

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.14
(346 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
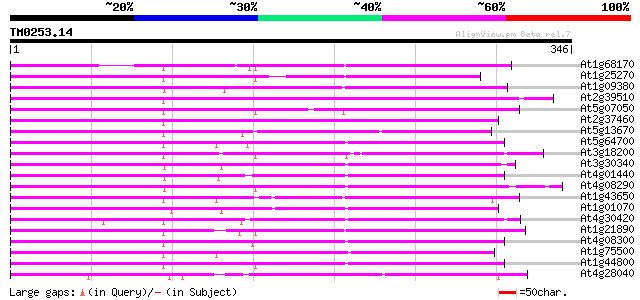
Score E
Sequences producing significant alignments: (bits) Value
At1g68170 MtN21-like protein 223 2e-58
At1g25270 220 1e-57
At1g09380 nodulin N21 like protein 220 1e-57
At2g39510 nodulin-like protein 207 7e-54
At5g07050 MtN21 nodulin protein-like 203 1e-52
At2g37460 nodulin-like protein 203 1e-52
At5g13670 unknown protein 202 2e-52
At5g64700 nodulin-like protein 186 1e-47
At3g18200 integral membrane protein, putative 186 2e-47
At3g30340 unknown protein 184 8e-47
At4g01440 predicted protein of unknown function 182 3e-46
At4g08290 nodulin-like protein 180 9e-46
At1g43650 nodulin-like protein 179 3e-45
At1g01070 unknown protein 177 1e-44
At4g30420 nodulin-like protein 175 3e-44
At1g21890 nodulin-like protein 174 5e-44
At4g08300 nodulin-like protein 174 8e-44
At1g75500 nodulin-like protein 172 2e-43
At1g44800 nodulin like protein 172 3e-43
At4g28040 Medicago nodulin N21-like protein 169 2e-42
>At1g68170 MtN21-like protein
Length = 329
Score = 223 bits (567), Expect = 2e-58
Identities = 126/331 (38%), Positives = 188/331 (56%), Gaps = 45/331 (13%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV++Q AG++I FKL GM+ +VL+AYR F++ FM+P+ + + P I
Sbjct: 1 MVVVQIATAGLNIFFKLAMEDGMNPSVLVAYRLLFATLFMIPICFIFQSVVIPSI----- 55
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
L + G+AL AT+ +A +E + + + G
Sbjct: 56 ----------------LTITGLALTSATFTSAAGVLTPLVTFIFAALLRMESVRLGSSVG 99
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNA---ATSH---IFGTSLAF 158
AKV G L GV GA++ FYR IEI LW T VNL+ N+P+++ AT+H I G L F
Sbjct: 100 LAKVFGTLFGVGGALVFIFYRGIEIRLWSTHVNLV-NQPRDSSRDATTHHISILGALLVF 158
Query: 159 GTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIK 218
G +S S+W ++Q ++S +F Y +A LM++M + + + A+C E D W+LGWNI+
Sbjct: 159 GGNISISLWFLLQVKISKQFGGPYWNATLMNMMGGVVAMLVALCWEHD-LDEWRLGWNIR 217
Query: 219 LFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVI 278
L T Y ++ SG+ + AW + +GPL+ S F+P+ LVIVA+ GS LDE L+LGS+I
Sbjct: 218 LLTIAYAAILISGMVVAVNAWCIESRGPLFVSVFSPVGLVIVALVGSFLLDETLHLGSII 277
Query: 279 GALLIVLGLYIVLWGKGKELKSNVEQKHKND 309
G ++IV LYIVLW K KE+KS + N+
Sbjct: 278 GTVIIVGALYIVLWAKNKEMKSMLTTSDHNE 308
>At1g25270
Length = 356
Score = 220 bits (560), Expect = 1e-57
Identities = 117/310 (37%), Positives = 185/310 (58%), Gaps = 31/310 (10%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV +QFIFAG+ ILFK+ G +L VL+AYR F++ FM+PLA +R+++P+ T ++L
Sbjct: 7 MVAVQFIFAGMFILFKITVDDGTNLKVLVAYRLSFATIFMLPLALIFQRKKRPEFTWRLL 66
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
AF+ GL GA I L++ G+A AT++ A +E L + + G
Sbjct: 67 LLAFVSGLLGAAIPNILYLPGMARTSATFSAASSIISPLITLVLGLVFRMETLRLGSNEG 126
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAATSH----IFGTSLAFGT 160
+AK++G L+G GA++ FY+ IEIH+W T V+L+K AT++ I G + G+
Sbjct: 127 RAKLVGTLLGACGALVFVFYKGIEIHIWSTHVDLLKGSHTGRATTNHHVSILGVLMVLGS 186
Query: 161 CLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLF 220
A++ + Y + +LM+ + + I A+C + D W +W+LGW+I L
Sbjct: 187 ----------NAKIGKELGGLYWNTSLMNGVGSLVCVIIALCSDHD-WEQWQLGWDINLL 235
Query: 221 TAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGA 280
+Y G+V SG+ L AW + KGPL+ + F+P+ LVIVA+ GS L+E L+LGS+IGA
Sbjct: 236 ATLYSGIVVSGMVVPLVAWCIATKGPLFVTVFSPIRLVIVALIGSFALEEPLHLGSIIGA 295
Query: 281 LLIVLGLYIV 290
+++V G+Y+V
Sbjct: 296 MIMVGGVYLV 305
>At1g09380 nodulin N21 like protein
Length = 374
Score = 220 bits (560), Expect = 1e-57
Identities = 123/329 (37%), Positives = 185/329 (55%), Gaps = 23/329 (6%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MVL+Q +AG++I K+ GM +L+AYR F++ P+A+F+ER+ +PKIT+++L
Sbjct: 12 MVLVQIGYAGMNITSKMAMEAGMKPLILVAYRQIFATIATFPVAFFLERKTRPKITLRIL 71
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAML----------------ERLNIKTKTG 104
Q F C + GAT Q L+ G+ + T A A+ E + IK +G
Sbjct: 72 VQVFFCSITGATGNQVLYFVGLQNSSPTIACALTNLLPAVTFLLAAIFRQETVGIKKASG 131
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHL------WPTIVNLMKNKPKNAATSHIFGTSLAF 158
+AKV+G LV V GAM+L+FY I + W N+ K+ + ++ G L
Sbjct: 132 QAKVIGTLVCVIGAMVLSFYHGHTIGIGESKIHWAYAENITKHGSSSGHSNFFLGPFLIM 191
Query: 159 GTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIK 218
+S++ W IIQ +MS F YTS LM +M IQ A+ + S W L ++
Sbjct: 192 AAAVSWAAWFIIQTKMSETFAAPYTSTLLMCLMGSIQCGAIALISDH-TISDWSLSSPLR 250
Query: 219 LFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVI 278
+A+Y GVVAS + + L +W ++ KGPLY S F+PL LV+VAI L+EKLY G+ +
Sbjct: 251 FISALYAGVVASALAFCLMSWAMQRKGPLYVSVFSPLLLVVVAIFSWALLEEKLYTGTFM 310
Query: 279 GALLIVLGLYIVLWGKGKELKSNVEQKHK 307
G+ L+V+GLY VLWGK +E+ E++ K
Sbjct: 311 GSALVVIGLYGVLWGKDREVSEKEEEREK 339
>At2g39510 nodulin-like protein
Length = 374
Score = 207 bits (527), Expect = 7e-54
Identities = 126/355 (35%), Positives = 200/355 (55%), Gaps = 23/355 (6%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
+V +QF +AG+SI+ K + GMS VL +YR+ ++ F+ P AYF++R+ +PK+T+ +
Sbjct: 12 VVSLQFGYAGLSIIAKFALNQGMSPHVLASYRHIVATIFIAPFAYFLDRKIRPKMTLSIF 71
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
F+ L GL TI QNL+ G+ AT+ AM LE++N+K
Sbjct: 72 FKILLLGLLEPTIDQNLYYTGMKYTSATFTAAMTNVLPAFAFIMAWIFRLEKVNVKKIHS 131
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHL-WPTIVNLMKNKPKNAATSHIF-GTSLAFGTCL 162
+AK+LG +V V GAM++T + I L W ++ ++ + G SL C+
Sbjct: 132 QAKILGTIVTVGGAMLMTVVKGPLIPLPWANPHDIHQDSSNTGVKQDLTKGASLIAIGCI 191
Query: 163 SYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFTA 222
++ ++ +QA +P + A + + I+STI A+ +ER N S W + + KL A
Sbjct: 192 CWAGFINLQAITLKSYPVELSLTAYICFLGSIESTIVALFIERGNPSAWAIHLDSKLLAA 251
Query: 223 VYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGALL 282
VY GV+ SGI + + +++ +GP++ ++FNPL +VIVAI GS+ L E ++LG ++GA++
Sbjct: 252 VYGGVICSGIGYYVQGVIMKTRGPVFVTAFNPLSMVIVAILGSIILAEVMFLGRILGAIV 311
Query: 283 IVLGLYIVLWGKGK-ELKSNVEQKHKNDPLEEVEPLEIVTTKQINGK-SADDKSV 335
IVLGLY VLWGK K E S+ K PL +IV + N K +D SV
Sbjct: 312 IVLGLYSVLWGKSKDEPSSSFSDMDKELPLSTP---QIVLPSKANAKMDTNDASV 363
>At5g07050 MtN21 nodulin protein-like
Length = 381
Score = 203 bits (517), Expect = 1e-52
Identities = 116/342 (33%), Positives = 194/342 (55%), Gaps = 31/342 (9%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
M+ +QF +AG++I+ K+ +TGMS VL+ YR+ ++A + P A+F ER+ +PKIT +
Sbjct: 1 MISLQFGYAGMNIITKISLNTGMSHYVLVVYRHAIATAVIAPFAFFFERKAQPKITFSIF 60
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
Q F+ GL G I QN + G+ T++ AM +E L++K
Sbjct: 61 MQLFILGLLGPVIDQNFYYMGLKYTSPTFSCAMSNMLPAMTFILAVLFRMEMLDLKKLWC 120
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHL-WPTIVNLMKNKPKNAATSH--------IFGTS 155
+AK+ G +V V+GAM++T Y+ + L W +++ + N +S + G+
Sbjct: 121 QAKIAGTVVTVAGAMLMTIYKGPIVELFWTKYMHIQDSSHANTTSSKNSSSDKEFLKGSI 180
Query: 156 LAFGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCME---RDNWSRWK 212
L L+++ ++QA++ + H S ++ + C T+ AV + N S W+
Sbjct: 181 LLIFATLAWASLFVLQAKILKTYAKHQLS---LTTLICFIGTLQAVAVTFVMEHNPSAWR 237
Query: 213 LGWNIKLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKL 272
+GW++ L A Y G+VAS I + + V++ +GP++A++F+PL +VIVA+ GS L EK+
Sbjct: 238 IGWDMNLLAAAYSGIVASSISYYVQGIVMKKRGPVFATAFSPLMMVIVAVMGSFVLAEKI 297
Query: 273 YLGSVIGALLIVLGLYIVLWGKGKELKSNVEQKHKNDPLEEV 314
+LG VIGA+LIV+GLY VLWGK KE + + + K D +V
Sbjct: 298 FLGGVIGAVLIVIGLYAVLWGKQKENQVTICELAKIDSNSKV 339
>At2g37460 nodulin-like protein
Length = 380
Score = 203 bits (516), Expect = 1e-52
Identities = 113/317 (35%), Positives = 179/317 (55%), Gaps = 16/317 (5%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV++Q AG+ IL K V + GMS VL+ YR+ ++ M P A++ +++ +PK+T+ +
Sbjct: 19 MVVLQVGLAGMDILSKAVLNKGMSNYVLVVYRHAVATIVMAPFAFYFDKKVRPKMTLMIF 78
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
F+ L GL I QNL+ G+ AT+ATAM LER+ ++
Sbjct: 79 FKISLLGLLEPVIDQNLYYLGMKYTTATFATAMYNVLPAITFVLAYIFGLERVKLRCIRS 138
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAATSHIFGTSLAFGTCLSY 164
KV+G L V GAMI+T + + L+ T N S I G L C SY
Sbjct: 139 TGKVVGTLATVGGAMIMTLVKGPVLDLFWTKGVSAHNTAGTDIHSAIKGAVLVTIGCFSY 198
Query: 165 SIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFTAVY 224
+ ++I+QA +P + A + +M I+ T A+ ME+ N S W +GW+ KL TA Y
Sbjct: 199 ACFMILQAITLRTYPAELSLTAWICLMGTIEGTAVALVMEKGNPSAWAIGWDTKLLTATY 258
Query: 225 VGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGALLIV 284
G+V S + + + V++ +GP++ ++F+PL ++IVAI ++ E++YLG V+GA++I
Sbjct: 259 SGIVCSALAYYVGGVVMKTRGPVFVTAFSPLCMIIVAIMSTIIFAEQMYLGRVLGAVVIC 318
Query: 285 LGLYIVLWGKGKELKSN 301
GLY+V+WGKGK+ K N
Sbjct: 319 AGLYLVIWGKGKDYKYN 335
>At5g13670 unknown protein
Length = 377
Score = 202 bits (515), Expect = 2e-52
Identities = 110/320 (34%), Positives = 190/320 (59%), Gaps = 26/320 (8%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
+V IQ ++A +SI+ KL + GMS VL+AYR +SA + P A +ER +PK+T K+L
Sbjct: 12 IVFIQCLYALMSIVAKLALNKGMSPHVLVAYRMAVASALITPFALILERNTRPKLTFKIL 71
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
Q + LF ++QNL+ G+ L AT+ +A+ LE++ I+ +
Sbjct: 72 LQIAILSLFEPVVEQNLYYSGMKLTTATFTSALCNALPAMTFIMACVFKLEKVTIERRHS 131
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHL-WPTIVNLMKNK------PKNAATSHIFGTSLA 157
+AK++G +V + GAM++TF + I L W + + PK A + G+ +
Sbjct: 132 QAKLVGTMVAIGGAMLMTFVKGNVIELPWTSNSRGLNGHTHAMRIPKQADIAR--GSIML 189
Query: 158 FGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNI 217
+C S+S ++I+QA++ A++ + ALM +M +++T+ + ER N S WK+ ++
Sbjct: 190 VASCFSWSCYIILQAKILAQYKAELSLTALMCIMGMLEATVMGLIWERKNMSVWKINPDV 249
Query: 218 KLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSV 277
L ++Y G+V SG+ + + W + +GP++ S+FNPL +V+VAI + EK+Y+G V
Sbjct: 250 TLLASIYGGLV-SGLAYYVIGWASKERGPVFVSAFNPLSMVLVAILSTFVFLEKVYVGRV 308
Query: 278 IGALLIVLGLYIVLWGKGKE 297
IG+++IV+G+Y+VLWGK K+
Sbjct: 309 IGSVVIVIGIYLVLWGKSKD 328
>At5g64700 nodulin-like protein
Length = 359
Score = 186 bits (473), Expect = 1e-47
Identities = 102/330 (30%), Positives = 182/330 (54%), Gaps = 26/330 (7%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
+ +IQ I+ + ++ K V + GM+ V + YR F++ F+ PLA+F ER+ P ++
Sbjct: 12 VTIIQVIYTIMFLISKAVFNGGMNTFVFVFYRQAFATIFLAPLAFFFERKSAPPLSFVTF 71
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
+ F+ LFG T+ +L ++ AT A A +ERL +K+ G
Sbjct: 72 IKIFMLSLFGVTLSLDLNGIALSYTSATLAAATTASLPAITFFLALLFGMERLKVKSIQG 131
Query: 105 KAKVLGPLVGVSGAMILTFYRS--IEIHLWPTIVNLMKNKPKN-------AATSHIFGTS 155
AK++G V + G +IL Y+ +++ L P + ++ +N +TS + G
Sbjct: 132 TAKLVGITVCMGGVIILAIYKGPLLKLPLCPHFYHGQEHPHRNNPGHVSGGSTSWLKGCV 191
Query: 156 LAFGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGW 215
L + + + +WL++Q R+ +P L +++ IQS + A+ +ERD S WKLGW
Sbjct: 192 LMITSNILWGLWLVLQGRVLKVYPSKLYFTTLHCLLSSIQSFVIAIALERDI-SAWKLGW 250
Query: 216 NIKLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLG 275
N++L +Y G + +G+ + L +WV+ +GP++ S F PL L+ ++ ++ L E + LG
Sbjct: 251 NLRLVAVIYCGFIVTGVAYYLQSWVIEKRGPVFLSMFTPLSLLFTLLSSAILLCEIISLG 310
Query: 276 SVIGALLIVLGLYIVLWGKGKELKSNVEQK 305
S++G LL+++GLY VLWGK +E K++ + K
Sbjct: 311 SIVGGLLLIIGLYCVLWGKSREEKNSGDDK 340
>At3g18200 integral membrane protein, putative
Length = 360
Score = 186 bits (471), Expect = 2e-47
Identities = 113/349 (32%), Positives = 183/349 (52%), Gaps = 26/349 (7%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
++ +QF FAG I+ ++ + G+S V YR + + P AYF E++ +P +T+ +L
Sbjct: 16 LITLQFCFAGFHIVSRVALNIGVSKVVYPVYRNLLALLLIGPFAYFFEKKERPPLTISLL 75
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
Q F L G T Q ++ G+ A T+A+AM LE +++ K G
Sbjct: 76 AQFFFLALIGITANQGFYLLGLYYATPTFASAMQNSVPAITFIMACALRLEHIDLVRKHG 135
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAATSH--IFGTSLAFGTCL 162
AKVLG LV + GA ++T YR I + +N+ K + + SH G G CL
Sbjct: 136 VAKVLGTLVSIGGATVITLYRGFPI--FDQGLNMQKEEVVGSDNSHSLTLGWLYLMGHCL 193
Query: 163 SYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERD--NWSRWKLGWNIKLF 220
S++ W+++QA + ++P T + IQ + A+ +E D NW + W +LF
Sbjct: 194 SWAGWMVLQAPVLKQYPAKLTLTSFTCFFGLIQFLVIALFVETDLNNWII--VSWE-ELF 250
Query: 221 TAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGA 280
T +Y G++ASG+ L W + GP++ + F PL ++VA L L ++LY G ++GA
Sbjct: 251 TILYAGIIASGLVVYLQTWCIYKSGPVFVAVFQPLQTLLVAAMAFLILGDQLYSGGIVGA 310
Query: 281 LLIVLGLYIVLWGKGKELKSNVEQKHKNDPLEEVEPLEIVTTKQINGKS 329
+ I+LGLY+VLWGK +E K +E+ + DP + L K+ N +S
Sbjct: 311 VFIMLGLYLVLWGKNEERKLALEES-QQDPESLTKHLLEAQHKKSNSES 358
>At3g30340 unknown protein
Length = 364
Score = 184 bits (466), Expect = 8e-47
Identities = 103/331 (31%), Positives = 176/331 (53%), Gaps = 23/331 (6%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
M +I + V+++FK + G++ V YR + F++P A F+ER +PK+T ++L
Sbjct: 15 MSMINIGLSVVNVMFKKMIDEGLNRMVATTYRLAVGTLFLIPFAIFLERHNRPKLTGRIL 74
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAML----------------ERLNIKTKTG 104
F L G ++ Q F+ G+ +T++ A E LNIK+ G
Sbjct: 75 CSLFFSALLGTSLVQYFFLIGLEYTSSTFSLAFSNMVPSVTFALALVFRQETLNIKSNVG 134
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEI---HLWPTIVNLMKNKPKNAATSHIFGTSLAFGTC 161
+AK+LG ++ + GA++LT Y+ + H + + G+ + +
Sbjct: 135 RAKLLGTMICICGALVLTLYKGTALSREHSTHMETHTRTDSTGAMTQKWAMGSIMLVISI 194
Query: 162 LSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFT 221
+ +S W I+QA++S +P YTS ++S IQS + ++ ER S W + ++
Sbjct: 195 IIWSSWFIVQAKISRVYPCQYTSTTILSFFGVIQSALLSLISERST-SMWVVKDKFQVLA 253
Query: 222 AVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGAL 281
+Y G+V SG+ +V +W LR +G ++ SSF PL V AI FL E++Y GSVIG++
Sbjct: 254 LLYSGIVGSGLCYVGMSWCLRQRGAVFTSSFIPLIQVFAAIFSFSFLHEQIYCGSVIGSM 313
Query: 282 LIVLGLYIVLWGKGKELKSNVEQKHKNDPLE 312
+I++GLYI+LWGK K+ ++V K +PL+
Sbjct: 314 VIIVGLYILLWGKSKDKSASVT---KQEPLD 341
>At4g01440 predicted protein of unknown function
Length = 365
Score = 182 bits (461), Expect = 3e-46
Identities = 105/327 (32%), Positives = 173/327 (52%), Gaps = 27/327 (8%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV+I + L K V G++ V+ YR S+ F+ P+A+F ER+ +P +T+ +L
Sbjct: 14 MVMINSALGLANALVKKVLDGGVNHMVIATYRLAISTLFLAPIAFFWERKTRPTLTLNIL 73
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAML----------------ERLNIKTKTG 104
Q F L GA++ Q F+ G++ AT A A + E+LN+K+K G
Sbjct: 74 VQLFFSALVGASLTQYFFLLGLSYTSATLACAFISMTPAITFVMALIFRVEKLNMKSKAG 133
Query: 105 KAKVLGPLVGVSGAMILTFYRSI------EIHLWPTIVNLMKNKPKNAATSHIFGTSLAF 158
V+G L+ + GA++LT Y+ + ++ I N KP+N I G L F
Sbjct: 134 MGMVMGALICIGGALLLTMYKGVPLTKLRKLETHQLINNNHAMKPEN----WIIGCVLLF 189
Query: 159 GTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIK 218
+ W++IQA+++ K+P Y+S ++S IQ + ++ RD + W L +
Sbjct: 190 AGSSCFGSWMLIQAKVNEKYPCQYSSTVVLSFFGTIQCALLSLIKSRDI-TAWILTDKLD 248
Query: 219 LFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVI 278
+ T VY G VA GI V T+W +R +GP++ S F P+ L+ + L L +++LGSV+
Sbjct: 249 IVTIVYAGAVAQGICTVGTSWCIRKRGPIFTSIFTPVGLIFATLFDFLILHRQIFLGSVV 308
Query: 279 GALLIVLGLYIVLWGKGKELKSNVEQK 305
G+ +++ GLYI L GK + +K E+K
Sbjct: 309 GSGVVIFGLYIFLLGKVRLMKEECEKK 335
>At4g08290 nodulin-like protein
Length = 384
Score = 180 bits (457), Expect = 9e-46
Identities = 107/362 (29%), Positives = 193/362 (52%), Gaps = 28/362 (7%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
M+ +QF AG I+ + G + V++ YR ++ + P A ER+ +PK+T+ VL
Sbjct: 17 MIFLQFGAAGTYIVIMATLNQGQNRYVVIVYRNLVAALVLAPFALIFERKVRPKMTLSVL 76
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAML----------------ERLNIKTKTG 104
++ G + Q G+ + ATY +A++ E++NI
Sbjct: 77 WKIMALGFLEPVLDQGFGYLGMNMTSATYTSAIMNILPSVTFIIAWILRMEKVNIAEVRS 136
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHL-WPTIVNLMKNKPKNAATSH---IFGTSLAFGT 160
KAK++G LVG+ GA+++T Y+ I L W +N N + H + GT L
Sbjct: 137 KAKIIGTLVGLGGALVMTLYKGPLIPLPWSNPNMDQQNGHTNNSQDHNNWVVGTLLILLG 196
Query: 161 CLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLF 220
C+++S + ++Q+ +P + +AL+ + +QS A+ +ER S W +GW+ +LF
Sbjct: 197 CVAWSGFYVLQSITIKTYPADLSLSALICLAGAVQSFAVALVVERHP-SGWAVGWDARLF 255
Query: 221 TAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGA 280
+Y G+V+SGI + + V++ +GP++ ++FNPL +++VA+ S L E+++ G VIG
Sbjct: 256 APLYTGIVSSGITYYVQGMVMKTRGPVFVTAFNPLCMILVALIASFILHEQIHFGCVIGG 315
Query: 281 LLIVLGLYIVLWGKGKELK-SNVEQKHKNDPLEEVEPLEIVTTKQINGKSADDKSVDDGN 339
+I GLY+V+WGKGK+ + S ++ KN ++ L I T + + K S+ D +
Sbjct: 316 AVIAAGLYMVVWGKGKDYEVSGLDILEKN----SLQELPITTKSEDDNKLV--SSISDNS 369
Query: 340 DI 341
++
Sbjct: 370 NV 371
>At1g43650 nodulin-like protein
Length = 341
Score = 179 bits (453), Expect = 3e-45
Identities = 107/335 (31%), Positives = 178/335 (52%), Gaps = 26/335 (7%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV +Q ++AG+ +L K+ S G + V + YR F++ + P A+F+E + ++ +L
Sbjct: 9 MVFVQIVYAGMPLLSKVAISQGTNPFVFVFYRQAFAALALSPFAFFLESSKSSPLSFILL 68
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
+ F L G T+ NL+ + AT+A A LE + +K G
Sbjct: 69 LKIFFISLCGLTLSLNLYYVAIENTTATFAAATTNAIPSITFVLALLFRLETVTLKKSHG 128
Query: 105 KAKVLGPLVGVSGAMILTFYRS---IEIHLWPTIVNLMKNKPKNAATSHIFGTSLAFGTC 161
AKV G +VG+ GA++ F + I + TI N KN+ I T LA TC
Sbjct: 129 VAKVTGSMVGMLGALVFAFVKGPSLINHYNSSTIPNGTVPSTKNSVKGSI--TMLAANTC 186
Query: 162 LSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFT 221
+ +W+I+Q+++ ++P AL + +CIQS ++AV + R N S WK+ + + L +
Sbjct: 187 --WCLWIIMQSKVMKEYPAKLRLVALQCLFSCIQSAVWAVAVNR-NPSVWKIEFGLPLLS 243
Query: 222 AVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGAL 281
Y G++ +G+ + L W + KGP++ + + PL L++ I S E YLGSV GA+
Sbjct: 244 MAYCGIMVTGLTYWLQVWAIEKKGPVFTALYTPLALILTCIVSSFLFKETFYLGSVGGAV 303
Query: 282 LIVLGLYIVLWGKGK--ELKSNVEQKHKNDPLEEV 314
L+V GLY+ LWGK K E++ E++ + + +EEV
Sbjct: 304 LLVCGLYLGLWGKTKEEEIQRYGEKQSQKEIIEEV 338
>At1g01070 unknown protein
Length = 365
Score = 177 bits (448), Expect = 1e-44
Identities = 107/322 (33%), Positives = 169/322 (52%), Gaps = 23/322 (7%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
MV+ V+ L K G++ V+ AYR S+ +VP AY +ER+ +P+IT +++
Sbjct: 20 MVMSNVAMGSVNALVKKALDVGVNHMVIGAYRMAISALILVPFAYVLERKTRPQITFRLM 79
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAMLERLN-----------------IKTKT 103
F+ GL GA++ Q F+ G++ AT + A++ L +KTK
Sbjct: 80 VDHFVSGLLGASLMQFFFLLGLSYTSATVSCALVSMLPAITFALALIFRTENVKILKTKA 139
Query: 104 GKAKVLGPLVGVSGAMILTFYRSIEI---HLWPTIVNLMKNKPKNAATSHIFGTS-LAFG 159
G KV+G L+ +SGA+ LTFY+ +I H N ++ A + + G L G
Sbjct: 140 GMLKVIGTLICISGALFLTFYKGPQISNSHSHSHGGASHNNNDQDKANNWLLGCLYLTIG 199
Query: 160 TCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKL 219
T L S+W++ Q +S K+P Y+S LMS+ A Q + ++ RD + W + +
Sbjct: 200 TVL-LSLWMLFQGTLSIKYPCKYSSTCLMSIFAAFQCALLSLYKSRDV-NDWIIDDRFVI 257
Query: 220 FTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIG 279
+Y GVV + V T W ++ G ++AS+F PL L+ + L L LYLGSVIG
Sbjct: 258 TVIIYAGVVGQAMTTVATTWGIKKLGAVFASAFFPLTLISATLFDFLILHTPLYLGSVIG 317
Query: 280 ALLIVLGLYIVLWGKGKELKSN 301
+L+ + GLY+ LWGK KE +S+
Sbjct: 318 SLVTITGLYMFLWGKNKETESS 339
>At4g30420 nodulin-like protein
Length = 373
Score = 175 bits (444), Expect = 3e-44
Identities = 109/339 (32%), Positives = 182/339 (53%), Gaps = 29/339 (8%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKIT---M 57
M +IQ +AGV++ + G+S V + YR F++ F+ P Y R+ K I+ +
Sbjct: 3 MTMIQLCYAGVTLFARATLVHGLSPRVFILYRQAFATIFIFPFLYLSRRKSKIAISSLDL 62
Query: 58 KVLFQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKT 101
K FL L G TI QNL++EG+ L ++ +A+ E+LN++
Sbjct: 63 KSFSLIFLVSLIGITINQNLYLEGLYLTSSSMGSAVGNIIPAITFLISFLAGYEKLNLRD 122
Query: 102 KTGKAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKN-----KPKNAATSHIFGTSL 156
G AK+ G ++ V+GA+ +T R +I + + + K+ K +N + + G
Sbjct: 123 IRGLAKIAGTILCVAGAISMTLLRGPKILNSESALPIAKSVLGHLKDQN---TWLIGCLF 179
Query: 157 AFGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWN 216
F + L +S WLI+Q +SA +P + + +A M + IQ + +E+D + W L
Sbjct: 180 LFSSTLCWSFWLILQVPISAYYPDNLSLSAWMCLFGTIQCAVVTFFLEKDP-NAWILHSY 238
Query: 217 IKLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGS 276
+ T +Y G+ AS + + + AW + +GP++++ FNPL VIV I +LF E++Y GS
Sbjct: 239 SEFATCLYAGIGASALSFTVQAWAIAKRGPVFSALFNPLCTVIVTILAALFFHEEIYTGS 298
Query: 277 VIGALLIVLGLYIVLWGKGKELKSNVEQKHKNDPLEEVE 315
+IG L ++LGLY VLWGK K++ N +Q+ ND EV+
Sbjct: 299 LIGGLGVILGLYTVLWGKAKDVMMNQDQR-DNDQKSEVK 336
>At1g21890 nodulin-like protein
Length = 389
Score = 174 bits (442), Expect = 5e-44
Identities = 110/354 (31%), Positives = 188/354 (53%), Gaps = 44/354 (12%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
M+ +QF +AG+ I+ + GM+ VL YR+ ++A + P A F ER+ +PK+T ++
Sbjct: 15 MISMQFGYAGMYIITMVSLKHGMNHYVLAVYRHAIATAVIAPFALFHERKIRPKMTFRIF 74
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
Q L G + QNL+ G+ AT+A+A LE +N K
Sbjct: 75 LQIALLGFIEPVLDQNLYYVGMTYTSATFASATANVLPAITFVLAIIFRLESVNFKKVRS 134
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMK------NKPKNAATSH-------- 150
AKV+G ++ VSGA+++T Y+ IV+ ++ A SH
Sbjct: 135 IAKVVGTVITVSGALLMTLYKG-------PIVDFIRFGGGGGGGSDGAGGSHGGAGAAAM 187
Query: 151 ----IFGTSLAFGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERD 206
I GT + G ++ + I+Q+ ++P + L+ +M ++ T ++ RD
Sbjct: 188 DKHWIPGTLMLLGRTFGWAGFFILQSFTLKQYPAELSLTTLICLMGTLEGTAVSLVTVRD 247
Query: 207 NWSRWKLGWNIKLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSL 266
S WK+G++ LF A Y GV+ SG+ + + V+R +GP++ ++FNPL +VI A G +
Sbjct: 248 -LSAWKIGFDSNLFAAAYSGVICSGVAYYVQGVVMRERGPVFVATFNPLCVVITAALGVV 306
Query: 267 FLDEKLYLGSVIGALLIVLGLYIVLWGKGKELK-SNVEQKHKNDPLEE-VEPLE 318
L E ++LGSVIG L I++GLY V+WGKGK+ + ++ ++ K P++ V+P++
Sbjct: 307 VLSESIHLGSVIGTLFIIVGLYTVVWGKGKDKRMTDDDEDCKGLPIKSPVKPVD 360
>At4g08300 nodulin-like protein
Length = 373
Score = 174 bits (440), Expect = 8e-44
Identities = 100/326 (30%), Positives = 176/326 (53%), Gaps = 22/326 (6%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
++ +QF +AG+ I+ + GM+ +L YR+ ++ + P A +ER+ +PK+T +
Sbjct: 15 IISLQFGYAGMYIITMVSFKHGMNHWILATYRHVVATIVIAPFALILERKIRPKMTWPLF 74
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
+ G + QNL+ G+ ATY++A +E +N+K
Sbjct: 75 LRILALGFLEPLLDQNLYYIGMKATSATYSSAFVNALPAITFIMAVIFRIETVNLKKTRS 134
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAAT-----SHIFGTSLAFG 159
AKV+G + V GAM++T Y+ I L+ T + + ++ + + GT G
Sbjct: 135 LAKVIGTAITVGGAMVMTLYKGPAIELFKTAHSSLHGGSSGTSSETTDQNWVTGTLAVMG 194
Query: 160 TCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKL 219
+ +++ + I+Q+ K+P + + M + +TI ++ M RD S WK+G +
Sbjct: 195 SITTWAGFFILQSFTLKKYPAELSLVMWICAMGTVLNTIASLIMVRDV-SAWKVGMDSGT 253
Query: 220 FTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIG 279
AVY GVV SG+ + + + V+R +GP++ +SF+P+ ++I A G L L EK++LGS+IG
Sbjct: 254 LAAVYSGVVCSGMAYYIQSIVIRERGPVFTTSFSPMCMIITAFLGVLVLAEKIHLGSIIG 313
Query: 280 ALLIVLGLYIVLWGKGKELKSNVEQK 305
A+ IV GLY V+WGK K+ +VE+K
Sbjct: 314 AIFIVFGLYSVVWGKAKDEVISVEEK 339
>At1g75500 nodulin-like protein
Length = 389
Score = 172 bits (436), Expect = 2e-43
Identities = 100/324 (30%), Positives = 165/324 (50%), Gaps = 26/324 (8%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
M+ +QF +AG ++ + + G+S V YR + ++P AYF+E++ +P IT+ L
Sbjct: 24 MLTLQFGYAGFHVVSRAALNMGISKLVFPVYRNIIALLLLLPFAYFLEKKERPAITLNFL 83
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
Q F L G T Q ++ G+ T+A++M +E++ I + G
Sbjct: 84 IQFFFLALIGITANQGFYLLGLDNTSPTFASSMQNSVPAITFLMAALLRIEKVRINRRDG 143
Query: 105 KAKVLGPLVGVSGAMILTFYRS---------IEIHLWPTIVNLMKNKPKNAATSHIFGTS 155
+K+LG + V+GA ++T Y+ + HL T ++ A + G
Sbjct: 144 ISKILGTALCVAGASVITLYKGPTIYTPASHLHAHLLTTNSAVLAPLGNAAPKNWTLGCI 203
Query: 156 LAFGTCLSYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGW 215
G CLS+S WL+ QA + +P + + IQ I A ERD+ + W
Sbjct: 204 YLIGHCLSWSGWLVFQAPVLKSYPARLSVTSYTCFFGIIQFLIIAAFCERDSQA-WVFHS 262
Query: 216 NIKLFTAVYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLG 275
+LFT +Y G+VASGI + + W + GP++ + + P+ ++VAI S+ L E+ YLG
Sbjct: 263 GWELFTILYAGIVASGIAFAVQIWCIDRGGPVFVAVYQPVQTLVVAIMASIALGEEFYLG 322
Query: 276 SVIGALLIVLGLYIVLWGKGKELK 299
+IGA+LI+ GLY VL+GK +E K
Sbjct: 323 GIIGAVLIIAGLYFVLYGKSEERK 346
>At1g44800 nodulin like protein
Length = 370
Score = 172 bits (435), Expect = 3e-43
Identities = 100/323 (30%), Positives = 173/323 (52%), Gaps = 19/323 (5%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFVERERKPKITMKVL 60
++ +QF +AG+ I+ + GM VL YR+ ++ M P A ER+ +PK+T+ +
Sbjct: 15 IISLQFGYAGMYIITMVSFKHGMDHWVLATYRHVVATVVMAPFALMFERKIRPKMTLAIF 74
Query: 61 FQAFLCGLFGATIQQNLFVEGVALAGATYATAM----------------LERLNIKTKTG 104
++ G+ + QNL+ G+ A+Y +A LE +N +
Sbjct: 75 WRLLALGILEPLMDQNLYYIGLKNTSASYTSAFTNALPAVTFILALIFRLETVNFRKVHS 134
Query: 105 KAKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAATSH--IFGTSLAFGTCL 162
AKV+G ++ V GAMI+T Y+ I + N + T + GT G+
Sbjct: 135 VAKVVGTVITVGGAMIMTLYKGPAIEIVKAAHNSFHGGSSSTPTGQHWVLGTIAIMGSIS 194
Query: 163 SYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFTA 222
+++ + I+Q+ +P + L+ + I + I ++ M RD S WK+G + A
Sbjct: 195 TWAAFFILQSYTLKVYPAELSLVTLICGIGTILNAIASLIMVRDP-SAWKIGMDSGTLAA 253
Query: 223 VYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGALL 282
VY GVV SGI + + + V++ +GP++ +SF+P+ ++I A G+L L EK++LGS+IGA+
Sbjct: 254 VYSGVVCSGIAYYIQSIVIKQRGPVFTTSFSPMCMIITAFLGALVLAEKIHLGSIIGAVF 313
Query: 283 IVLGLYIVLWGKGKELKSNVEQK 305
IVLGLY V+WGK K+ + +++K
Sbjct: 314 IVLGLYSVVWGKSKDEVNPLDEK 336
>At4g28040 Medicago nodulin N21-like protein
Length = 359
Score = 169 bits (428), Expect = 2e-42
Identities = 105/342 (30%), Positives = 183/342 (52%), Gaps = 33/342 (9%)
Query: 1 MVLIQFIFAGVSILFKLVASTGMSLTVLMAYRYFFSSAFMVPLAYFV--ERERKPKITMK 58
+V++QF AGV++ K G++ TV + YR ++ F+ P+++ +E KP + ++
Sbjct: 12 LVMLQFTSAGVALFTKAAFMEGLNPTVFVVYRQAIATLFICPISFISAWRKENKPSLGVR 71
Query: 59 VLFQAFLCGLFGATIQQNLFVEGVALAGATYATAMLERL--------------NIKTKTG 104
+ L + G T+ QN + +G+ L+ ++ A AM + +IK ++
Sbjct: 72 GFWWVALTAVIGVTVNQNAYFKGIDLSSSSMACAMTNLIPAVTFIISIIVGFESIKRRSM 131
Query: 105 K--AKVLGPLVGVSGAMILTFYRSIEIHLWPTIVNLMKNKPKNAATSHIFGTSLAFGTCL 162
K AKV+G V V GAM +TF R P ++N + N+ T+ + G +
Sbjct: 132 KSVAKVIGTGVCVGGAMAMTFLRG------PKLLNALLNQDN---TAWLLGCFFLLISTF 182
Query: 163 SYSIWLIIQARMSAKFPWHYTSAALMSVMACIQSTIFAVCMERDNWSRWKLGWNIKLFTA 222
++S+WLI+Q +++ P H ++A +A I S + A+ + + WKL +KL
Sbjct: 183 AWSLWLILQVPIASHCPDHLYTSACTCFIATIASFLVALALGNTHLPPWKLDSFLKLSCC 242
Query: 223 VYVGVVASGIPWVLTAWVLRLKGPLYASSFNPLFLVIVAIAGSLFLDEKLYLGSVIGALL 282
+Y G + I + L AW++ KGP++++ FNPL VIV G+L+L E+ YLGS++GAL
Sbjct: 243 IYSGFQLA-ISFFLQAWIVSQKGPVFSALFNPLSAVIVTFFGALYLKEQTYLGSLLGALA 301
Query: 283 IVLGLYIVLWGKGKELKS-----NVEQKHKNDPLEEVEPLEI 319
I+LGLYIVLWGK ++ + +E +H ++ + I
Sbjct: 302 IILGLYIVLWGKSEDYQEESTDLKLENEHNTSSQSDIVSIMI 343
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,164,597
Number of Sequences: 26719
Number of extensions: 286015
Number of successful extensions: 1196
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1019
Number of HSP's gapped (non-prelim): 68
length of query: 346
length of database: 11,318,596
effective HSP length: 100
effective length of query: 246
effective length of database: 8,646,696
effective search space: 2127087216
effective search space used: 2127087216
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0253.14


