score = 57.0 bits (136), Expect = 5e-08
Identities = 90/325 (27%), Positives = 116/325 (35%), Gaps = 72/325 (22%)
Query: 13 PAQPHQFENRHFDPGQNQ-PPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPP 71
PAQP + PG PP Y + GS + GY YDQ PP
Sbjct: 337 PAQPGY--GGYMQPGAYPGPPQYGQSPYGSYP-------QQTSAGY------YDQSSVPP 381
Query: 72 GRSIQDGQNV---SSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGY----Y 124
+ G+ + S+ G++ P D Y Y +S GYG AG GY Y
Sbjct: 382 SQQSAQGEYDYYGQQQSQQPSSGGSSAPPTDTTGYNYYQHAS---GYGQAGQ-GYQQDGY 437
Query: 125 NNYQQQPNQTYSQPVG-----AYQNTGAPYQPISSFQN---TGSYAGSANYSSTYYNPAP 176
Y Y Q G Y +T P Q + Q + + +G A Y +T P P
Sbjct: 438 GAYNASQQSGYGQAAGYDQQGGYGSTTNPSQEEDASQAAPPSSAQSGQAGYGTTGQQP-P 496
Query: 177 ADYQTTGVYQNSTGYANQ-APPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQQQQQ 235
A Q STG A APP + YSS P Y + A+ Y Q QQ
Sbjct: 497 A--------QGSTGQAGYGAPPTSQAGYSS---QPAAAYNSGYGAPPPASKPPTYGQSQQ 545
Query: 236 NPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNS----QPQQSYPPY-- 289
+P APG S S S + P A SGY P + Q Q Y Y
Sbjct: 546 SPG-----------APG----SYGSQSGYAQP---AASGYGQPPAYGYGQAPQGYGSYGG 587
Query: 290 WRQDSSASAVPSFQPAAINSGDHDG 314
+ Q ++ S A +G G
Sbjct: 588 YTQPAAGGGYSSDGSAGATAGGGGG 612
Score = 36.2 bits (82), Expect = 0.096
Identities = 42/160 (26%), Positives = 53/160 (32%), Gaps = 17/160 (10%)
Query: 25 DPGQNQPPSYAPTTP---GSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNV 81
D Q PPS A + G+ Q ST GY P + P + G
Sbjct: 471 DASQAAPPSSAQSGQAGYGTTGQQPPAQGSTGQAGYGAPPTSQAGYSSQPAAAYNSGYGA 530
Query: 82 SSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGA 141
A G + Q A SY S S GY +GY Q P Y Q
Sbjct: 531 PPPASKPPTYGQSQ--QSPGAPGSYGSQS---GYAQPAASGY----GQPPAYGYGQAPQG 581
Query: 142 YQNTGAPYQP-----ISSFQNTGSYAGSANYSSTYYNPAP 176
Y + G QP SS + G+ AG + + AP
Sbjct: 582 YGSYGGYTQPAAGGGYSSDGSAGATAGGGGGTPASQSAAP 621
Score = 35.0 bits (79), Expect = 0.21
Identities = 52/238 (21%), Positives = 76/238 (31%), Gaps = 39/238 (16%)
Query: 171 YYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQY 230
Y P+ + G GY P Y P+ +Y +++ +SV
Sbjct: 322 YQARPPSSWAPPGGPPAQPGYGGYMQPGAYPGPPQYGQSPYGSYPQQTSAGYYDQSSVPP 381
Query: 231 QQQQQNPWADYYNQTEV-------SCAPGTENLSVTSSSTFGGPIPAA----TSGYATPN 279
QQ DYY Q + S AP T+ G A GY N
Sbjct: 382 SQQSAQGEYDYYGQQQSQQPSSGGSSAPPTDTTGYNYYQHASGYGQAGQGYQQDGYGAYN 441
Query: 280 SQPQQSY----------------PPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTS 323
+ Q Y P +D+S +A PS + SG GY G Q
Sbjct: 442 ASQQSGYGQAAGYDQQGGYGSTTNPSQEEDASQAAPPS----SAQSG-QAGYGTTGQQPP 496
Query: 324 QIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTP 381
T Q Y +P ++ Y + + ++ + PPP PP Q +P
Sbjct: 497 AQGSTG--QAGYGAPPTSQAGYSS-----QPAAAYNSGYGAPPPASKPPTYGQSQQSP 547
>At3g06290 hypothetical protein
Length = 1713
Score = 53.5 bits (127), Expect = 6e-07
Identities = 52/207 (25%), Positives = 91/207 (43%), Gaps = 38/207 (18%)
Query: 729 TSAPDPATVRPEEVLEKAL-----LMVQNSQKNYL----YKCDQLKSIRQDLTVQRIRNQ 779
T+ + +RP +L+ + L+ + +N+L + D++++IR DL +Q I NQ
Sbjct: 529 TAEREAILIRPMPILQNTMEYLLSLLDRPYNENFLGMYNFLWDRMRAIRMDLRMQHIFNQ 588
Query: 780 LTVKVYETHARFAMEA-GDLSEY-------------------NQCQSQLTALYAD----G 815
+ + E R + A +L EY N+ +L +Y D G
Sbjct: 589 EAITLLEQMIRLHIIAMHELCEYTKGEGFSEGFDAHLNIEQMNKTSVELFQMYDDHRKKG 648
Query: 816 IE-GSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYV 874
I + EF Y L + + L + ++ E ++ V A V A +GN++
Sbjct: 649 ITVPTEKEFRGYYALLKL----DKHQLSLDLANMTPEIRQTSEVLFARNVARACRTGNFI 704
Query: 875 AFFRLYKEAPNLNTCLMDLYVEKMRYK 901
AFFRL ++A L CLM + K+R +
Sbjct: 705 AFFRLARKASYLQACLMHAHFSKVRLR 731
>At5g61910 unknown protein
Length = 738
Score = 47.0 bits (110), Expect = 5e-05
Identities = 77/319 (24%), Positives = 120/319 (37%), Gaps = 51/319 (15%)
Query: 13 PAQPHQFENRHFDPGQNQPPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPG 72
PA P E F + +Y T GS+ S +S ANG + Y+ Q G
Sbjct: 340 PATPS--EKDQFAVPYSDNKNYPSTLSGSEHPS----ASAANGSVYRSEF-YNSASQKEG 392
Query: 73 RSIQDGQ---------NVSSVAGNSSNLG----TANVPQDYNAYTSYPSSSNPYGYGS-- 117
+ Q + S+V+ + ++ +V Q + YP+ + +G S
Sbjct: 393 EASQQHEIPAGTYHHPEASTVSNTTKSMQPDMQAVSVAQSHTETAGYPTPA--HGEASQP 450
Query: 118 -AGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPA- 175
AG GY + QP N QP + ++T SYAG+ +YS Y A
Sbjct: 451 PAGAIGYTH-----------QPQSVAGNYSTHSQPGNVEESTQSYAGTDSYSQQQYYAAM 499
Query: 176 -PADYQTTGVY---QNSTGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQ 231
P G Y + GY+ Q + + + Y+ PH T S A T Y
Sbjct: 500 GPTTQLHAGGYIQKPHEIGYSQQP----HDAATGYSQQPHDAATGYSQQPHDAATG--YS 553
Query: 232 QQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPYWR 291
QQ Y QT + T+ ++ P AAT GY + QP + + +
Sbjct: 554 QQPHAASTGYSQQTYAAATGYTQQPHAAAAGYTQQPHAAAT-GY---SQQPHAAATAHAQ 609
Query: 292 QDSSASAVPSFQPAAINSG 310
Q +A+ + Q A+ +G
Sbjct: 610 QPYAAATAHAQQLHAVATG 628
>At1g10390 unknown protein
Length = 1041
Score = 46.2 bits (108), Expect = 9e-05
Identities = 100/421 (23%), Positives = 156/421 (36%), Gaps = 69/421 (16%)
Query: 13 PAQPHQFENRHFDPGQNQPPSYAPTTPGSQAVS---WAVQSST-ANGGYSNPTYQYDQHP 68
P+QP+ F P Q S PT P S + S +A Q+ T A+ + T + P
Sbjct: 415 PSQPNPFSP---SPAFGQT-SANPTNPFSSSTSTNPFAPQTPTIASSSFGTATSNFGSSP 470
Query: 69 QPPGRSIQ----DGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSA---GYT 121
S +SV G+SS GT + + ++ S+ +GSA G T
Sbjct: 471 FGVTSSSNLFGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFGSSSSIFGSAPGQGAT 530
Query: 122 GYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQT 181
+ N SQP + +T + Q S+F TGS G SS APA Q
Sbjct: 531 PAFGN---------SQPSTLFNSTPSTGQTGSAFGQTGSAFGQFGQSS-----APAFGQN 576
Query: 182 TGVYQNSTGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQQ--QQQNPWA 239
+ + STG+ N +SS ++ ++ +P + A T Q Q Q N +
Sbjct: 577 SIFNKPSTGFGNM--------FSSSSTLTTSSSSPFGQTMPAGVTPFQSAQPGQASNGFG 628
Query: 240 -DYYNQTEVSCAPGTE-NLSVTSSSTFG-GPIP---------AATSGYATPNSQPQQSY- 286
+ + Q + + G L + FG P P A T+ + T + PQ S
Sbjct: 629 FNNFGQNQAANTTGNAGGLGIFGQGNFGQSPAPLNSVVLQPVAVTNPFGTLPAMPQISIN 688
Query: 287 -----PPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDL 341
P SS V P I+S + H + P + + P
Sbjct: 689 QGGNSPSIQYGISSMPVVDKPAPVRISSLLTSRHLLHRRVRLPARKYRPGENGPKVPFFT 748
Query: 342 KSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTN 401
D++ + + + L++P P+ +V P+Q S D + K Q PT
Sbjct: 749 -------DDEESSSTPKADALFIP-----RENPRALVIRPVQQWSSRDKSILPKEQRPTA 796
Query: 402 P 402
P
Sbjct: 797 P 797
Score = 41.6 bits (96), Expect = 0.002
Identities = 49/195 (25%), Positives = 80/195 (40%), Gaps = 21/195 (10%)
Query: 109 SSNPYGYGSAGYT-GYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANY 167
SSNP+G S G + + Q N + + P AP P + + +GS+ +
Sbjct: 4 SSNPFGQSSGTSPFGSQSLFGQTSNTSSNNPF-------APATPFGTSAPFAAQSGSSIF 56
Query: 168 SSTYYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTS 227
ST A ++ T A+ +P + N S ++ + P ++ P SSG
Sbjct: 57 GSTSTGVFGAPQTSSPFASTPTFGASSSPAFGN-STPAFGASPASS--PFGGSSGFGQKP 113
Query: 228 VQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYP 287
+ + Q NP+ + Q++ P N S SS+ FG A +TP S S P
Sbjct: 114 LGFSTPQSNPFGNSTQQSQ----PAFGNTSFGSSTPFGATNTPAFGAPSTP-SFGATSTP 168
Query: 288 PYWRQDSSASAVPSF 302
+ AS+ P+F
Sbjct: 169 SF-----GASSTPAF 178
Score = 35.0 bits (79), Expect = 0.21
Identities = 114/513 (22%), Positives = 180/513 (34%), Gaps = 111/513 (21%)
Query: 19 FENRHFDPG-----QNQPPSYAPTTPG---SQAVSWAVQSSTANGGYSNPTYQYDQHPQP 70
F N F G N P A TP S ++ S+ A G S P + P
Sbjct: 217 FGNTGFGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASSTPAFGASSTPAF 276
Query: 71 PGRSIQD--GQNVSSVA-GNSSNLGTANVPQDYNAYTSYPS------SSNPY----GYGS 117
G S N SS + G+S G + +A+ S PS +S P G+G
Sbjct: 277 GGSSTPSFGASNTSSFSFGSSPAFGQSTSAFGSSAFGSTPSPFGGAQASTPTFGGSGFGQ 336
Query: 118 AGYTGYYNNYQQQP--------NQTYSQPVGAYQNTGA---------------PYQ---- 150
+ + G + P T +QP G ++ A YQ
Sbjct: 337 STFGGQQGGSRAVPYAPTVEADTGTGTQPAGKLESISAMPAYKEKNYEELRWEDYQRGDK 396
Query: 151 --PISSFQNTGSYA-GSANYSSTYYNPAPADYQT----TGVYQNSTG---YANQAPPWNN 200
P+ + Q+ G+ G + ++P+PA QT T + +ST +A Q P +
Sbjct: 397 GGPLPAGQSPGNAGFGISPSQPNPFSPSPAFGQTSANPTNPFSSSTSTNPFAPQTPTIAS 456
Query: 201 GSY----SSYASHPHTNYTPDSN--SSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTE 254
S+ S++ S P T SN SG++TT+ + + T S G+
Sbjct: 457 SSFGTATSNFGSSPF-GVTSSSNLFGSGSSTTTSVFGSSSA------FGTTTPSPLFGSS 509
Query: 255 NL-SVTSSSTFGGPIPAATSGYATPNSQPQQSY--PPYWRQDSSA-------------SA 298
+ SSS+ G P + A NSQP + P Q SA S+
Sbjct: 510 STPGFGSSSSIFGSAPGQGATPAFGNSQPSTLFNSTPSTGQTGSAFGQTGSAFGQFGQSS 569
Query: 299 VPSFQPAAINSGDHDGYWNHGAQTSQIHQTN-------------PIQPNYQSPLDLKSSY 345
P+F +I + G+ N + +S + ++ P Q +
Sbjct: 570 APAFGQNSIFNKPSTGFGNMFSSSSTLTTSSSSPFGQTMPAGVTPFQSAQPGQASNGFGF 629
Query: 346 DNFQDQQK---TVSSQGTNLYLPPP-PPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTN 401
+NF Q T ++ G ++ P P VV P+ +P
Sbjct: 630 NNFGQNQAANTTGNAGGLGIFGQGNFGQSPAPLNSVVLQPVAVTNPFGT-------LPAM 682
Query: 402 PRIASNLTYGQPKIEKDSSTTNTALKPAYLAVS 434
P+I+ N P I+ S+ KPA + +S
Sbjct: 683 PQISINQGGNSPSIQYGISSMPVVDKPAPVRIS 715
Score = 33.9 bits (76), Expect = 0.47
Identities = 73/326 (22%), Positives = 112/326 (33%), Gaps = 76/326 (23%)
Query: 29 NQPPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVAGNS 88
N P AP+TP S+ S+ + G S P + P + A NS
Sbjct: 150 NTPAFGAPSTP-----SFGATSTPSFGASSTPAFGATNTP-------------AFGASNS 191
Query: 89 SNLGTANVP----QDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQT---------- 134
+ G N P A+ S ++ G+GS G G N + T
Sbjct: 192 PSFGATNTPAFGASPTPAFGSTGTTFGNTGFGSGGAFGASNTPAFGASGTPAFGASGTPA 251
Query: 135 ---YSQPVGAYQNT---GAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNS 188
S P +T GA P +T S+ G++N SS + +PA Q+T + S
Sbjct: 252 FGASSTPAFGASSTPAFGASSTPAFGGSSTPSF-GASNTSSFSFGSSPAFGQSTSAF-GS 309
Query: 189 TGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQQ--QQQNPWADYYN-QT 245
+ + + P+ S TP SG ++ QQ + P+A T
Sbjct: 310 SAFGSTPSPFGGAQAS----------TPTFGGSGFGQSTFGGQQGGSRAVPYAPTVEADT 359
Query: 246 EVSCAPGTENLSVTSSSTF------------------GGPIPAATS----GYATPNSQPQ 283
P + S+++ + GGP+PA S G+ SQP
Sbjct: 360 GTGTQPAGKLESISAMPAYKEKNYEELRWEDYQRGDKGGPLPAGQSPGNAGFGISPSQPN 419
Query: 284 QSYP-PYWRQDSSASAVPSFQPAAIN 308
P P + Q S+ P + N
Sbjct: 420 PFSPSPAFGQTSANPTNPFSSSTSTN 445
>At5g04170 EF - hand Calcium binding protein - like
Length = 354
Score = 45.4 bits (106), Expect = 2e-04
Identities = 40/139 (28%), Positives = 55/139 (38%), Gaps = 24/139 (17%)
Query: 104 TSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQ-----PVGAYQNTGAPY-------QP 151
+ YP +S YGYG G N P YS P G+ T +PY +P
Sbjct: 2 SGYPPTSQGYGYGYGG-----GNQPPPPQPPYSSGGNNPPYGS-STTSSPYAVPYGASKP 55
Query: 152 ISSFQNTGSYAGSANYS----STYYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYA 207
SS + +Y GS++Y S Y P+P DY + G PP + Y SY
Sbjct: 56 QSSSSSAPTY-GSSSYGAPPPSAPYAPSPGDYNKPPKEKPYGGGYGAPPPSGSSDYGSYG 114
Query: 208 SHPHTNYTPDSNSSGAATT 226
+ P + P + G T
Sbjct: 115 AGPRPS-QPSGHGGGYGAT 132
Score = 37.0 bits (84), Expect = 0.056
Identities = 41/165 (24%), Positives = 58/165 (34%), Gaps = 21/165 (12%)
Query: 24 FDPGQNQPPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQH-------PQPPGRSIQ 76
+ G N PP + TT AV + S + S PTY + P P
Sbjct: 28 YSSGGNNPPYGSSTTSSPYAVPYGA-SKPQSSSSSAPTYGSSSYGAPPPSAPYAPSPGDY 86
Query: 77 DGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYS 136
+ G + DY +Y + P S P G+G Y P S
Sbjct: 87 NKPPKEKPYGGGYGAPPPSGSSDYGSYGAGPRPSQPSGHG--------GGYGATPPHGVS 138
Query: 137 QPVGAYQNTGAPYQPISSFQ--NTGSYAGSANYSSTYYNPAPADY 179
G+Y GAP +P SS G Y A+Y S + + P+ +
Sbjct: 139 D-YGSY--GGAPPRPASSGHGGGYGGYPPQASYGSPFASLIPSGF 180
>At1g20110 unknown protein
Length = 601
Score = 44.7 bits (104), Expect = 3e-04
Identities = 59/246 (23%), Positives = 83/246 (32%), Gaps = 41/246 (16%)
Query: 101 NAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGS 160
N Y S P + GYGSA Y+ Y NY + P A T +P QP S T
Sbjct: 47 NTYASAPPFTG--GYGSADYSNYSQNYTPYGQNSEHVPPSAPSFT-SPSQPPPSPPATSL 103
Query: 161 YAGSANYSSTYYNPAP------------ADYQTTGVYQNSTGYANQAPPWNNGSYSSYAS 208
+ N ST+ P P + +T YQ T P++ S Y+S
Sbjct: 104 ---NPNSYSTFNQPPPPPTIHPQPLSSYGSFDSTAPYQQPTSQHMYYSPYDQHQTSGYSS 160
Query: 209 HPHTNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPI 268
P + P N + A +S Y A Y+ S P E SV
Sbjct: 161 APPPSSAPAPNPNPAPYSSSLYS-------APPYSSGGSSIPPSYEKPSVKFDQ------ 207
Query: 269 PAATSGYATPNSQPQQSYPPYWRQDSSASAVPSFQPA------AINSGDHDGYWNHGAQT 322
SGY N + + S + P+F+ + A G + Y + G
Sbjct: 208 ----SGYDGYNRSRSDLGSDLYGKRSDSGEYPAFEDSYGDGVYAYQGGKVEPYGSRGTAP 263
Query: 323 SQIHQT 328
+ T
Sbjct: 264 KSSNST 269
>At4g08380 extensin-like protein
Length = 437
Score = 43.9 bits (102), Expect = 5e-04
Identities = 91/371 (24%), Positives = 120/371 (31%), Gaps = 69/371 (18%)
Query: 29 NQPPSYAPTTPGSQAVSWA-----------VQSSTANGGYSNPTYQYDQHPQPPGRSIQD 77
+ PP YA + P S V + V SS YS P Y Y P P
Sbjct: 113 SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYVYSSPPPYAYSPPPYAYSPPPSP------- 165
Query: 78 GQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQ 137
S + P Y AY+ PS PY Y S Y Y P YS
Sbjct: 166 -------YVYKSPPYVYSSPPPY-AYSPPPS---PYVYKSPPYV-----YSSPPPYAYSP 209
Query: 138 PVGAYQNTGAPY---QPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNS---TGY 191
P AY +PY P + + YA S S Y P Y + Y S + Y
Sbjct: 210 PPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPY 269
Query: 192 ANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQTEVSCAP 251
++PP+ Y+S P Y+P S S Y P+A P
Sbjct: 270 VYKSPPY------VYSSPPPYAYSPP--PSPYVYKSPPYVYSSPPPYA--------YSPP 313
Query: 252 GTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPY-WRQDSSASAVPSFQPAAINSG 310
+ + + + P P A Y+ P S PPY + + P P S
Sbjct: 314 PSPYVYKSPPYVYSSPPPYA---YSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSP 370
Query: 311 DHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPL 370
+ +S T P SP D ++ SS +Y PPP
Sbjct: 371 PY-------VYSSPPPYTYSPPPYAYSP--PPPCPDVYKPPPYVYSSPPPYVYNPPPSSP 421
Query: 371 PPPPQQVVSTP 381
PP P S+P
Sbjct: 422 PPSPSYSYSSP 432
Score = 41.6 bits (96), Expect = 0.002
Identities = 53/195 (27%), Positives = 68/195 (34%), Gaps = 23/195 (11%)
Query: 29 NQPPSYAPTTPGSQAVSWA---VQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVA 85
+ PP YA + P S V + V SS YS P Y Y P P ++ V
Sbjct: 176 SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPYAYSPPPSP-----YVYKSPPYVY 230
Query: 86 GNSSNLGTANVPQDYNAYTSYP---SSSNPYGYGSAGYTGYYNN----YQQQPNQTYSQP 138
+ + P Y Y S P SS PY Y Y + Y P YS P
Sbjct: 231 SSPPPYAYSPPPSPY-VYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPP 289
Query: 139 VGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNS---TGYANQA 195
Y PY + + YA S S Y P Y + Y S + Y ++
Sbjct: 290 PSPYVYKSPPY----VYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKS 345
Query: 196 PPWNNGSYSSYASHP 210
PP+ S YA P
Sbjct: 346 PPYVYSSPPPYAYSP 360
Score = 34.3 bits (77), Expect = 0.36
Identities = 69/299 (23%), Positives = 97/299 (32%), Gaps = 36/299 (12%)
Query: 103 YTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYA 162
Y P S PY Y S P TYS P Y PY + + YA
Sbjct: 28 YPYSPPSPPPYVYSSP------------PPYTYSPPPSPYVYKSPPY----VYSSPPPYA 71
Query: 163 GSANYSSTYYNPAPADYQTTGVYQNS---TGYANQAPPWNNGSYSSYASHPHTNYTPDSN 219
S S Y P Y + Y S + Y ++PP+ S YA P + +
Sbjct: 72 YSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKS 131
Query: 220 SSGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATP- 278
++ Y P+A Y P + + + + P P A S +P
Sbjct: 132 PPYVYSSPPPYVYSSPPPYA-YSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPY 190
Query: 279 --NSQP--QQSYPPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPN 334
S P S PPY + P P S + + + + P
Sbjct: 191 VYKSPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKSPP----YVYSSPPPYAYSPPPSPYV 246
Query: 335 YQSPLDLKSS---YDNFQDQQKTVSSQGTNLYLPPPPPL--PPPPQQVVSTP--IQSAP 386
Y+SP + SS Y V +Y PPP PPP V +P + S+P
Sbjct: 247 YKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSP 305
>At1g73840 proline-rich like protein precursor
Length = 388
Score = 40.4 bits (93), Expect = 0.005
Identities = 73/284 (25%), Positives = 105/284 (36%), Gaps = 67/284 (23%)
Query: 129 QQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNS 188
QQP Q++ Q + N A +SSFQ GS AN T AP T
Sbjct: 81 QQPQQSH-QSIPPKPNVQAH---MSSFQGGGSVHEPAN---TMQPQAPIRKHPTPQPMP- 132
Query: 189 TGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQQQQQ----NPWADYYNQ 244
PP + S +S S P ++ A TS+ + Q Q P A ++
Sbjct: 133 -----MPPPPPSVSANSAQSQPRFSHPQRQGHLNPAVTSMSHPQSSQVQNAPPPASHHPT 187
Query: 245 TEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPYWRQDSSASAVPSFQP 304
++ P +L + +SST P + G P+ QQS P Y Q A
Sbjct: 188 SQ---QPPFHHLDIPASSTQLQQQPMHSGG--GPHVAQQQSRP-YHHQYGQAQ------- 234
Query: 305 AAINSGDHDGYWNHGAQTSQIHQTN----------------------PIQPNYQSPLDLK 342
+G + G+ +HGA T + Q P QP YQ +
Sbjct: 235 ----TGPNTGFQHHGAPTQHLSQPMYHSGNRPPASGGPQFPQGQPHLPSQPTYQGGGQYR 290
Query: 343 SSYDNFQ-----DQQKTVS-----SQGTNL-YLPPPPPLPPPPQ 375
Y+N Q Q + S S+ +N+ +LP P+PPP Q
Sbjct: 291 GDYNNNQLAGLMAQDRGPSWMAGQSESSNITHLPGLGPVPPPSQ 334
Score = 37.0 bits (84), Expect = 0.056
Identities = 30/127 (23%), Positives = 53/127 (41%), Gaps = 8/127 (6%)
Query: 321 QTSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPP--LPPPPQQVV 378
Q Q HQ+ P +PN Q+ + + + T+ Q P P P +PPPP V
Sbjct: 82 QPQQSHQSIPPKPNVQAHMSSFQGGGSVHEPANTMQPQAPIRKHPTPQPMPMPPPPPSVS 141
Query: 379 STPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQPKIEK---DSSTTNTALKPAYLAVSL 435
+ QS P + Q NP + S +++ +S T+ +P + + +
Sbjct: 142 ANSAQSQPRFSH---PQRQGHLNPAVTSMSHPQSSQVQNAPPPASHHPTSQQPPFHHLDI 198
Query: 436 PKSTEKI 442
P S+ ++
Sbjct: 199 PASSTQL 205
Score = 29.6 bits (65), Expect = 8.9
Identities = 50/216 (23%), Positives = 75/216 (34%), Gaps = 31/216 (14%)
Query: 15 QPHQFENRHFDPGQNQPPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRS 74
QP +H P P P+ + A S S G+ NP HPQ
Sbjct: 117 QPQAPIRKHPTPQPMPMPPPPPSVSANSAQSQPRFSHPQRQGHLNPAVTSMSHPQ----- 171
Query: 75 IQDGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSS-----NPYGYGSAGYTGYYNN--Y 127
V + +S+ T+ P + + P+SS P G + + Y
Sbjct: 172 ---SSQVQNAPPPASHHPTSQQPPFH--HLDIPASSTQLQQQPMHSGGGPHVAQQQSRPY 226
Query: 128 QQQPNQTYSQPVGAYQNTGAPYQPIS-----SFQNTGSYAGSANYSSTYYNPAPADYQTT 182
Q Q + P +Q+ GAP Q +S S + G + P+ YQ
Sbjct: 227 HHQYGQAQTGPNTGFQHHGAPTQHLSQPMYHSGNRPPASGGPQFPQGQPHLPSQPTYQGG 286
Query: 183 GVYQ---NSTGYA-----NQAPPWNNG-SYSSYASH 209
G Y+ N+ A ++ P W G S SS +H
Sbjct: 287 GQYRGDYNNNQLAGLMAQDRGPSWMAGQSESSNITH 322
>At1g07310 unknown protein
Length = 352
Score = 39.3 bits (90), Expect = 0.011
Identities = 47/178 (26%), Positives = 65/178 (36%), Gaps = 28/178 (15%)
Query: 31 PPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDGQNVSSVAGNSSN 90
PP + P P SQ + + S+ Y +P+ P PP I
Sbjct: 142 PPQHPPPRPQSQPLDY--YSAPQGNHYYSPS---PPPPPPPQAPI--------------- 181
Query: 91 LGTANVPQ-DYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPY 149
TA PQ DY ++ PS S PY + Y+GYY Y P ++ Y P
Sbjct: 182 --TAPSPQRDYREFSQSPSPS-PYAFTDHYYSGYY--YPPPPPRSMYDRASNYGLPSGPS 236
Query: 150 QPISSFQNTG-SYAGSANYSSTYYNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSY 206
P+ +F + A + Y P P+ T Y QAPP + SSY
Sbjct: 237 APVDAFSSIDHKQPPLAPPRFSNYGPPPSGPSAPVDAFLVTEYKPQAPPMGS-RLSSY 293
Score = 34.7 bits (78), Expect = 0.28
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 358 QGTNLYLPPPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPT 400
QG + Y P PPP PPPPQ ++ P +P D + S+ P+
Sbjct: 162 QGNHYYSPSPPP-PPPPQAPITAP---SPQRDYREFSQSPSPS 200
>At4g28300 predicted proline-rich protein
Length = 496
Score = 38.5 bits (88), Expect = 0.019
Identities = 49/180 (27%), Positives = 67/180 (37%), Gaps = 9/180 (5%)
Query: 128 QQQPNQTYSQPVGAY-QNTGAPYQ---PISSFQNTGSYAGSANYSSTYYNPAPADYQTTG 183
Q Q +Q Y P QNT AP P S Q + + +P+ A Q+
Sbjct: 232 QPQQHQYYMPPPPTQLQNTPAPVPVSTPPSQLQAPPAQSQFMPPPPAPSHPSSAQTQSFP 291
Query: 184 VYQNSTGYANQAPPWNNGSYSSYASHPHTNYTP-DSNSSGAATTSVQYQQQQQNPWADYY 242
YQ + QA P ++G Y +Y+ P N P +S S S QQ+ A Y
Sbjct: 292 QYQQNWPPQPQARPQSSGGYPTYSPAPPGNQPPVESLPSSMQMQSPYSGPPQQSMQAYGY 351
Query: 243 NQTEVSCAPGTENLSVTSSSTFGGPIPAA---TSGYATPNSQ-PQQSYPPYWRQDSSASA 298
AP + S T G +P+ SGYA + + YPP Q A
Sbjct: 352 GAAPPPQAPPQQTKMSYSPQTGDGYLPSGPPPPSGYANAMYEGGRMQYPPPQPQQQQQQA 411
>At4g23140 receptor-like protein kinase 5 (RLK5)
Length = 674
Score = 38.5 bits (88), Expect = 0.019
Identities = 17/28 (60%), Positives = 20/28 (70%)
Query: 364 LPPPPPLPPPPQQVVSTPIQSAPSLDAK 391
LPPPPP PPP + +VSTP S+ SL K
Sbjct: 256 LPPPPPPPPPRESLVSTPPISSSSLPGK 283
>At4g20020 DAG-like protein
Length = 406
Score = 38.1 bits (87), Expect = 0.025
Identities = 48/172 (27%), Positives = 57/172 (32%), Gaps = 19/172 (11%)
Query: 49 QSSTANGGYSN--PTYQYDQHPQPPGRSIQDGQNVSSVAGNSSNLGTANVPQDYNAYTSY 106
+S +GG N QY Q P G G A GT P Y
Sbjct: 206 RSGGGSGGPQNFQRNTQYGQQPPMQGGGGSYGPQ-QGYATPGQGQGTQAPPPFQGGYNQG 264
Query: 107 PSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSAN 166
P S P Y YN Q P Y G Q G+P P Q++ GS N
Sbjct: 265 PRSPPP------PYQAGYNQGQGSPVPPYQ--AGYNQVQGSPVPPYQGTQSSYGQGGSGN 316
Query: 167 YS----STYYNPAPADY--QTTGVYQNSTGYAN--QAPPWNNGSYSSYASHP 210
YS Y P +Y Q G + ++G N AP N Y S P
Sbjct: 317 YSQGPQGGYNQGGPRNYNPQGAGNFGPASGAGNLGPAPGAGNPGYGQGYSGP 368
>At5g51300 unknown protein
Length = 804
Score = 37.7 bits (86), Expect = 0.033
Identities = 44/164 (26%), Positives = 55/164 (32%), Gaps = 21/164 (12%)
Query: 11 LVPAQPHQFENRHFDPGQNQPP---SYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQH 67
L P P + H GQ+ PP Y P P V + G NP+ Q
Sbjct: 617 LPPPPPGSYHPVH---GQHMPPYGMQYPPPPPH-------VTQAPPPGTTQNPSSSEPQQ 666
Query: 68 PQPPGRSIQDGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSN----PYGYGSAGYTGY 123
PPG G SS+ N +P Y SYPS N P A T +
Sbjct: 667 SFPPGVQADSGAATSSIPPNVYGSSVTAMP-GQPPYMSYPSYYNAVPPPTPPAPASSTDH 725
Query: 124 YNNYQQQP---NQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGS 164
N P N + S P + AP+ P T Y+ S
Sbjct: 726 SQNMGNMPWANNPSVSTPDHSQGLVNAPWAPNPPMPPTVGYSQS 769
>At4g32760 unknown protein
Length = 675
Score = 37.7 bits (86), Expect = 0.033
Identities = 61/238 (25%), Positives = 89/238 (36%), Gaps = 34/238 (14%)
Query: 67 HPQPPGRSIQDGQNVSSVAGNSSNLGTANVPQ-DYNAYTSYPSSSNPY------GYGSAG 119
H QP ++ + G S+ G + +G + Q Y S P SS P YG+
Sbjct: 399 HQQPNSQAGEAGLQQSN--GFAPQVGYSQFEQPSYGQGVSSPWSSQPAQQPVQPSYGAQD 456
Query: 120 YTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQP-ISSFQNTGSYAGSANYSSTYYNPAPAD 178
T + + Q YS +G+P+ P + Q ++A N + NP P
Sbjct: 457 STAFPPPPWEAQLQDYSPSA----ESGSPFSPGMHPTQTAFTHAQPVNNN----NPYPQI 508
Query: 179 YQTTGVYQNSTGYANQAPPWNNG--SYSSYASHPHTN-YTPDS-NSSGAATTSVQYQQQQ 234
QT N++ YA Q P + S Y P Y P+ N + + Q QQQQ
Sbjct: 509 PQTGPPVNNNSPYA-QMPQTGQAVANISPYPQIPQNGVYMPNQPNQALGSGYQPQQQQQQ 567
Query: 235 QNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPYWRQ 292
Q A YY Q + + +G + GY +Q QQ PY Q
Sbjct: 568 QMMMAQYYAQQQ-------QLQQQQQQQAYGNQM----GGYGYGYNQQQQGSSPYLDQ 614
>At2g43170 unknown protein
Length = 504
Score = 37.7 bits (86), Expect = 0.033
Identities = 44/208 (21%), Positives = 70/208 (33%), Gaps = 22/208 (10%)
Query: 223 AATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNS-- 280
A TTS + AD ++ ++ P T+ GP P+ ++ + S
Sbjct: 3 APTTSNSVEMDLLGSLADVFSSNALAIVPADSIYVETNGQANAGPAPSFSTSQPSTQSFD 62
Query: 281 QPQQSYPPYWRQDSSASAVP------SFQP------AAINSGDHDGYWNHGAQTSQIHQT 328
P P + + P SFQP + ++ D + G S +
Sbjct: 63 DPFGDSPFKAFTSTDTDSTPQQNFGASFQPPPPAFTSEVSHPDTAHNFGFGDSFSAVANP 122
Query: 329 NPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSL 388
+P N Q P S+ F +Q S G ++ PP PP Q S P P
Sbjct: 123 DPASQNVQPP----SNSPGFPQEQFATSQSGIDILAGILPPSGPPVQSGPSIPTSQFPPS 178
Query: 389 DAKRV----SKMQIPTNPRIASNLTYGQ 412
S+ + T P + +GQ
Sbjct: 179 GNNMYEGFHSQPPVSTAPNLPGQTPFGQ 206
>At2g38410 unknown protein
Length = 671
Score = 37.7 bits (86), Expect = 0.033
Identities = 37/143 (25%), Positives = 53/143 (36%), Gaps = 22/143 (15%)
Query: 85 AGNSSNLGTANVPQDYNAYTSY--PSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAY 142
AG+ N T PQ + SY P + GY + Q Q Q YSQP +
Sbjct: 459 AGSDQN--THIYPQPQPRFDSYVAPWAQQQQPQQPQAQQGYSQHQQHQQQQGYSQPQHSQ 516
Query: 143 QNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNSTGYANQAPPWNNGS 202
Q G Y + Q Y+ S + P+ + Y PPW + S
Sbjct: 517 QQQG--YSQLQQPQPQQGYSQSQPQAQVQMQPST---------RPQNPYEYPPPPWASTS 565
Query: 203 YSSYASHPHTNYTPDSNSSGAAT 225
++Y YTP +N+S + T
Sbjct: 566 ANAY-------YTPRANASASYT 581
>At1g10270 unknown protein
Length = 913
Score = 37.7 bits (86), Expect = 0.033
Identities = 60/249 (24%), Positives = 82/249 (32%), Gaps = 42/249 (16%)
Query: 2 ANEGTNTETLVPAQPHQFENRHFDPGQNQPPSYAPTTPG-SQAVSWAVQS--STANGGYS 58
AN G N + Q ++ QPPS++ PG Q SW+ QS S+ +G
Sbjct: 666 ANNGQNPSWSNTSDNQQQQSWSNQTAGQQPPSWSRQAPGYQQQQSWSQQSGWSSPSGHQQ 725
Query: 59 NPTYQYDQHPQP-----PGRSIQDGQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSNPY 113
+ T Q QP PG Q Q + G L Q P +
Sbjct: 726 SWTNQTAGQQQPWANQTPG---QQQQWANQTPGQQQQLANQTPGQQQQWANQTPGQQQQW 782
Query: 114 GYGSAGYTGYYNNYQQQP--NQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTY 171
NN QQP NQ T + QP ++ Q TG G N
Sbjct: 783 --------ANQNNGHQQPWANQNTGHQQSWANQTPSQQQPWAN-QTTGQQQGWGN----- 828
Query: 172 YNPAPADYQTTGVYQNSTGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQYQ 231
QTTG Q +ANQ G S + + + S+ V +
Sbjct: 829 --------QTTGQQQQ---WANQTA----GQQSGWTAQQQWSNQTASHQQSQWLNPVPGE 873
Query: 232 QQQQNPWAD 240
Q PW++
Sbjct: 874 VANQTPWSN 882
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,342,341
Number of Sequences: 26719
Number of extensions: 1148191
Number of successful extensions: 7305
Number of sequences better than 10.0: 256
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 201
Number of HSP's that attempted gapping in prelim test: 5236
Number of HSP's gapped (non-prelim): 1124
length of query: 1003
length of database: 11,318,596
effective HSP length: 109
effective length of query: 894
effective length of database: 8,406,225
effective search space: 7515165150
effective search space used: 7515165150
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0260.13


