

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.6
(499 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
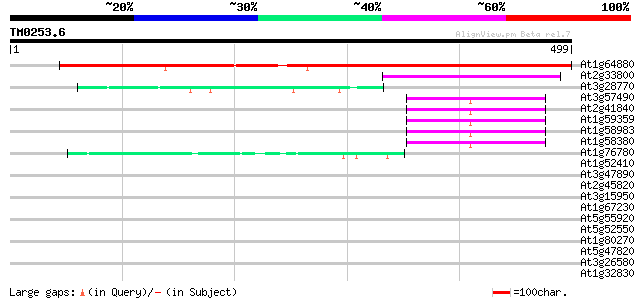
At1g64880 unknown protein 612 e-175
At2g33800 30S ribosomal protein S5 89 4e-18
At3g28770 hypothetical protein 46 4e-05
At3g57490 40S ribosomal protein S2 homolog 44 2e-04
At2g41840 40S ribosomal protein S2 44 3e-04
At1g59359 ribosomal protein S2, putative 44 3e-04
At1g58983 ribosomal protein S2, putative 44 3e-04
At1g58380 unknown protein 44 3e-04
At1g76780 putative heat shock protein 42 8e-04
At1g52410 myosin-like protein 41 0.002
At3g47890 putative protein 40 0.004
At2g45820 remorin 40 0.004
At3g15950 unknown protein 39 0.005
At1g67230 unknown protein 38 0.011
At5g55920 nucleolar protein-like 37 0.019
At5g52550 unknown protein 37 0.019
At1g80270 unknown protein (At1g80270) 37 0.019
At5g47820 kinesin-like protein 37 0.025
At3g26580 unknown protein 37 0.033
At1g32830 putaive protein on FARE2.4 (cds2) 36 0.056
>At1g64880 unknown protein
Length = 515
Score = 612 bits (1579), Expect = e-175
Identities = 301/459 (65%), Positives = 378/459 (81%), Gaps = 12/459 (2%)
Query: 45 YSGKSLYSTSSDARIVQDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIE 104
+ S S S R+V++L+A+VEK++ RE+ ER R GLD DIDAE EEDY+G+ P IE
Sbjct: 65 FFSSSTMSPSEADRVVRELIAEVEKEKQREREERQRQGLDYKDIDAEDEEDYLGIEPFIE 124
Query: 105 KLEKEKLKECPELMRYEEPTDSDSDEDEYEASQ--KKFDDFERKFKRHQDLLKNFTDADT 162
KL+K+ LK+ EL R EE +DSDS+ DE + + KK D F +KF+RH++LL+ T ++T
Sbjct: 125 KLKKQNLKDDGELNRREESSDSDSEYDEIDLDEERKKEDIFNKKFQRHKELLQTLTKSET 184
Query: 163 LDDAFKWMNRIDKFEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALRLVQW 222
LD+AF+WMN++DKFE+KHF+LRPEYRVIGELMNRLKV + K+KF+LQ KLNRA+RLV+W
Sbjct: 185 LDEAFRWMNKLDKFEEKHFKLRPEYRVIGELMNRLKVA-EGKEKFVLQQKLNRAMRLVEW 243
Query: 223 KEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQG--QGDAADDDDEQEFDDMK 280
KEAFDP+NP NYGVI+ D +Q G E+E++ + + D DDDDE+EFDDMK
Sbjct: 244 KEAFDPNNPANYGVIER-------DNVQGGGEEREERLVADGPKDDNDDDDDEEEFDDMK 296
Query: 281 EKDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEKKRRPLYRK 340
E+D+I+L KL+AIDKKLE KL+EL++TFGKKGK LEEEIR+LAE+RN LTEKKR+PLYRK
Sbjct: 297 ERDDILLEKLNAIDKKLEIKLSELDHTFGKKGKRLEEEIRELAEDRNALTEKKRQPLYRK 356
Query: 341 GFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQ 400
G+DV +I+V + CKVTKGG+V +YTA++ CGNY G++G+AKAK A+QKAYEKCFQ
Sbjct: 357 GYDVHVIDVKKVCKVTKGGRVERYTALMVCGNYEGIIGYAKAKAETGQSAMQKAYEKCFQ 416
Query: 401 NLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVV 460
NLH+VERHEEHTIAHAIQTS+KKTK+YLWPAPT TGMKAGR+V+TIL LAGFKN+KSKV+
Sbjct: 417 NLHYVERHEEHTIAHAIQTSYKKTKLYLWPAPTTTGMKAGRVVKTILLLAGFKNIKSKVI 476
Query: 461 GSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKYLL 499
GSRN +NTVKAV KALNA+ETP+DVQEKFGRTVVEKYLL
Sbjct: 477 GSRNSYNTVKAVLKALNAVETPKDVQEKFGRTVVEKYLL 515
>At2g33800 30S ribosomal protein S5
Length = 303
Score = 89.4 bits (220), Expect = 4e-18
Identities = 52/159 (32%), Positives = 82/159 (50%)
Query: 332 KKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVAL 391
K ++ R GF+ +++ V R KV KGG+ +K+ A++ G+ G VG AK V A+
Sbjct: 137 KPKKDKIRDGFEERVVQVRRVTKVVKGGKQLKFRAIVVVGDKQGNVGVGCAKAKEVVAAV 196
Query: 392 QKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAG 451
QK+ +N+ V + T H + + KV L PA TG+ AG V+ +L +AG
Sbjct: 197 QKSAIDARRNIVQVPMTKYSTFPHRSEGDYGAAKVMLRPASPGTGVIAGGAVRIVLEMAG 256
Query: 452 FKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFG 490
+N K +GS N N +A A+ + RDV ++ G
Sbjct: 257 VENALGKQLGSNNALNNARATLAAVQQMRQFRDVAQERG 295
>At3g28770 hypothetical protein
Length = 2081
Score = 46.2 bits (108), Expect = 4e-05
Identities = 65/307 (21%), Positives = 119/307 (38%), Gaps = 42/307 (13%)
Query: 61 QDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRY 120
+DL A+ +++ +EK E + D + ED + +K EK+K +E +
Sbjct: 1056 RDLKAKKKEEETKEKKESENHKSKKKE-DKKEHEDNKSMKKEEDKKEKKKHEESKSRKKE 1114
Query: 121 EEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTD------------ADTLDDAFK 168
E+ D + ED+ +++KK D E+K +H L+K +D ++ +
Sbjct: 1115 EDKKDMEKLEDQ-NSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKS 1173
Query: 169 WMNRIDKFE-----DKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALRLVQWK 223
N +DK E D+ + E + E + +K + +NK + + + K
Sbjct: 1174 QKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNK 1233
Query: 224 EAFDPDNPDNYGVIQHEQLGPTSDALQN---------AGFEKEKQTIQGQGDAADDDDEQ 274
D N + E + S +N A ++ K I Q D+ D
Sbjct: 1234 PKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSD 1293
Query: 275 EFDDMKEKDNIILAKLDA---------IDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEE 325
D E N IL + D+ D+K + +AE KK K +EE ++
Sbjct: 1294 SQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAE-----NKKQKETKEEKNKPKDD 1348
Query: 326 RNELTEK 332
+ T++
Sbjct: 1349 KKNTTKQ 1355
Score = 35.8 bits (81), Expect = 0.056
Identities = 70/321 (21%), Positives = 133/321 (40%), Gaps = 25/321 (7%)
Query: 24 NVVRNSVRSQLGCEGTALSSSYSGKSLYSTSSDARIVQDLLAQVEKDRLREKNERMRAGL 83
N VRN + +G S K S DA+ V+ + + R+G
Sbjct: 766 NRVRNKEEN---VQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGE 822
Query: 84 DTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSDSDEDEYEASQKKFDDF 143
D + D E +DY V E EK + + +E + D+ E K +
Sbjct: 823 DNKE-DKEESKDYQSV----EAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESM 877
Query: 144 ERKFKRHQDLLKNFTDADTLDDAFKWMNRIDKFEDKHFRLRPEYRVIG---ELMNRLKVV 200
++K + Q K+ T + D A + K + + + + + G E + +
Sbjct: 878 KKKREEVQRNDKSSTK-EVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTS 936
Query: 201 TDQKDKFILQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKE--- 257
+ QK K + K ++ KE + +N Q + T+ + +N+ ++E
Sbjct: 937 SKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKS-ENSKLKEENKD 995
Query: 258 -KQTIQGQGDAADDDDEQEFDDMKEK--DNIILAKLDAIDKKLEEKLAELEYTFGKKGKV 314
K+ + + A+ + +++E+++ K K + K + DKK EEK +E +K K
Sbjct: 996 NKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSE-----ERKSKK 1050
Query: 315 LEEEIRDL-AEERNELTEKKR 334
+EE RDL A+++ E T++K+
Sbjct: 1051 EKEESRDLKAKKKEEETKEKK 1071
Score = 32.0 bits (71), Expect = 0.81
Identities = 53/285 (18%), Positives = 103/285 (35%), Gaps = 36/285 (12%)
Query: 32 SQLGCEGTALSSSYSGKSLYSTSSDARIVQDLLAQVEKDRLREKNERMRAGLDTADIDAE 91
+Q + T + S K+ +D++ +Q + D KNE + A
Sbjct: 1261 NQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDE--SKNEILMQADSQATTQRN 1318
Query: 92 GEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQ 151
EED + E ++++ KE +P D + + +K+ + E K +Q
Sbjct: 1319 NEEDRKKQTSVAENKKQKETKE-----EKNKPKDDKKNTTKQSGGKKESMESESKEAENQ 1373
Query: 152 DLLKNFTDADTLDDAFKWMNRIDKFEDKHFRLRPEYRVIGELMNRLKVVTD--------- 202
+ T AD+ + + + + D D H + + E N + + D
Sbjct: 1374 QKSQATTQADSDESKNEILMQADSQADSHSDSQAD---SDESKNEILMQADSQATTQRNN 1430
Query: 203 ----QKDKFILQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEK 258
+K + +NK + + + K D N + E + S +N +K +
Sbjct: 1431 EEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTEQSGGKKESMESESKEAENQ--QKSQ 1488
Query: 259 QTIQGQGDAADDDDEQEFD-----------DMKEKDNIILAKLDA 292
T QG+ D + ++ + D D E N IL + D+
Sbjct: 1489 ATTQGESDESKNEILMQADSQADTHANSQGDSDESKNEILMQADS 1533
>At3g57490 40S ribosomal protein S2 homolog
Length = 276
Score = 43.9 bits (102), Expect = 2e-04
Identities = 35/131 (26%), Positives = 57/131 (42%), Gaps = 8/131 (6%)
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ + R
Sbjct: 99 KQTRAGQRTRFKAFIVVGDSNGHVGLGVKCSKEVATAIRGAIILAKLSVVPIRRGYWGNK 158
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 159 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 218
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 219 GNFVKATFDCL 229
>At2g41840 40S ribosomal protein S2
Length = 285
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 108 KQTRAGQRTRFKAFVVVGDGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNK 167
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 168 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 227
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 228 GNFVKATFDCL 238
>At1g59359 ribosomal protein S2, putative
Length = 284
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 107 KQTRAGQRTRFKAFVVVGDGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNK 166
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 167 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 226
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 227 GNFVKATFDCL 237
>At1g58983 ribosomal protein S2, putative
Length = 284
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 107 KQTRAGQRTRFKAFVVVGDGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNK 166
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 167 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 226
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 227 GNFVKATFDCL 237
>At1g58380 unknown protein
Length = 284
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 107 KQTRAGQRTRFKAFVVVGDGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNK 166
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 167 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 226
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 227 GNFVKATFDCL 237
>At1g76780 putative heat shock protein
Length = 1871
Score = 42.0 bits (97), Expect = 8e-04
Identities = 72/314 (22%), Positives = 126/314 (39%), Gaps = 34/314 (10%)
Query: 52 STSSDARIVQDLLAQVEKDRLREKNERMRAGLDTADIDAEG-EEDYMGVGPLIEKLEKEK 110
++ ++ I D+L V+K R K++ + + + EG E M G I + K
Sbjct: 481 ASEAEKNIKNDILKPVQK-RSEGKHKIQKTFQEETNKQPEGYNEKIMETGKKINEDGTRK 539
Query: 111 LKECPELMRYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDDAFKWM 170
++E +EP S+ + E + K +D E+K K + ++D +
Sbjct: 540 VQEMIRQQELDEPARSEKENRSRELVKSKTNDEEKKEKEIAGTERKEKESDR-----PKI 594
Query: 171 NRIDKFEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALRLVQWKEAFDPDN 230
R + D+ + ++ + GE+ ++ +K+ F + + R +R D +
Sbjct: 595 LREQEVADEVAEDKTKFSIYGEVKEEEEIAGKEKE-FGSDDDIARIVR--------DTEQ 645
Query: 231 PDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKL 290
D+ + E+ D +Q E EK G+G A + + E + K KL
Sbjct: 646 LDSNAMKGQEE----KDMIQELVLE-EKVCDGGKGIIAVAETKAENNKSKRVQETEEQKL 700
Query: 291 DAIDK--KLEEKLAELEYT---------FGKKGKVLEEEIRDLAEERNELTEKKR--RPL 337
D D K +KL E E + GKK E+ I+D A E E+ EK
Sbjct: 701 DKEDTCGKHFQKLIEGEISDHGEVEDVEKGKKRTEAEKRIKDRAREAEEIKEKDLGVSGR 760
Query: 338 YRKGFDVKLINVNR 351
Y KG +K + NR
Sbjct: 761 YIKGTTIKELVENR 774
Score = 35.4 bits (80), Expect = 0.074
Identities = 65/308 (21%), Positives = 124/308 (40%), Gaps = 47/308 (15%)
Query: 70 DRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSDSD 129
++ + +ER+ AD + E + + P IE +EC E R+EE +++
Sbjct: 96 EKSHQIDERIEEEKGLADSNKESVDSSLRKPPDIEG------RECHEQTRHEE---QENN 146
Query: 130 EDEYEASQKKFDDF-ERKFKRHQDLLKNFTD------------ADTLDDAFKWMNRIDKF 176
+ +A DDF R F+ ++ + D D + D + + I++
Sbjct: 147 KQLVQAESDDSDDFGSRAFEEIEEQESDVLDRTSTSGAMEKEMTDDVGDGLRKVQGIEEP 206
Query: 177 EDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALRLV---QWKEAFDPDNPDN 233
E + + V GE +K K + +K NR ++ E F P N D
Sbjct: 207 ERHNEESKISEMVDGETSGH------EKKKVVKMDKKNRDVKEEVDGAMGEGFRP-NIDR 259
Query: 234 YGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLDAI 293
V+ +++ T + FE +K + D ++ + E N+I
Sbjct: 260 TQVVGDDEIAETEK--NDEEFESDKLEAD-EVDKINEGGNTKVRRHSEDRNLI------- 309
Query: 294 DKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEKK-RRPLYRKGFDVKLINVNRT 352
KL+EK E +++ +KG EE +++L EE+ E R + G ++++ V+
Sbjct: 310 --KLQEK--EEQHSKEQKGHSKEENMKELVEEKTPEAETTIRNDILGPGQEIEVPEVDTL 365
Query: 353 CKVTKGGQ 360
K + G+
Sbjct: 366 GKTSDEGK 373
Score = 32.0 bits (71), Expect = 0.81
Identities = 30/128 (23%), Positives = 60/128 (46%), Gaps = 18/128 (14%)
Query: 52 STSSDARIVQDLLAQ--VEKDRLREKNERMRAGLDTADIDAEGE----EDYMGVGPLIEK 105
+T +++R + + Q E++RL+E+ D E E E G +I++
Sbjct: 1721 ATDNESRKIHQIKEQGTSEQERLKEQGRIKELVEDRTHFCREKENRETEYEDGSSKMIQE 1780
Query: 106 LEKEKLKECPELMRYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDD 165
++KE+ E P D ++ ED+ E + +F+D E ++ + TD+D+ +D
Sbjct: 1781 IDKEESIE---------PVDRETSEDDEEELEIEFEDEEEDWEAE---VIQETDSDSDND 1828
Query: 166 AFKWMNRI 173
+ + RI
Sbjct: 1829 KIRQIKRI 1836
Score = 31.6 bits (70), Expect = 1.1
Identities = 30/119 (25%), Positives = 50/119 (41%), Gaps = 22/119 (18%)
Query: 223 KEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEK 282
KE + P+ I+++ LGP G E E + G +D+ KEK
Sbjct: 332 KELVEEKTPEAETTIRNDILGP--------GQEIEVPEVDTLGKTSDEG--------KEK 375
Query: 283 DNIILAKL------DAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEKKRR 335
NI+ ++ + ID K+ E A G + E + + A+E +++ EKK R
Sbjct: 376 QNIVKKEIKNGDATEEIDAKMGEVFASNIADTGMNSEDFESDKLESADEVDKMVEKKDR 434
Score = 28.5 bits (62), Expect = 9.0
Identities = 30/124 (24%), Positives = 55/124 (44%), Gaps = 25/124 (20%)
Query: 201 TDQKDKFILQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQT 260
+DQ +K+ QNK+ + D D + Y + S+ ++N E K+
Sbjct: 1520 SDQAEKYAKQNKIQEVMN--------DEDKKEEYHI---------SERVRN---EMAKRI 1559
Query: 261 IQGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIR 320
+Q + A D ++ + +E + K ++ ++L ELE + KKG V E+E+
Sbjct: 1560 LQVESKANDGSSKKNETEGQESTGLRGRK----KRENHQELVELETSDQKKG-VKEDEVV 1614
Query: 321 DLAE 324
AE
Sbjct: 1615 GKAE 1618
>At1g52410 myosin-like protein
Length = 755
Score = 40.8 bits (94), Expect = 0.002
Identities = 57/268 (21%), Positives = 114/268 (42%), Gaps = 19/268 (7%)
Query: 61 QDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKE--KLKECPELM 118
Q LL ++E++ E + ++ D + EE ++E +E+E E E +
Sbjct: 181 QSLLEEIEREFEAATKELEQLKVNDFTGDKDDEEHSAKRKSMLEAIEREFEAAMEGIEAL 240
Query: 119 RYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDD--AFKWMNRIDKF 176
+ + T S DE++ ++ ER+F+ + L + + AD ++ A K + +++
Sbjct: 241 KVSDSTGSGDDEEQSAKRLSMLEEIEREFEGLEQLRASDSTADNNEEEHAAKGQSLLEEI 300
Query: 177 EDKHFRLRPEYRVIGELMNRLKV--VTDQKDKFILQNKLNRALRLVQWKEAFDPD----N 230
E E+ E + +L+V T+ K+ F + + + + EA D N
Sbjct: 301 E-------REFEAATESLKQLQVDDSTEDKEHFTAAKRQSLLEEIEREFEAATKDLKQLN 353
Query: 231 PDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKL 290
G EQ + L++ E E TI + A+D E ++ +E+ + L
Sbjct: 354 DFTEGSADDEQSAKRNKMLEDIEREFEAATIGLEQLKANDFSEG--NNNEEQSAKRKSML 411
Query: 291 DAIDKKLEEKLAELEYTFGKKGKVLEEE 318
+ I+++ E + L+ + LEEE
Sbjct: 412 EEIEREFEAAIGGLKQIKVDDSRNLEEE 439
>At3g47890 putative protein
Length = 1528
Score = 39.7 bits (91), Expect = 0.004
Identities = 46/201 (22%), Positives = 81/201 (39%), Gaps = 20/201 (9%)
Query: 109 EKLKECPELMRYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDDAFK 168
E ++EC + E P D + E + +Q K E + QD L++ +
Sbjct: 118 EVVQECHRALSIENPIDP-AKESLQDETQLKILTPEARIVHVQDELRSLIQKSNISSLST 176
Query: 169 WMNRIDKFEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNR-----------AL 217
WMN + K E+K FRL P R+ + + V T + ++ NK A
Sbjct: 177 WMNNLGKGEEK-FRLIPIRRMAEDPIESNLVQTRRPNEIKKANKSIEEIRKEVEVRVAAA 235
Query: 218 RLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDD--EQE 275
RL+Q K +N G + ++ T + + +G ++ + G AD D +
Sbjct: 236 RLLQQK-----SECENVGAVDNKGSDATLGSGKRSGERRKHGNARRNGSTADRRDRVKSY 290
Query: 276 FDDMKEKDNIILAKLDAIDKK 296
+D M ++ L ++ D K
Sbjct: 291 WDSMSKEMKKQLLRVKLSDLK 311
>At2g45820 remorin
Length = 190
Score = 39.7 bits (91), Expect = 0.004
Identities = 26/89 (29%), Positives = 47/89 (52%), Gaps = 6/89 (6%)
Query: 252 AGFEKEKQTI------QGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELE 305
A EKEK+T + + A++ +++ D+ +N A ++A +K+EEKL + +
Sbjct: 74 ADLEKEKKTSFIKAWEESEKSKAENRAQKKISDVHAWENSKKAAVEAQLRKIEEKLEKKK 133
Query: 306 YTFGKKGKVLEEEIRDLAEERNELTEKKR 334
+G+K K I LAEE+ + E K+
Sbjct: 134 AQYGEKMKNKVAAIHKLAEEKRAMVEAKK 162
>At3g15950 unknown protein
Length = 622
Score = 39.3 bits (90), Expect = 0.005
Identities = 59/286 (20%), Positives = 106/286 (36%), Gaps = 26/286 (9%)
Query: 61 QDLLAQVEKDRLREKNERMRAGLDTADID----AEGEEDYMGVGPLIEKLEKEKLKECPE 116
Q +L ++E+D E GL+ D EE ++E++E+E +
Sbjct: 37 QSMLDEIERDF-----EAATKGLEQLKADDLTGINDEEHAAKRQKMLEEIEREFEEATKG 91
Query: 117 LMRYEEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNF-TDADTLDDAFKWMNRIDK 175
L T S DE + Q D+ ER+F+ LK +A T+ D + K
Sbjct: 92 LEELRHSTSSTDDEAQSAKRQNMLDEIEREFEAATSGLKELKINAHTVKDDVDDKEQDAK 151
Query: 176 FEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALRLVQWKEAFDPDNPDNYG 235
+ + E+ + E +L+ + D K + R L + + F+ ++
Sbjct: 152 RQSMLDAIEREFEAVTESFKQLEDIADNKAEGD-DESAKRQSMLDEIEREFEAAT-NSLK 209
Query: 236 VIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAA--------------DDDDEQEFDDMKE 281
+ + D+ ++A + I+ + +AA D DD++ K
Sbjct: 210 QLNLDDFSEGDDSAESARRNSMLEAIEREFEAATKGLEELKANDSTGDKDDDEHVARRKI 269
Query: 282 KDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERN 327
I + +A K LEE E E K+ +LE R+ N
Sbjct: 270 MLEAIEREFEAATKGLEELKNESEQAENKRNSMLEAFEREFEAATN 315
>At1g67230 unknown protein
Length = 1132
Score = 38.1 bits (87), Expect = 0.011
Identities = 58/262 (22%), Positives = 110/262 (41%), Gaps = 54/262 (20%)
Query: 91 EGEEDYMGVGPLIEKLEKEKLKECPELMRY-EEPTDSDSDEDEYEASQKKFDDFERKFKR 149
E +E + + L+EK+ E + E+ + +E ++ + EY Q + + K +
Sbjct: 443 EDKEIILNLKALVEKVSGENQAQLSEINKEKDELRVTEEERSEYLRLQTELKEQIEKCRS 502
Query: 150 HQDLLKNFTDADTLDDAFKWMNRIDKFEDKHFRLRPEYRVIGELMNRLKVVTDQKDKFIL 209
Q+LL+ +A + + FE + L IG N LK +TDQK+
Sbjct: 503 QQELLQK--------EAEDLKAQRESFEKEWEELDERKAKIG---NELKNITDQKE---- 547
Query: 210 QNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAAD 269
KL R + L + E+L A N E+E +T++
Sbjct: 548 --KLERHIHL------------------EEERLKKEKQA-ANENMERELETLEVA----- 581
Query: 270 DDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEER-NE 328
+ F + E + +L+ KK E + ++L + + + LE +++ + EE+ E
Sbjct: 582 ---KASFAETMEYERSMLS------KKAESERSQLLHDIEMRKRKLESDMQTILEEKERE 632
Query: 329 LTEKKRRPLYRKGFDVKLINVN 350
L KK+ L+ + + +L N+N
Sbjct: 633 LQAKKK--LFEEEREKELSNIN 652
>At5g55920 nucleolar protein-like
Length = 682
Score = 37.4 bits (85), Expect = 0.019
Identities = 39/120 (32%), Positives = 54/120 (44%), Gaps = 26/120 (21%)
Query: 224 EAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKD 283
+ F D +N E LG D L+ +G E E+ ++ DA DDD+
Sbjct: 108 QLFSDDEEEN----DEETLG--DDFLEGSGDEDEEGSLDADSDADSDDDD---------- 151
Query: 284 NIILAKLDAIDKKL----EEKLAELEYTFGKKGKVLEEEIRDLA---EERNELTEKKRRP 336
I+AK DAID+ L ++ AELE F K+ V +EE A EL E+ R P
Sbjct: 152 --IVAKSDAIDRDLAMQKKDAAAELE-DFIKQDDVHDEEPEHDAFRLPTEEELEEEARGP 208
Score = 32.7 bits (73), Expect = 0.48
Identities = 29/108 (26%), Positives = 55/108 (50%), Gaps = 5/108 (4%)
Query: 230 NPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNI-ILA 288
N D + V + +++ + + G + +T++ ++DDDDE E + EK ++ +
Sbjct: 552 NMDGFFVAKLKKMSNVKQSSEE-GDDDAVETVEQAEVSSDDDDEAEAIEETEKPSVPVRQ 610
Query: 289 KLDAIDKKLEEKLAE-LEYTFGKKGKVLEEEIRDLAEERNELTEKKRR 335
+ +KK +EKLA+ E GKK K + E ++ E +KK+R
Sbjct: 611 PKERKEKKNKEKLAKSKEDKRGKKDKKSKSE--NVEEPSKPRKQKKKR 656
>At5g52550 unknown protein
Length = 360
Score = 37.4 bits (85), Expect = 0.019
Identities = 37/104 (35%), Positives = 58/104 (55%), Gaps = 17/104 (16%)
Query: 245 TSDALQNAGFEKEKQTI-QGQGDAADDDD-------EQEFDDM---KEKDNIILAKLDAI 293
TS A++ A EK+KQ +GQ +AAD++D +QE D++ K+ +N K + +
Sbjct: 46 TSAAIR-AELEKKKQMKKEGQLEAADEEDSADAAKKKQERDELERIKQAEN----KKNRL 100
Query: 294 DKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEKKRRPL 337
+K + A + KK + LEE+ R LAEE + EKK+R L
Sbjct: 101 EKSIATSAAIMAELEKKKLRKLEEQKR-LAEEGAAIAEKKKRRL 143
>At1g80270 unknown protein (At1g80270)
Length = 596
Score = 37.4 bits (85), Expect = 0.019
Identities = 29/117 (24%), Positives = 57/117 (47%), Gaps = 12/117 (10%)
Query: 253 GFEKEKQTIQGQGDAADDDDEQEFD-DMKEKDNIILAKLDAIDKKLEEKLAELEYTF-GK 310
GF + + + GQG + D+DE + D +E++ + L + D K +E+K +EL T
Sbjct: 80 GFSELEGSKSGQGSTSSDEDEGKLSADEEEEEELDLIETDVSRKTVEKKQSELFKTIVSA 139
Query: 311 KGKVLEEEIRDLAEERNELT---------EKKRRPLYRKGFDV-KLINVNRTCKVTK 357
G + + EE NE+T + +RR +Y + + + + N+ ++T+
Sbjct: 140 PGLSIGSALDKWVEEGNEITRVEIAKAMLQLRRRRMYGRALQMSEWLEANKKIEMTE 196
>At5g47820 kinesin-like protein
Length = 1035
Score = 37.0 bits (84), Expect = 0.025
Identities = 29/109 (26%), Positives = 56/109 (50%), Gaps = 14/109 (12%)
Query: 232 DNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLD 291
D G ++ + L + +++++ + + T GD+ + D+E + + K N + +L
Sbjct: 456 DIVGSVRPDGLKRSLHSIESSNYPMVEATT---GDSREIDEEAKEWEHKLLQNSMDKELY 512
Query: 292 AIDKKLEEKLAE-----------LEYTFGKKGKVLEEEIRDLAEERNEL 329
++++LEEK +E L+ FGKK +E+E R + EERN L
Sbjct: 513 ELNRRLEEKESEMKLFDGYDPAALKQHFGKKIAEVEDEKRSVQEERNRL 561
>At3g26580 unknown protein
Length = 350
Score = 36.6 bits (83), Expect = 0.033
Identities = 24/80 (30%), Positives = 44/80 (55%), Gaps = 9/80 (11%)
Query: 257 EKQTIQGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLE 316
++++ G G D+++E+E+ D+ EK+ AK K+LE K EL+Y ++G E
Sbjct: 91 DEESSDGGGGGGDEEEEEEYFDV-EKERKRRAKEFHDTKELERKAEELQYKIDEEGDDSE 149
Query: 317 E--------EIRDLAEERNE 328
E E++ +A+E+ E
Sbjct: 150 EKKRMRVKRELQKVAQEQAE 169
>At1g32830 putaive protein on FARE2.4 (cds2)
Length = 707
Score = 35.8 bits (81), Expect = 0.056
Identities = 41/217 (18%), Positives = 94/217 (42%), Gaps = 11/217 (5%)
Query: 60 VQDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMR 119
++ ++ Q K + ++ G I +G+E+ G EKLE+E +E E +
Sbjct: 367 IETVVHQSRKRKSMSFGKKATKGQKNKRIPKQGDEEMEGEE---EKLEEEGKEEEEEKVE 423
Query: 120 Y-EEPTDSDSDEDEYEASQKKFDDFERKFKRHQDLLKNFTDADTLDDAFKWMNRIDKFED 178
Y + + + +E E + + K+ D+ + +R + + + D +D K N + E
Sbjct: 424 YRDHHSTCNVEETEKQENPKQCDE---EMEREEGKEEKVEEHDEYNDVLKEENVKEHDEH 480
Query: 179 KHFRLRPEYRVIGELMNRLKVVTDQKDKFILQNKLNRALR---LVQWKEAFDPDNPDNYG 235
+ EY ++ + N T +K+ L+ K + + + E + ++ + Y
Sbjct: 481 DEIEDQEEYAILSDDENNGTAPT-EKESHPLKEKTTEVPKEETVEEHDEHDETEDQEAYV 539
Query: 236 VIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDD 272
++ ++ T+ + + +KE+ T + DD+D
Sbjct: 540 ILSDDEDNGTAPTEKESQPQKEETTEVPRETKKDDED 576
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,426,342
Number of Sequences: 26719
Number of extensions: 534879
Number of successful extensions: 3701
Number of sequences better than 10.0: 193
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 156
Number of HSP's that attempted gapping in prelim test: 3247
Number of HSP's gapped (non-prelim): 409
length of query: 499
length of database: 11,318,596
effective HSP length: 103
effective length of query: 396
effective length of database: 8,566,539
effective search space: 3392349444
effective search space used: 3392349444
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0253.6


