

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
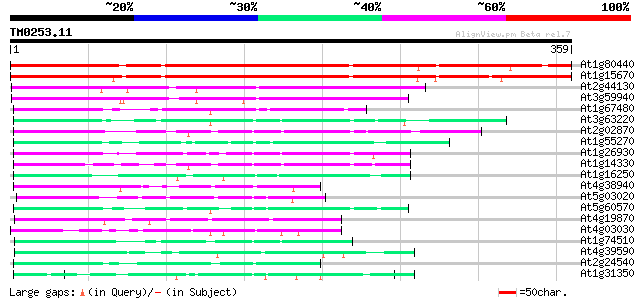
Query= TM0253.11
(359 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g80440 unknown protein 360 e-100
At1g15670 unknown protein 343 6e-95
At2g44130 unknown protein 164 5e-41
At3g59940 unknown protein 153 1e-37
At1g67480 unknown protein 74 1e-13
At3g63220 unknown protein 73 2e-13
At2g02870 unknown protein 73 2e-13
At1g55270 unknown protein (At1g55270) 71 8e-13
At1g26930 unknown protein 71 8e-13
At1g14330 unknown protein 69 5e-12
At1g16250 unknown protein 67 2e-11
At4g38940 unknown protein 61 1e-09
At5g03020 unknown protein 60 2e-09
At5g60570 putative protein 58 7e-09
At4g19870 unknown protein 55 6e-08
At4g03030 OR23peptide 54 1e-07
At1g74510 unknown protein 53 2e-07
At4g39590 unknown protein 53 3e-07
At2g24540 unknown protein 52 7e-07
At1g31350 unknown protein 52 7e-07
>At1g80440 unknown protein
Length = 354
Score = 360 bits (923), Expect = e-100
Identities = 176/366 (48%), Positives = 247/366 (67%), Gaps = 19/366 (5%)
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELI LP+DVAR+CL+R SY+QFP + + C+ WN E+ +F +R+ + ++Q++L+ +
Sbjct: 1 MELIPNLPDDVARECLLRSSYQQFPVIASVCRAWNREVSLSQFLHQRKASRHSQELLILS 60
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
QAR D G K P YR+SV E +G W+EL PP GLP+FC++ VG
Sbjct: 61 QARVDPAGSG----KIIATPEYRISVLESGSGLWTEL--PPIPGQTKGLPLFCRLVSVGS 114
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
+L+V+GG DP +W+A +SVF+++FL++ WR GA MPG R+FF C SD DRTV VAGGH+
Sbjct: 115 DLIVLGGLDPITWQAHDSVFVFSFLTSKWRVGATMPGVRRSFFGCASDSDRTVLVAGGHN 174
Query: 181 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
EEK AL SA+ YDV+ D+W LPDMARERDECKAVF AG+ V+GGY TE QG+F ++
Sbjct: 175 EEKCALTSAMVYDVSEDKWTFLPDMARERDECKAVFH--AGRFHVIGGYATEEQGQFSKT 232
Query: 241 AEEFDVDTWQWGPVEEEFLD---CGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKV 297
AE FDV TW+WGP+ E+FLD P TC+ GGD +Y C GGDV+ + WQ V ++
Sbjct: 233 AESFDVSTWEWGPLTEDFLDDTGDTVSPPTCVAGGD-LYACWGGDVMMFLNDKWQKVGQI 291
Query: 298 PSEIRNVACMGAWERSLLLIGS----SGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQS 353
P+++ NV + L++IG+ +G+GE +G++ DL S +W KL + + GHVQ+
Sbjct: 292 PADVYNVTYVAVRPGMLIVIGNGKALAGYGEATVGYICDLSSSRWVKL---ETHGGHVQA 348
Query: 354 YCLLEI 359
C LE+
Sbjct: 349 GCFLEV 354
>At1g15670 unknown protein
Length = 359
Score = 343 bits (881), Expect = 6e-95
Identities = 180/367 (49%), Positives = 244/367 (66%), Gaps = 16/367 (4%)
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELI LPE VA +CL+R SY+QFP + + CK W EI +F R R+ +G++Q+++V +
Sbjct: 1 MELIPDLPETVAYECLLRSSYKQFPLMASVCKLWQREISLSDFFRHRKASGHSQELVVLS 60
Query: 61 QARFD--SDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGV 118
QAR D + G K PVYR+SV E TG SEL PP +GLP+FC++ V
Sbjct: 61 QARVDPVKELVSGN--KTIPTPVYRISVLELGTGLRSEL--PPVPGHSNGLPLFCRLVSV 116
Query: 119 GYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG 178
G +LVV+ G DP +W+ S+SVF+++FL++TWR G MPGGPR+FFAC SD R V+VAGG
Sbjct: 117 GSDLVVLCGLDPVTWRTSDSVFVFSFLTSTWRVGKSMPGGPRSFFACASDSQRNVFVAGG 176
Query: 179 HDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFE 238
HDE+KNA+ SAL YDVA D W LPDM RERDEC A+F AGK V+GGY TE QG+F
Sbjct: 177 HDEDKNAMMSALVYDVAEDRWAFLPDMGRERDECTAIFH--AGKFHVIGGYSTEEQGQFS 234
Query: 239 RSAEEFDVDTWQWGPVEEEFL--DCGTCPRTCLDG--GDAMYMCRGGDVVALQGNTWQTV 294
++AE FDV TW+W P EEFL + P C G GD +Y C D++ ++ +TW V
Sbjct: 235 KTAESFDVTTWRWSPQGEEFLSSEMTMWPPICAAGENGD-LYACCRRDLMMMKDDTWYKV 293
Query: 295 AKVPSEIRNVACMGAWERS--LLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQ 352
+P+++ NV+ + A RS L++IGS+ +GEP +G+ D+ + +W KL + Y GHVQ
Sbjct: 294 GNLPADVCNVSYV-AIRRSGNLVVIGSARYGEPSVGYNWDMSNSRWLKLETHDKYEGHVQ 352
Query: 353 SYCLLEI 359
+ C LEI
Sbjct: 353 AGCFLEI 359
>At2g44130 unknown protein
Length = 409
Score = 164 bits (416), Expect = 5e-41
Identities = 95/283 (33%), Positives = 141/283 (49%), Gaps = 23/283 (8%)
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKIL---- 57
ELI GLP ++A +CL+RV ++ A+ + C+ W + + F + RR G T+ +L
Sbjct: 18 ELIPGLPSELALECLVRVPFQFQSAMRSVCRSWRSLLSDSSFIQERRRCGKTELLLCLVQ 77
Query: 58 -VTAQARFDSDTCGGLLV-----------KATTNPVYRLSVFEPETGNWSELTMPPEFDS 105
+T L+V + P + LSV+ W + P E
Sbjct: 78 PLTPPIPASKSVDETLMVDEKKSEDESHPRVFCTPRFGLSVYNAAMSTWHRVAFPEE--- 134
Query: 106 GSGLPMFCQIAGV--GYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFF 163
+P+FC+ + +++++GGWDPE+ + + V++ F WRRGA M R+FF
Sbjct: 135 -EQIPLFCECVVLQDAGKILLIGGWDPETLQPTRDVYVLEFAGRKWRRGAPMKES-RSFF 192
Query: 164 ACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKL 223
AC S VYVAGGHD++KNAL+SA YDV DEW + M RDEC+ G +
Sbjct: 193 ACASVSPTKVYVAGGHDDQKNALRSAEVYDVEKDEWSSVTPMTEGRDECQGFAVGMGLRF 252
Query: 224 RVVGGYCTEMQGRFERSAEEFDVDTWQWGPVEEEFLDCGTCPR 266
V+ GY TE QGRF E +D T W ++ + T PR
Sbjct: 253 CVLSGYGTESQGRFRSDGEIYDPATDSWSRIDNVWRFPDTSPR 295
>At3g59940 unknown protein
Length = 418
Score = 153 bits (387), Expect = 1e-37
Identities = 93/285 (32%), Positives = 136/285 (47%), Gaps = 38/285 (13%)
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQ 61
+LI GLPE++A +CL+RV ++ ++ + C+ W I S F + R G + +L Q
Sbjct: 14 DLIPGLPEELAIECLVRVPFQFHSSIKSVCRSWKCVISSRSFIKERIGFGKAESLLCLVQ 73
Query: 62 ARFDSDTC----GG--------------------LLVKATTNPVYRLSVFEPETGNWSEL 97
+ GG L + T P+Y LSV+ W +
Sbjct: 74 PLTSPPSPAMMEGGEMSQKKKEEEEGESQMTQQLLQPRITGTPLYGLSVYNATLDTWHRV 133
Query: 98 TMPPEFDSGSGLPMFCQIAGV--GYELVVMGGWDPESWKASNSVFIYNFLSAT-----WR 150
+P +P+FC+ + +++++GGWDPE+ + VF+ +F + +R
Sbjct: 134 AIPER------IPLFCECVAIQDAGKVLLIGGWDPETLQPVRDVFVLDFFAGEGSGRRFR 187
Query: 151 RGADMPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERD 210
RG M R+FFAC S VYVAGGHD++KNAL+SA YDV DEW LP M RD
Sbjct: 188 RGRPMSAA-RSFFACASVGSTKVYVAGGHDDQKNALRSAEVYDVEKDEWSMLPPMTEGRD 246
Query: 211 ECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVE 255
EC V+ GY TE QG+F E +D T W +E
Sbjct: 247 ECHGFSMATDPGFCVLSGYGTETQGQFRSDGEIYDPITNSWSTIE 291
>At1g67480 unknown protein
Length = 376
Score = 73.9 bits (180), Expect = 1e-13
Identities = 71/235 (30%), Positives = 99/235 (41%), Gaps = 38/235 (16%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI GLP+DVA+ CL V +FP++ + CK W +QS EF RR+ G ++ L
Sbjct: 39 LIPGLPDDVAKQCLALVPRARFPSMGSVCKKWRFVVQSKEFITVRRLAGMLEEWLYVL-- 96
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
+ GG + W M S LP A G+++
Sbjct: 97 ---TMNAGG------------------KDNRWE--VMDCLGQKLSSLPPMPGPAKTGFKV 133
Query: 123 VVMGG---------WDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTV 173
VV+ G S AS V+ Y+ +W R AD+ R FAC ++ + V
Sbjct: 134 VVVDGKLLVIAGCCMINGSLVASADVYQYDTCLNSWSRLADLEVA-RYDFAC-AEVNGHV 191
Query: 174 YVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGG 228
YV GGH + +L SA YD W + + R R C A GKL V+GG
Sbjct: 192 YVVGGHGVDGESLSSAEVYDPETCTWTFIESLRRPRWGCFA--SAFNGKLYVMGG 244
>At3g63220 unknown protein
Length = 345
Score = 73.2 bits (178), Expect = 2e-13
Identities = 83/328 (25%), Positives = 130/328 (39%), Gaps = 49/328 (14%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
L+ G+PE VA CL V P + + W I+S E R R+ +++ +L
Sbjct: 4 LLDGIPEAVALRCLAHVPLHLHPNLELVSRSWRAAIRSHELFRVRKELRSSEHLLCV--C 61
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
FD P V+ P W LT+P L F + G
Sbjct: 62 AFD--------------PENIWQVYSPNCDRW--LTLPLLPSRIRHLAHFGAVTTAGMLF 105
Query: 123 VVMGG--------WDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVY 174
V+ GG D + A++ V+ Y+F+ W A M PR FAC Q + V
Sbjct: 106 VLGGGSDAVSPVTGDHDGTFATDQVWSYDFVQRQWTPRASML-VPRAMFACCVLQGKIV- 163
Query: 175 VAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDE-CKAVFRGGAGKLRVVGGYCTEM 233
VAGG + ++ A YD ND W +PD+ + + C + GK+ V+ + +
Sbjct: 164 VAGGFTTCRKSISGAEMYDPENDVWTSIPDLHQTHNSACSGLVVN--GKVHVLHKGLSTV 221
Query: 234 QGRFERSAEEFDVDTWQW--GPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTW 291
Q E +DV + W GP+ + D +Y+ G V +G+TW
Sbjct: 222 Q-VLESVKLGWDVKDYGWPQGPM--------------VVVEDVLYVMSHGLVFKQEGDTW 266
Query: 292 QTVAKVPSEIRNVA-CMGAWERSLLLIG 318
+ VA R + M + +L++G
Sbjct: 267 KMVASASEFKRRIGMAMTSLSDEVLIVG 294
>At2g02870 unknown protein
Length = 467
Score = 73.2 bits (178), Expect = 2e-13
Identities = 75/305 (24%), Positives = 125/305 (40%), Gaps = 44/305 (14%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQK-ILVTAQ 61
LI+ + D + DCLIR S + ++ + + + + ++S E R RR G + + + Q
Sbjct: 117 LINEIGRDNSIDCLIRCSRSDYGSIASLNRNFRSLVKSGEIYRLRRQNGFVEHWVYFSCQ 176
Query: 62 ARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSEL-TMPPEFDSGSGLPMFC---QIAG 117
+ F+P W +L TMP S + C +
Sbjct: 177 -------------------LLEWVAFDPVERRWMQLPTMP------SSVTFMCADKESLA 211
Query: 118 VGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAG 177
VG +L+V+G D +S+ ++ Y+ L+ +W G M PR F S + ++ AG
Sbjct: 212 VGTDLLVLGKDD----FSSHVIYRYSLLTNSWSSGMKM-NSPRCLFGSASLGEIAIF-AG 265
Query: 178 GHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRF 237
G D + L A Y+ W+ LP M + R C VF GK V+GG +
Sbjct: 266 GCDSQGKILDFAEMYNSELQTWITLPRMNKPRKMCSGVFMD--GKFYVIGG-IGGADSKG 322
Query: 238 ERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKV 297
EE+D++T +W + + + PR+ D D +VA+ N
Sbjct: 323 LTCGEEYDLETKKWTQIPDL-----SPPRSRADQADMSPAAEAPPLVAVVNNQLYAADHA 377
Query: 298 PSEIR 302
E+R
Sbjct: 378 DMEVR 382
>At1g55270 unknown protein (At1g55270)
Length = 434
Score = 71.2 bits (173), Expect = 8e-13
Identities = 75/281 (26%), Positives = 113/281 (39%), Gaps = 29/281 (10%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
L+ GLP+D+A CLIRV + + CK W F +R++ G +++ +
Sbjct: 78 LLPGLPDDLAVACLIRVPRAEHRKLRLVCKRWYRLASGNFFYSQRKLLGMSEEWVYV--- 134
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSEL-TMPPEFDSGSGLPMFCQIAGVGYE 121
F D G + + F+P + W L +P E+ G C + G
Sbjct: 135 -FKRDRDGKI----------SWNTFDPISQLWQPLPPVPREYSEAVGFG--CAVLS-GCH 180
Query: 122 LVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDE 181
L + GG DP +F YN + W R DM R FF C + +YVAGG E
Sbjct: 181 LYLFGGKDPLRGSMRRVIF-YNARTNKWHRAPDML-RKRHFFGCCV-INNCLYVAGGECE 237
Query: 182 E-KNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
+ L+SA YD + W + DM+ V L+ +G + M
Sbjct: 238 GIQRTLRSAEVYDPNKNRWSFIADMSTAMVPLIGVVYDKKWFLKGLGSHQLVM------- 290
Query: 241 AEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDAMYMCRGG 281
+E +D + W PV + + P T L+G CR G
Sbjct: 291 SEAYDPEVNSWSPVSDGMVAGWRNPCTSLNGRLYGLDCRDG 331
>At1g26930 unknown protein
Length = 421
Score = 71.2 bits (173), Expect = 8e-13
Identities = 66/256 (25%), Positives = 107/256 (41%), Gaps = 31/256 (12%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI G+ D + CLIR S + ++ + + + I+S E R RR+ G + + +
Sbjct: 71 LIPGMNRDDSLSCLIRCSRADYCSIASVNRSLRSLIRSGEIYRLRRLQGTLEHWVYFS-- 128
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
C + F+P + W L P+ + + VG +L
Sbjct: 129 ------CH----------LNEWEAFDPRSKRWMHLPSMPQNECFRYADK--ESLAVGTDL 170
Query: 123 VVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEE 182
+V G W+ S+ ++ Y+ L+ +W M PR F S + V +AGG D
Sbjct: 171 LVFG-WEVSSYV----IYRYSLLTNSWSTAKSM-NMPRCLFGSASYGEIAV-LAGGCDSS 223
Query: 183 KNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCT--EMQGRFERS 240
L +A Y+ + W+ LP M + R C VF GK V+GG E + +
Sbjct: 224 GRILDTAELYNYEDQTWLVLPGMNKRRKMCSGVFMD--GKFYVIGGIGVGEENEPKVLTC 281
Query: 241 AEEFDVDTWQWGPVEE 256
EEFD+ T +W + E
Sbjct: 282 GEEFDLKTRKWTEIPE 297
>At1g14330 unknown protein
Length = 441
Score = 68.6 bits (166), Expect = 5e-12
Identities = 71/258 (27%), Positives = 108/258 (41%), Gaps = 37/258 (14%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI+ + D + CLIR S + ++ + + + + +++ E R RR Q +V
Sbjct: 94 LINDIGRDNSISCLIRCSRSGYGSIASLNRSFRSLVKTGEIYRLRR-----QNQIVEHWV 148
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSEL-TMPPEFDSGSGLPMFC---QIAGV 118
F LL NP R W L TMP SG+ C + V
Sbjct: 149 YFSCQ----LLEWVAFNPFER---------RWMNLPTMP------SGVTFMCADKESLAV 189
Query: 119 GYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG 178
G +L+V+G D +S+ ++ Y+ L+ +W G M PR F S + ++ AGG
Sbjct: 190 GTDLLVLGKDD----YSSHVIYRYSLLTNSWSSGMRM-NSPRCLFGSASLGEIAIF-AGG 243
Query: 179 HDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFE 238
D SA Y+ W LP M + R C VF GK V+GG +
Sbjct: 244 FDSFGKISDSAEMYNSELQTWTTLPKMNKPRKMCSGVFMD--GKFYVIGGIGGN-DSKVL 300
Query: 239 RSAEEFDVDTWQWGPVEE 256
EEFD++T +W + E
Sbjct: 301 TCGEEFDLETKKWTEIPE 318
>At1g16250 unknown protein
Length = 383
Score = 67.0 bits (162), Expect = 2e-11
Identities = 67/267 (25%), Positives = 104/267 (38%), Gaps = 41/267 (15%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
+I GLP+D+A C+ ++S+ + +GW ++ ++ + G +
Sbjct: 8 IIPGLPDDLALRCIAKLSHGYHGVLECVSRGWRDLVRGADYSCYKARNGWS--------- 58
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSG---SGLPMFCQIAGVG 119
G L T + ++PE W L G SG C V
Sbjct: 59 -------GSWLFVLTERSKNQWVAYDPEADRWHPLPRTRAVQDGWHHSGFACVC----VS 107
Query: 120 YELVVMGGWDPESWKA--------SNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDR 171
L+V+GG S + + V ++ W+ A M PRT FACTS +
Sbjct: 108 NCLLVIGGCYAPSVSSFPHQKPVVTKDVMRFDPFKKQWKMVASMRT-PRTHFACTSVSGK 166
Query: 172 TVYVAGGHD-EEKNALKSALAYDVANDEWVPLPDMARERDECKAV-FRGGAGKLRVVGGY 229
VYVAGG + + SA YD D W LP M R + +C + +RG L G+
Sbjct: 167 -VYVAGGRNLTHSRGIPSAEVYDPVADRWEELPAMPRPQMDCSGLSYRGCFHVLSDQVGF 225
Query: 230 CTEMQGRFERSAEEFDVDTWQWGPVEE 256
+ S+E F+ W VE+
Sbjct: 226 AE------QNSSEVFNPRDMTWSTVED 246
>At4g38940 unknown protein
Length = 370
Score = 60.8 bits (146), Expect = 1e-09
Identities = 57/209 (27%), Positives = 92/209 (43%), Gaps = 32/209 (15%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LIS LPE++ D + RV +P ++ + + + + SPE +RR G T++ L A +
Sbjct: 20 LISLLPEEIVVDIVARVPRCYYPTLSQVSRRFRSLVASPEIYKRRSFFGCTEQCLYIAIS 79
Query: 63 RFDSDTC---------GGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFC 113
+ + G TT +RL V P T+PP +PM
Sbjct: 80 KDQTSDIHWFTLCRKPNGQQFSGTTASDHRL-VHIP--------TLPP-------MPMHG 123
Query: 114 QIAGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTV 173
G+G + VMGG+ +WK ++SV + + + T + +MP F T DR +
Sbjct: 124 SYVGIGSNIFVMGGFC--NWKITSSVSLIDCRTHTAQTLPNMP--KAVAFPVTELIDRKI 179
Query: 174 YVAGGHD---EEKNALKSALAYDVANDEW 199
YV GG D K+ + + YD + W
Sbjct: 180 YVIGGSDTLSPMKSPSRIMMVYDTDTEMW 208
>At5g03020 unknown protein
Length = 347
Score = 59.7 bits (143), Expect = 2e-09
Identities = 54/200 (27%), Positives = 85/200 (42%), Gaps = 21/200 (10%)
Query: 5 SGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQARF 64
S LP+DVA DC R+S +P ++ KG+ T I SPE R G + L
Sbjct: 18 SSLPDDVALDCRARISRFHYPTLSLVSKGFRTLIASPELEATRSFIGKPENHL------- 70
Query: 65 DSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYELVV 124
C L + NP++ +F P + +P + P + + G ++ +
Sbjct: 71 ----CVCLRLYKNPNPLW--FIFSPIPKQKLKPIVP--WFPNQQYPQYPTVVSNGSQIYI 122
Query: 125 MGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGH--DEE 182
+GG+ + SN V I+++ + WRR M PR + A S D +YV GG
Sbjct: 123 IGGFVRR--RRSNRVSIFDYRTYQWRRLPKM-RQPRVYPA-ASVIDGKIYVIGGFRGSMP 178
Query: 183 KNALKSALAYDVANDEWVPL 202
+ S YD + W P+
Sbjct: 179 TDIENSGEVYDPKTNTWEPI 198
Score = 37.0 bits (84), Expect = 0.017
Identities = 25/91 (27%), Positives = 42/91 (45%), Gaps = 9/91 (9%)
Query: 173 VYVAGGHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGA---GKLRVVGGY 229
+Y+ GG + + + ++ +D +W LP M + R V+ + GK+ V+GG+
Sbjct: 120 IYIIGGFVRRRRSNRVSI-FDYRTYQWRRLPKMRQPR-----VYPAASVIDGKIYVIGGF 173
Query: 230 CTEMQGRFERSAEEFDVDTWQWGPVEEEFLD 260
M E S E +D T W P+ LD
Sbjct: 174 RGSMPTDIENSGEVYDPKTNTWEPILLTSLD 204
>At5g60570 putative protein
Length = 393
Score = 58.2 bits (139), Expect = 7e-09
Identities = 68/255 (26%), Positives = 99/255 (38%), Gaps = 35/255 (13%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
++ GL +DVA +CL V +P+++ K +N I S R+ G + ++
Sbjct: 49 VLPGLIDDVALNCLAWVPRSDYPSLSCVNKKYNKLINSGHLFALRKELGIVEYLVFMV-- 106
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
D G L+ F P W L P + + + V EL
Sbjct: 107 ---CDPRGWLM-------------FSPMKKKWMVLPKMPCDECFNHADK--ESLAVDDEL 148
Query: 123 VVMGG--WDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
+V G + WK Y+ S W + M PR FA + VAGG D
Sbjct: 149 LVFGRELFQFAIWK-------YSLRSRCWVKCEGM-HRPRCLFA-SGSLGGIAIVAGGTD 199
Query: 181 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
N L SA YD ++ W LP+M R C F GK V+GG +
Sbjct: 200 MNGNILASAELYDSSSGRWEMLPNMHSPRRLCSGFFMD--GKFYVIGGMSSPNVS--VTF 255
Query: 241 AEEFDVDTWQWGPVE 255
EEFD++T +W +E
Sbjct: 256 GEEFDLETRKWRKIE 270
>At4g19870 unknown protein
Length = 400
Score = 55.1 bits (131), Expect = 6e-08
Identities = 54/214 (25%), Positives = 89/214 (41%), Gaps = 15/214 (7%)
Query: 4 ISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVT--AQ 61
+S LP+++ +CL R+S +P ++ K + + I S E R NT++ + +
Sbjct: 30 LSSLPDEIVENCLARISRSYYPTLSIVSKSFRSIISSTELYVARSHLRNTEECVYVCLSD 89
Query: 62 ARFDSDTCGGLLVKATTNPVYRLSVFE---PETGNWSELTMPPEFDSGSGLPMFCQIAGV 118
F+ L V NP S+ E + +L +P S + P+ V
Sbjct: 90 KSFEFPKWFTLWV----NPNQANSMVEKKRKKKKTIGKLLVP--IPSSNLSPVSKSAIAV 143
Query: 119 GYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG 178
G E+ V+GG S++V I + S TWR M + F C D +YV GG
Sbjct: 144 GSEIYVIGG--KVDGALSSAVRILDCRSNTWRDAPSMTVARKRPFICL--YDGKIYVIGG 199
Query: 179 HDEEKNALKSALAYDVANDEWVPLPDMARERDEC 212
+++ + A +D+ W L D E C
Sbjct: 200 YNKLSESEPWAEVFDIKTQTWECLSDPGTEIRNC 233
>At4g03030 OR23peptide
Length = 434
Score = 54.3 bits (129), Expect = 1e-07
Identities = 62/227 (27%), Positives = 92/227 (40%), Gaps = 42/227 (18%)
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
+ LI GL DV R L V Y + + CK R T N +L
Sbjct: 43 LTLIPGLSNDVGRLILSFVPYPHISRIKSTCKSC----------RDNSNTNNLSHLLCI- 91
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
F D + +P + +F+P T +W L + P GL F +A +G
Sbjct: 92 ---FPQDP--------SISPPF---LFDPVTLSWRSLPLMPCNPHVYGLCNFVAVA-LGP 136
Query: 121 ELVVMGG--WDPESWKA-----SNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRT- 172
+ V+GG +D S+ ++SVF Y+F+ + W R + M PR FAC +
Sbjct: 137 YVYVLGGSAFDTRSYPLDVPLPTSSVFRYSFVKSVWERLSPMMS-PRGSFACAAMPGSCG 195
Query: 173 -VYVAGGHDEEK------NALKSALAYDVANDEWVPLPDMARERDEC 212
+ VAGG + + S YDV DEW + ++ R R C
Sbjct: 196 RIIVAGGGSRHTLFGAAGSRMSSVEMYDVEKDEWRVMNELPRFRAGC 242
>At1g74510 unknown protein
Length = 451
Score = 53.1 bits (126), Expect = 2e-07
Identities = 50/216 (23%), Positives = 85/216 (39%), Gaps = 27/216 (12%)
Query: 4 ISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQAR 63
++ L ++ +CL S F ++ + + + + I+ E R RR G + + +
Sbjct: 96 VTRLDQNALLNCLAHCSLSDFGSIASTNRTFRSLIKDSELYRLRRAKGIVEHWIYFSCRL 155
Query: 64 FDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYELV 123
+ + ++P W L +P + + + VG EL+
Sbjct: 156 LEWEA------------------YDPNGDRW--LRVPKMTFNECFMCSDKESLAVGTELL 195
Query: 124 VMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEEK 183
V G S+ ++ Y+ L+ TW G M PR F S + V +AGG D
Sbjct: 196 VFG-----KEIMSHVIYRYSILTNTWTSGMQM-NVPRCLFGSASLGEIAV-IAGGCDPRG 248
Query: 184 NALKSALAYDVANDEWVPLPDMARERDECKAVFRGG 219
L SA Y+ EW +P M + R C +VF G
Sbjct: 249 RILSSAELYNSETGEWTVIPSMNKARKMCSSVFMDG 284
Score = 32.3 bits (72), Expect = 0.41
Identities = 30/108 (27%), Positives = 42/108 (38%), Gaps = 12/108 (11%)
Query: 116 AGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSD-QDRTVY 174
A +G V+ GG DP + +S +YN + W +P + C+S D Y
Sbjct: 232 ASLGEIAVIAGGCDPRG-RILSSAELYNSETGEW---TVIPSMNKARKMCSSVFMDGNFY 287
Query: 175 VAGGHDE-EKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAG 221
GG E L YD+ W +P+M ER GG G
Sbjct: 288 CIGGIGEGNSKMLLCGEVYDLKKKTWTLIPNMLPERSS------GGGG 329
>At4g39590 unknown protein
Length = 402
Score = 52.8 bits (125), Expect = 3e-07
Identities = 70/271 (25%), Positives = 110/271 (39%), Gaps = 42/271 (15%)
Query: 4 ISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKIL-VTAQA 62
I LP D+ +C RVS +PA++ K + + + SPE R + T+K L + +
Sbjct: 38 IDSLPNDLLLNCFARVSRMYYPALSRVSKRFRSIVTSPEIYNTRSLLNRTEKCLYLCLRF 97
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMP-PEFDSGSGLPMFCQIAGVGYE 121
FD++T L + N VF L +P P++ L + + VG
Sbjct: 98 PFDNNTHWFTLYQ-NPNRTVSDKVF---------LQIPSPQYP----LTLSSNLVAVGSN 143
Query: 122 LVVMGGWDPE-------SWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVY 174
+ +GG + + S+ V I + S TWR G M R+ + TS D +Y
Sbjct: 144 IYRIGGTVGDDSCPLGFDREPSSKVSILDCRSHTWRDGPRMRLNRRS--STTSVVDGKIY 201
Query: 175 VAGGHDEEKNALKSALAYDVANDEW--VPLPDMARERDEC---KAVFR-GGAGKLRVVGG 228
V GG ++ N +D W V P + + +E +AV G GKL + G
Sbjct: 202 VTGGTEDTDNPSHWIEVFDPKTQSWGTVTNPHIVKVWEEVCYRRAVKSIGHDGKLYLSGD 261
Query: 229 YCTEMQGRFERSAEEFDVDTWQWGPVEEEFL 259
+D D +W VEE +L
Sbjct: 262 -----------KYVVYDPDEGKWNSVEEHWL 281
>At2g24540 unknown protein
Length = 372
Score = 51.6 bits (122), Expect = 7e-07
Identities = 51/200 (25%), Positives = 79/200 (39%), Gaps = 23/200 (11%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFR-RRRRVTGNTQKILVTAQ 61
LISGLP D+A CL+R+ Y + WN I +P F ++ ++ ++ + V A
Sbjct: 28 LISGLPNDIAELCLLRLPYPYHALYRSVSSSWNKTITNPRFLFSKQSLSISSPYLFVFA- 86
Query: 62 ARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSEL-TMPPEFDS-GSGLPMFCQIAGVG 119
F+ T + + +G W L MP F S + C
Sbjct: 87 --FNKSTAR-----------IQWQSLDLASGRWFVLPPMPNSFTKISSPHALSCASMPRQ 133
Query: 120 YELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGH 179
+L V+GG D + S +Y L+ W + M PRT+F + + + V G
Sbjct: 134 GKLFVLGGGD-----VNRSAVVYTALTNRWSCISPMM-SPRTYFVSGNVNGKIMAVGGSV 187
Query: 180 DEEKNALKSALAYDVANDEW 199
A +YD ND W
Sbjct: 188 GGNGEATTEVESYDPDNDTW 207
>At1g31350 unknown protein
Length = 395
Score = 51.6 bits (122), Expect = 7e-07
Identities = 66/278 (23%), Positives = 105/278 (37%), Gaps = 46/278 (16%)
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI GLP+ ++ CL V ++A C W + SPEF + L
Sbjct: 35 LIEGLPDHISEICLSLVHRPSL--LSAVCTRWRRLLYSPEF--------PSFPSLYALFV 84
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDS------------GSGLP 110
SDT NP R F P + W L PP LP
Sbjct: 85 DSTSDT-------GRVNPSVRFMCFNPVSSKWYPLPPPPPDPPLHRILYRHPSFISFNLP 137
Query: 111 MFCQIAGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQD 170
+ C ++ G +L+++ G + + A + I++ +S++W G + G PR + A T D
Sbjct: 138 IQC-VSAAG-KLILIAGSNQQLSPAISHPLIFDPISSSWSSGPRI-GSPRRWCA-TGACD 193
Query: 171 RTVYVAGGHDEE--KNALKSALAYDVAND-------EWVPLPDMARERDECKAVFRGGAG 221
+Y+A G + KS D+ W L DM R +A+ G
Sbjct: 194 GAIYIASGISSQFSSTVAKSVEKLDLTEQNRNNHRFNWEKLRDMRDLRFSREAIDAVGYR 253
Query: 222 KLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVEEEFL 259
+ ++ ++G + +DV W P+ EE L
Sbjct: 254 RKLLM----VNVKGDAVKEGAIYDVVKDDWEPMPEEML 287
Score = 48.9 bits (115), Expect = 4e-06
Identities = 51/226 (22%), Positives = 76/226 (33%), Gaps = 35/226 (15%)
Query: 36 TEIQSPEFRRRRRVTGNTQKILVTAQARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWS 95
T +SP +RRR VTGN L+ S+ C L V+R S+ W
Sbjct: 14 TAAESPPAKRRRTVTGNENSALIEGLPDHISEICLSL--------VHRPSLLSAVCTRWR 65
Query: 96 ELTMPPEFDSGSGLPMFCQIAGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADM 155
L PEF S L Y L V D S +N +S+ W
Sbjct: 66 RLLYSPEFPSFPSL----------YALFVDSTSDTGRVNPSVRFMCFNPVSSKWYPLPPP 115
Query: 156 PGGPRTF---------------FACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWV 200
P P C S + + +AG + + A+ L +D + W
Sbjct: 116 PPDPPLHRILYRHPSFISFNLPIQCVSAAGKLILIAGSNQQLSPAISHPLIFDPISSSWS 175
Query: 201 PLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDV 246
P + R C GA + + G ++ +S E+ D+
Sbjct: 176 SGPRIGSPRRWCATGACDGA--IYIASGISSQFSSTVAKSVEKLDL 219
Score = 28.5 bits (62), Expect = 5.9
Identities = 30/126 (23%), Positives = 50/126 (38%), Gaps = 22/126 (17%)
Query: 184 NALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEE 243
+A+K YDV D+W P+P +E +RG + Y + + + +
Sbjct: 264 DAVKEGAIYDVVKDDWEPMP------EEMLVGWRGPVAAMEEEILYSVDER---RGTVRK 314
Query: 244 FDVDTWQWGPV-----EEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVAKVP 298
+D + +W V EE L T + D G + G +V + VA P
Sbjct: 315 YDDEKREWREVVVVEGGEEMLKGAT--QVTADSGKLCVVTGDGKIVVVD------VAAEP 366
Query: 299 SEIRNV 304
++I NV
Sbjct: 367 AKIWNV 372
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,146,833
Number of Sequences: 26719
Number of extensions: 416244
Number of successful extensions: 1117
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 882
Number of HSP's gapped (non-prelim): 207
length of query: 359
length of database: 11,318,596
effective HSP length: 100
effective length of query: 259
effective length of database: 8,646,696
effective search space: 2239494264
effective search space used: 2239494264
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0253.11


