

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.10
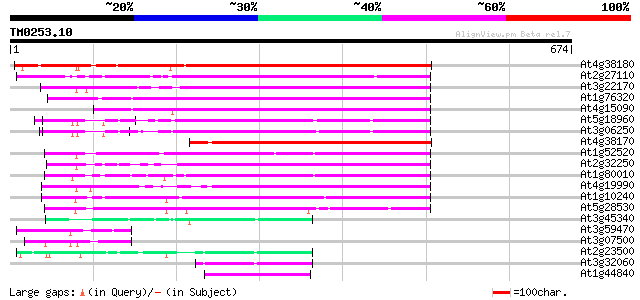
(674 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 417 e-116
At2g27110 Mutator-like transposase 302 5e-82
At3g22170 far-red impaired response protein, putative 301 6e-82
At1g76320 putative phytochrome A signaling protein 293 2e-79
At4g15090 unknown protein 293 3e-79
At5g18960 FAR1 - like protein 265 8e-71
At3g06250 unknown protein 263 3e-70
At4g38170 hypothetical protein 251 7e-67
At1g52520 F6D8.26 250 2e-66
At2g32250 Mutator-like transposase 249 3e-66
At1g80010 hypothetical protein 246 3e-65
At4g19990 putative protein 244 1e-64
At1g10240 unknown protein 238 6e-63
At5g28530 far-red impaired response protein (FAR1) - like 199 3e-51
At3g45340 putative protein 65 9e-11
At3g59470 unknown protein 57 4e-08
At3g07500 hypothetical protein 57 4e-08
At2g23500 Mutator-like transposase 55 2e-07
At3g32060 hypothetical protein 52 1e-06
At1g44840 hypothetical protein 52 1e-06
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 417 bits (1071), Expect = e-116
Identities = 212/525 (40%), Positives = 323/525 (61%), Gaps = 34/525 (6%)
Query: 6 IQGHQLMDD-----------NG-IGSSSKDFTTSILDTSDIGGELNSKPEIGMEFESIEK 53
I+ H L DD NG +G+S F + D+ L+ +P G+EFES E
Sbjct: 28 IEHHALDDDDMLDSPIMPCGNGLVGNSGNYFPNQEEEACDL---LDLEPYDGLEFESEEA 84
Query: 54 VREFYNSFDKKNGFGVRVRSSKPKR---AVL----VCCNEGQHKVKISRTEEIKDSTNQT 106
+ FYNS+ ++ GF RV SS+ R A++ VC EG + RT++ +
Sbjct: 85 AKAFYNSYARRIGFSTRVSSSRRSRRDGAIIQRQFVCAKEGFRNMNEKRTKD-----REI 139
Query: 107 KRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVAAKS 166
KR + R GC+ASL V + KW++ F DHNH +V P V +R H+++S AK+
Sbjct: 140 KRPRTITRVGCKASLSVKMQDS-GKWLVSGFVKDHNHELVPPDQVHCLRSHRQISGPAKT 198
Query: 167 LVEKFEEEGI-PTGKVAAIFNDGDST----FTNRDCWNYIRNVRRKNLDVGDAQAVFDYC 221
L++ + G+ P ++A+ + FT DC NY+RN R+K+++ G+ Q + DY
Sbjct: 199 LIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSIE-GEIQLLLDYL 257
Query: 222 KRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYSMPFAP 281
++ +N NFFY++Q ED + N+FW D ++ + + FGD +TFDTTY++N+Y +PFAP
Sbjct: 258 RQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAP 317
Query: 282 FVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAALAKV 341
F G+N+H Q IL+GCA + +E+EASFVWLF TWL AM P+SI TD D ++AA+ V
Sbjct: 318 FTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHV 377
Query: 342 FPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWMRIMVEYN 401
FP +RHR C WHI+KK EKL+H++ K F+ D +C+ + S++ FE W ++ +Y
Sbjct: 378 FPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYE 437
Query: 402 LKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTLQEFVLKF 461
L+++EWLQ +Y R W+P+Y R TFFA M+ T RS+SIN++FD +++AST L +F +
Sbjct: 438 LRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINASTNLSQFFKLY 497
Query: 462 EKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIFL 506
EKA+ESRLE E K +Y++ + +L T S +E+ A LYT ++F+
Sbjct: 498 EKALESRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYTRKLFM 542
>At2g27110 Mutator-like transposase
Length = 851
Score = 302 bits (773), Expect = 5e-82
Identities = 169/498 (33%), Positives = 282/498 (55%), Gaps = 27/498 (5%)
Query: 9 HQLMDDNGIGSSSKDFTTSILDTSDIGGELN-SKPEIGMEFESIEKVREFYNSFDKKNGF 67
H++ D+ + S ++ ++ + E+ ++P +GMEF S ++ + FY+ + ++ GF
Sbjct: 15 HEIGDEGDVEPSDCSGQNNMDNSLGVQDEIGIAEPCVGMEFNSEKEAKSFYDEYSRQLGF 74
Query: 68 GVRVRSSKPKRAVLVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGT 127
+SK L+ +G V R S+ ++KR+ S C+A + +
Sbjct: 75 -----TSK-----LLPRTDGSVSV---REFVCSSSSKRSKRRLS---ESCDAMVRIEL-Q 117
Query: 128 TKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFND 187
KW++ F +H H + S + +R + + + KS + E +P+G + + D
Sbjct: 118 GHEKWVVTKFVKEHTHGLASSNMLHCLRPRRHFANSEKSSYQ--EGVNVPSGMMY-VSMD 174
Query: 188 GDSTFTNRDCWNYIRNVRRKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLF 247
+S R N K DA + +Y KR Q EN FFYA+Q DED++M N+F
Sbjct: 175 ANS----RGARNASMATNTKRTIGRDAHNLLEYFKRMQAENPGFFYAVQLDEDNQMSNVF 230
Query: 248 WVDARSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASF 307
W D+RSR++Y FGD +T DT Y+ N++ +PFAPF G+N+H Q+IL+GCAL+ DES+ SF
Sbjct: 231 WADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTGVNHHGQAILFGCALILDESDTSF 290
Query: 308 VWLFKTWLQAMGGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYH 367
+WLFKT+L AM + P+S++TDQD A++ A +VFP +RH + W ++++ EKLAH+
Sbjct: 291 IWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPGARHCINKWDVLREGQEKLAHVCL 350
Query: 368 KQSIFKRDMKRCIRGSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTF 427
F+ ++ CI + +I+ FE W ++ +Y+L +EWL LY R W+P+Y R +F
Sbjct: 351 AYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRHEWLNSLYNARAQWVPVYFRDSF 410
Query: 428 FAGMNTTQRSESINAFFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILS 487
FA + +Q +FFD +V+ TTL F +E+A+ES E E + + ++ + +L
Sbjct: 411 FAAVFPSQGYS--GSFFDGYVNQQTTLPMFFRLYERAMESWFEMEIEADLDTVNTPPVLK 468
Query: 488 TGSKLEEHAGHLYTPEIF 505
T S +E A +L+T +IF
Sbjct: 469 TPSPMENQAANLFTRKIF 486
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 301 bits (772), Expect = 6e-82
Identities = 161/480 (33%), Positives = 259/480 (53%), Gaps = 31/480 (6%)
Query: 38 LNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKR-------AVLVCCNEGQHK 90
+N +P GMEFES + FY + + GF +++S+ + A C G +
Sbjct: 66 MNLEPLNGMEFESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTKR 125
Query: 91 -----VKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVM 145
R + K + + ++ C+AS+ V R KW+I SF +HNH +
Sbjct: 126 EYDKSFNRPRARQSKQDPENMAGRRTCAKTDCKASMHVKR-RPDGKWVIHSFVREHNHEL 184
Query: 146 VSPKSVSYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVR 205
+ ++VS + K + AK E V ++ +D S+F R
Sbjct: 185 LPAQAVSE-QTRKIYAAMAKQFAEY--------KTVISLKSDSKSSFEKG---------R 226
Query: 206 RKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVIT 265
+++ GD + + D+ R Q NSNFFYA+ +D R+ N+FWVDA+SR +Y F DV++
Sbjct: 227 TLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVS 286
Query: 266 FDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPIS 325
DTTY NKY MP A FVG+N H Q ++ GCAL+ DES A++ WL +TWL+A+GG+ P
Sbjct: 287 LDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAPKV 346
Query: 326 IITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHS 385
+IT+ D+ M + + ++FP +RH L LWH++ K E L + + F ++CI S
Sbjct: 347 LITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGK 406
Query: 386 IQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFD 445
+ F +W + + + LK+++W+ LY+ R+ W P Y AGM+T+QR++SINAFFD
Sbjct: 407 DEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFD 466
Query: 446 SFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
++ T++QEFV ++ ++ R E E K + E +K + + S E+ +YTP +F
Sbjct: 467 KYMHKKTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPFEKSVSEVYTPAVF 526
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 293 bits (750), Expect = 2e-79
Identities = 156/460 (33%), Positives = 257/460 (54%), Gaps = 6/460 (1%)
Query: 46 MEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVCCNEGQHKVKISRTEEIKDSTNQ 105
MEFE+ E FY + K GFG SS+ RA + ++ ++ D+ N
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSKQQSDDAINP 60
Query: 106 TKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVAAK 165
++ + GC+AS+ V R KW + SF +HNH ++ P+ Y R H+ +
Sbjct: 61 R----ASPKIGCKASMHVKR-RPDGKWYVYSFVKEHNHDLL-PEQAHYFRSHRNTELVKS 114
Query: 166 SLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDVGDAQAVFDYCKRKQ 225
+ ++ P + D F + N RR LD GDA+ + ++ R Q
Sbjct: 115 NDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQ 174
Query: 226 VENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGL 285
EN FF+A+ ED + N+FWVDA+ Y+ F DV++F+T+Y +KY +P FVG+
Sbjct: 175 EENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGV 234
Query: 286 NNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAALAKVFPES 345
N+H Q +L GC LL D++ ++VWL ++WL AMGG+KP ++TDQ+ A+KAA+A V PE+
Sbjct: 235 NHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPET 294
Query: 346 RHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWMRIMVEYNLKEN 405
RH CLWH++ + P L + Q F + + +CI S S + F+ W++++ +++L++
Sbjct: 295 RHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDV 354
Query: 406 EWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTLQEFVLKFEKAV 465
W++ LY+ R+ W P + R FAG++ RSES+N+ FD +V T+L+EF+ + +
Sbjct: 355 PWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLML 414
Query: 466 ESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
E R E E K ++++ H++ L + S E+ +Y+ EIF
Sbjct: 415 EDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIF 454
>At4g15090 unknown protein
Length = 768
Score = 293 bits (749), Expect = 3e-79
Identities = 147/411 (35%), Positives = 238/411 (57%), Gaps = 8/411 (1%)
Query: 101 DSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKM 160
+S+ + R+ + ++ C+AS+ V R KW+I F DHNH ++ P + R + +
Sbjct: 50 ESSGSSSRRSTVKKTDCKASMHVKR-RPDGKWIIHEFVKDHNHELL-PALAYHFRIQRNV 107
Query: 161 SVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTN------RDCWNYIRNVRRKNLDVGDA 214
+A K+ ++ T K+ + + N D + + R L+ GD+
Sbjct: 108 KLAEKNNIDILHAVSERTKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDS 167
Query: 215 QAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNK 274
Q + +Y KR + EN FFYAI +ED R+ NLFW DA+SR Y F DV++FDTTY
Sbjct: 168 QVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFN 227
Query: 275 YSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAM 334
+P A F+G+N+HSQ +L GCAL+ DES +FVWL KTWL+AMGG+ P I+TDQD +
Sbjct: 228 DKLPLALFIGVNHHSQPMLLGCALVADESMETFVWLIKTWLRAMGGRAPKVILTDQDKFL 287
Query: 335 KAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWM 394
+A++++ P +RH LWH+++K PE +H+ + F +CI S + F+ W
Sbjct: 288 MSAVSELLPNTRHCFALWHVLEKIPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWW 347
Query: 395 RIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTL 454
+++ ++ L+ +EWL L++ R+ W+P + F AGM+T+QRSES+N+FFD ++ TL
Sbjct: 348 KMVSQFGLENDEWLLWLHEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITL 407
Query: 455 QEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
+EF+ ++ +++R E E ++++ HK L + S E+ YT IF
Sbjct: 408 KEFLRQYGVILQNRYEEESVADFDTCHKQPALKSPSPWEKQMATTYTHTIF 458
>At5g18960 FAR1 - like protein
Length = 788
Score = 265 bits (676), Expect = 8e-71
Identities = 156/475 (32%), Positives = 255/475 (52%), Gaps = 57/475 (12%)
Query: 40 SKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRS---SKPKRAV----LVCCNEG-QHKV 91
++P G+EF S + +FY ++ + GF VR+ SK ++ VC EG QH
Sbjct: 209 TEPYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQHPS 268
Query: 92 KISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSV 151
R GC A + + R + W++ N DHNH + K
Sbjct: 269 ----------------------RMGCGAYMRIKRQDSGG-WIVDRLNKDHNHDLEPGKKN 305
Query: 152 SYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDV 211
+ ++K ++G TG + ++ + F N N+I+ R +
Sbjct: 306 D-------------AGMKKIPDDG--TGGLDSVDLIELNDFGN----NHIKKTRENRIGK 346
Query: 212 GDAQAVFDYCKRKQVENSNFFYAIQCD-EDSRMVNLFWVDARSRLSYQLFGDVITFDTTY 270
+ DY + +Q E+ FFYA++ D + +++FW D+R+R + FGD + FDT+Y
Sbjct: 347 EWYPLLLDYFQSRQTEDMGFFYAVELDVNNGSCMSIFWADSRARFACSQFGDSVVFDTSY 406
Query: 271 KTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQ 330
+ YS+PFA +G N+H Q +L GCA++ DES+ +F+WLF+TWL+AM G++P SI+ DQ
Sbjct: 407 RKGSYSVPFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRSIVADQ 466
Query: 331 DLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFE 390
DL ++ AL +VFP + HR W I +K E L S FK + ++CI + +I F+
Sbjct: 467 DLPIQQALVQVFPGAHHRYSAWQIREKERENLIPF---PSEFKYEYEKCIYQTQTIVEFD 523
Query: 391 EEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDA 450
W ++ +Y L+++ WL+ +Y+ RE+W+P Y R++FFAG+ + +I FF + +DA
Sbjct: 524 SVWSALINKYGLRDDVWLREIYEQRENWVPAYLRASFFAGIPI---NGTIEPFFGASLDA 580
Query: 451 STTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
T L+EF+ ++E+A+E R E ERKE++ S + L T +EE LYT +F
Sbjct: 581 LTPLREFISRYEQALEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTVF 635
Score = 49.3 bits (116), Expect = 7e-06
Identities = 40/128 (31%), Positives = 59/128 (45%), Gaps = 29/128 (22%)
Query: 31 TSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVCCNEGQHK 90
+ D G +P +G+EF++ E+ REFYN++ + GF VR GQ
Sbjct: 32 SEDEEGGSGVEPYVGLEFDTAEEAREFYNAYAARTGFKVRT---------------GQ-- 74
Query: 91 VKISRTEEIKDSTNQTKR-KCS------TIRSGCEASLIVSRGTTKSKWMIKSFNNDHNH 143
+ SRT D T ++R CS R+GC A + V R T KW++ +HNH
Sbjct: 75 LYRSRT----DGTVSSRRFVCSKEGFQLNSRTGCTAFIRVQRRDT-GKWVLDQIQKEHNH 129
Query: 144 VMVSPKSV 151
+ SV
Sbjct: 130 ELGGEGSV 137
>At3g06250 unknown protein
Length = 764
Score = 263 bits (671), Expect = 3e-70
Identities = 155/474 (32%), Positives = 248/474 (51%), Gaps = 58/474 (12%)
Query: 40 SKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRS---SKPKRAV----LVCCNEG-QHKV 91
++P G+EF S + +FY ++ + GF VR+ SK ++ VC EG QH
Sbjct: 188 TEPYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSKEGFQHPS 247
Query: 92 KISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSV 151
R GC A + + R + W++ N DHNH + K
Sbjct: 248 ----------------------RMGCGAYMRIKRQDSGG-WIVDRLNKDHNHDLEPGKKN 284
Query: 152 SYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDV 211
+ M KK+ T V + D N D N+I + R +
Sbjct: 285 AGM---KKI-----------------TDDVTGGLDSVDLIELN-DLSNHISSTRENTIGK 323
Query: 212 GDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYK 271
+ DY + KQ E+ FFYAI+ D + +++FW D+RSR + FGD + FDT+Y+
Sbjct: 324 EWYPVLLDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYR 383
Query: 272 TNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQD 331
YS+PFA F+G N+H Q +L G AL+ DES+ +F WLF+TWL+AM G++P S++ DQD
Sbjct: 384 KGDYSVPFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQD 443
Query: 332 LAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEE 391
L ++ A+A+VFP + HR W I K E L ++ FK + ++C+ S + F+
Sbjct: 444 LPIQQAVAQVFPGTHHRFSAWQIRSKERENLRSFPNE---FKYEYEKCLYQSQTTVEFDT 500
Query: 392 EWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDAS 451
W ++ +Y L++N WL+ +Y+ RE W+P Y R++FF G++ + + F+ + +++
Sbjct: 501 MWSSLVNKYGLRDNMWLREIYEKREKWVPAYLRASFFGGIHV---DGTFDPFYGTSLNSL 557
Query: 452 TTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
T+L+EF+ ++E+ +E R E ERKE++ S + L T +EE LYT IF
Sbjct: 558 TSLREFISRYEQGLEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTIF 611
Score = 48.1 bits (113), Expect = 2e-05
Identities = 36/115 (31%), Positives = 55/115 (47%), Gaps = 29/115 (25%)
Query: 36 GELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVCCNEGQHKVKISR 95
G+ +P +G+EF++ E+ R++YNS+ + GF VR GQ + SR
Sbjct: 22 GDSGLEPYVGLEFDTAEEARDYYNSYATRTGFKVRT---------------GQ--LYRSR 64
Query: 96 TEEIKDSTNQTKR-KCS------TIRSGCEASLIVSRGTTKSKWMIKSFNNDHNH 143
T D T ++R CS R+GC A + V R T KW++ +HNH
Sbjct: 65 T----DGTVSSRRFVCSKEGFQLNSRTGCPAFIRVQRRDT-GKWVLDQIQKEHNH 114
>At4g38170 hypothetical protein
Length = 531
Score = 251 bits (642), Expect = 7e-67
Identities = 118/291 (40%), Positives = 182/291 (61%), Gaps = 5/291 (1%)
Query: 217 VFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNK-Y 275
V +Y KR+Q+EN F YAI+ D N+FW D RL+Y FGD + FDTTY+ K Y
Sbjct: 7 VLNYLKRRQLENPGFLYAIEDD----CGNVFWADPTCRLNYTYFGDTLVFDTTYRRGKRY 62
Query: 276 SMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMK 335
+PFA F G N+H Q +L+GCAL+ +ESE+SF WLF+TWLQAM P SI + D ++
Sbjct: 63 QVPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQ 122
Query: 336 AALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWMR 395
A+++VF ++R R I ++ EKLAH++ F+ + C+ + + FE W
Sbjct: 123 VAVSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDS 182
Query: 396 IMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTLQ 455
I+ Y +++N+WLQ +Y R+ W+ ++ R TF+ ++T + S +N+FF FVDASTT+Q
Sbjct: 183 IVRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQ 242
Query: 456 EFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIFL 506
+ ++EKA++S E E K +YE+ + + ++ T S +E+ A LYT F+
Sbjct: 243 MLIKQYEKAIDSWREKELKADYEATNSTPVMKTPSPMEKQAASLYTRAAFI 293
>At1g52520 F6D8.26
Length = 703
Score = 250 bits (638), Expect = 2e-66
Identities = 147/474 (31%), Positives = 242/474 (51%), Gaps = 30/474 (6%)
Query: 42 PEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKR-------AVLVCCNEGQHKVKIS 94
P +GMEFES + +YN + + GF VRV++S KR AVL C ++G ++
Sbjct: 85 PAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSSQGFKRI--- 141
Query: 95 RTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYM 154
N R R+GC A +I R +W + DHNH++ S
Sbjct: 142 ---------NDVNRVRKETRTGCPA-MIRMRQVDSKRWRVVEVTLDHNHLLGCKLYKSVK 191
Query: 155 RCHKKMS--VAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDVG 212
R K +S V+ ++ + + G N ++ N+ N + NL G
Sbjct: 192 RKRKCVSSPVSDAKTIKLYRACVVDNGS-----NVNPNSTLNKKFQNSTGSPDLLNLKRG 246
Query: 213 DAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKT 272
D+ A+++Y R Q+ N NFFY + +++ ++ N+FW DA S++S FGDVI D++Y +
Sbjct: 247 DSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYIS 306
Query: 273 NKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDL 332
K+ +P F G+N+H ++ L C L E+ S+ WL K WL M + P +I+TD+
Sbjct: 307 GKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVM-KRSPQTIVTDRCK 365
Query: 333 AMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEE 392
++AA+++VFP S R L HI++K PEKL + H ++ + + + + FE
Sbjct: 366 PLEAAISQVFPRSHQRFSLTHIMRKIPEKLGGL-HNYDAVRKAFTKAVYETLKVVEFEAA 424
Query: 393 WMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDAST 452
W ++ + + ENEWL+ LY+ R W P+Y + TFFAG+ E++ FF+ +V T
Sbjct: 425 WGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQT 484
Query: 453 TLQEFVLKFEKAVESRLEAERKENYESRH-KSRILSTGSKLEEHAGHLYTPEIF 505
L+EF+ K+E A++ + E + ES+ + L T E +YT ++F
Sbjct: 485 PLKEFLDKYELALQKKHREETLSDIESQTLNTAELKTKCSFETQLSRIYTRDMF 538
>At2g32250 Mutator-like transposase
Length = 684
Score = 249 bits (637), Expect = 3e-66
Identities = 140/468 (29%), Positives = 236/468 (49%), Gaps = 54/468 (11%)
Query: 45 GMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKR-------AVLVCCNEGQHKVKISRTE 97
GM+FES E FY + + GFG+ +++S+ + + C G + K
Sbjct: 41 GMDFESKEAAYYFYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGTKREK----- 95
Query: 98 EIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCH 157
+T R C ++GC+A L + R + KW+I +F +HNH + C
Sbjct: 96 ----ATAINPRSCP--KTGCKAGLHMKRKEDE-KWVIYNFVKEHNHEI----------CP 138
Query: 158 KKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDVGDAQAV 217
V+ + + P G +A I+ + L+ D + +
Sbjct: 139 DDFYVSVRG-------KNKPAGALA------------------IKKGLQLALEEEDLKLL 173
Query: 218 FDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYSM 277
++ Q + FFYA+ D D R+ N+FW+DA+++ Y F DV+ FDT Y N Y +
Sbjct: 174 LEHFMEMQDKQPGFFYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRI 233
Query: 278 PFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAA 337
PFAPF+G+++H Q +L GCAL+ + SE+++ WLF+TWL+A+GG+ P +ITDQD +
Sbjct: 234 PFAPFIGVSHHRQYVLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSDI 293
Query: 338 LAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHSIQSFEEEWMRIM 397
+ +VFP+ RH CLW ++ K E L + F C+ S + + FE W ++
Sbjct: 294 VVEVFPDVRHIFCLWSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMI 353
Query: 398 VEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFVDASTTLQEF 457
++ L ENEW+Q L++ R+ W+P Y AG++ +RS SI + FD ++++ T ++F
Sbjct: 354 GKFELNENEWVQLLFRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKDF 413
Query: 458 VLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
+ K ++ R + E K++ E + K L + E+ +YT F
Sbjct: 414 FELYMKFLQYRCDVEAKDDLEYQSKQPTLRSSLAFEKQLSLIYTDAAF 461
>At1g80010 hypothetical protein
Length = 696
Score = 246 bits (628), Expect = 3e-65
Identities = 154/488 (31%), Positives = 248/488 (50%), Gaps = 43/488 (8%)
Query: 42 PEIGMEFESIEKVREFYNSFDKKNGFGVRVRSS------KPKRAVLVCCNEGQHKVKISR 95
P GMEFES + FYNS+ ++ GF +RV+SS K KR ++CCN K+
Sbjct: 66 PTPGMEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGFKL---- 121
Query: 96 TEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMR 155
+KD+ ++ K R+GC+A +I R +W + DHNH P+ +
Sbjct: 122 ---LKDAHSRRKET----RTGCQA-MIRLRLIHFDRWKVDQVKLDHNHSF-DPQRAHNSK 172
Query: 156 CHKKMSVAAKSLVEKFEEEGIPTGKVAAI-----------------FNDGDSTFTNRDCW 198
HKK S +A S K E P +V I + G+++ + D
Sbjct: 173 SHKKSSSSA-SPATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSLD-- 229
Query: 199 NYIRNVRRKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQ 258
+ ++ RR L G +A+ D+ + Q+ + NF Y + +D + N+FW+DAR+R +Y
Sbjct: 230 -HFQSSRRLELR-GGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVFWIDARARAAYS 287
Query: 259 LFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAM 318
FGDV+ FDTT +N Y +P FVG+N+H +IL GC LL D+S ++VWLF+ WL M
Sbjct: 288 HFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCM 347
Query: 319 GGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKR 378
G+ P IT+Q AM+ A+++VFP + HRL L H++ + + + +F + R
Sbjct: 348 LGRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQL-QDSDLFPMALNR 406
Query: 379 CIRGSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSE 438
+ G ++ FE W +++ + + NE ++ +++ RE W P+Y + TF AG T
Sbjct: 407 VVYGCLKVEEFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGN 466
Query: 439 SINAF-FDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAG 497
F F +V +T+L+EF+ +E ++ + E + ES L T E
Sbjct: 467 VAAPFIFSGYVHENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLKTTHPYESQMA 526
Query: 498 HLYTPEIF 505
++T EIF
Sbjct: 527 KVFTMEIF 534
>At4g19990 putative protein
Length = 672
Score = 244 bits (622), Expect = 1e-64
Identities = 146/480 (30%), Positives = 241/480 (49%), Gaps = 61/480 (12%)
Query: 39 NSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKR-------AVLVCCNEGQHKV 91
N + + G EFES E+ EFY + GF +++S+ R A VC G K
Sbjct: 18 NLEIDEGREFESKEEAFEFYKEYANSVGFTTIIKASRRSRMTGKFIDAKFVCTRYGSKKE 77
Query: 92 KISR---TE--EIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMV 146
I T+ I + + + S+ ++ C+A L V R +W+++S +HNH +
Sbjct: 78 DIDTGLGTDGFNIPQARKRGRINRSSSKTDCKAFLHVKR-RQDGRWVVRSLVKEHNHEIF 136
Query: 147 SPKSVSYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRR 206
+ ++ S ++ + +EK AI ++ V+
Sbjct: 137 TGQADSLRE------LSGRRKLEKLN---------GAI----------------VKEVKS 165
Query: 207 KNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITF 266
+ L+ GD + + NFF +Q + N+FWVDA+ R Y F DV++
Sbjct: 166 RKLEDGDVERLL-----------NFFTDMQS-----LRNIFWVDAKGRFDYTCFSDVVSI 209
Query: 267 DTTYKTNKYSMPFAPFVGLNNHSQSILYGCALL-QDESEASFVWLFKTWLQAMGGKKPIS 325
DTT+ N+Y +P F G+N+H Q +L G LL DES++ FVWLF+ WL+AM G +P
Sbjct: 210 DTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHGCRPRV 269
Query: 326 IITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQSIFKRDMKRCIRGSHS 385
I+T D +K A+ +VFP SRH +W + + PEKL H+ + ++ I GS
Sbjct: 270 ILTKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGHVIRLEKKLVDEINDAIYGSCQ 329
Query: 386 IQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFD 445
+ FE+ W ++ +++++N WLQ LY+ RE W+P+Y + AGM T QRS+S+N+ D
Sbjct: 330 SEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVSLAGMCTAQRSDSVNSGLD 389
Query: 446 SFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIF 505
++ TT + F+ +++K ++ R E E K E+ +K L + S + +YT E+F
Sbjct: 390 KYIQRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLKSPSPFGKQMAEVYTREMF 449
>At1g10240 unknown protein
Length = 680
Score = 238 bits (608), Expect = 6e-63
Identities = 147/485 (30%), Positives = 244/485 (50%), Gaps = 28/485 (5%)
Query: 39 NSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPK--------RAVLVCCNEGQHK 90
N+ P +G F + + EFY++F K+ GF +R ++ K R VC G
Sbjct: 45 NAIPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRHRTEGKDGVGKGLTRRYFVCHRAGNTP 104
Query: 91 VKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTT--KSKWMIKSFNNDHNHVMVSP 148
+K + + Q R+ S R GC+A L +S+ T ++W + F N HNH ++ P
Sbjct: 105 IKT-----LSEGKPQRNRRSS--RCGCQAYLRISKLTELGSTEWRVTGFANHHNHELLEP 157
Query: 149 KSVSYMRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFN------DGDSTFTNRDCWNYIR 202
V ++ ++ +S A KS + F + GI ++ + G FT +D N ++
Sbjct: 158 NQVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEKCVEPGFLPFTEKDVRNLLQ 217
Query: 203 NVRRKNLDVGDAQAVF-DYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFG 261
+ K LD D F C+ + ++ NF + D + ++ N+ W A S SY+LFG
Sbjct: 218 SF--KKLDPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDKLENIAWSYASSIQSYELFG 275
Query: 262 DVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGK 321
D + FDTT++ + MP +VG+NN+ +GC LL+DE+ S+ W + + M GK
Sbjct: 276 DAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQAFTGFMNGK 335
Query: 322 KPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKL-AHIYHKQSIFKRDMKRCI 380
P +I+TD ++ +K A+A P ++H LC+W ++ KFP A + + + +K + R +
Sbjct: 336 APQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFNAGLGERYNDWKAEFYR-L 394
Query: 381 RGSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESI 440
S++ FE W ++ + L N + LY R W Y RS F AGM T RS++I
Sbjct: 395 YHLESVEEFELGWRDMVNSFGLHTNRHINNLYASRSLWSLPYLRSHFLAGMTLTGRSKAI 454
Query: 441 NAFFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLY 500
NAF F+ A T L FV + V+ + +A ++ + ++ L TG+ +E HA +
Sbjct: 455 NAFIQRFLSAQTRLAHFVEQVAVVVDFKDQATEQQTMQQNLQNISLKTGAPMESHAASVL 514
Query: 501 TPEIF 505
TP F
Sbjct: 515 TPFAF 519
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 199 bits (507), Expect = 3e-51
Identities = 128/491 (26%), Positives = 233/491 (47%), Gaps = 40/491 (8%)
Query: 42 PEIGMEFESIEKVREFYNSFDKKNGFGVR-VRSSKPK-----RAVLVCCNEGQHKVKISR 95
P +G F + ++ E+Y++F +K+GF +R RS++ + R VC G ++ +
Sbjct: 70 PYVGQIFTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYRSGFNQPR--- 126
Query: 96 TEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTK--SKWMIKSFNNDHNHVMVSPKSVSY 153
K + + R+ ++R GC+ L +++ S W + F+N HNH ++ V
Sbjct: 127 ----KKANVEHPRERKSVRCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQVRL 182
Query: 154 MRCHKKMSVAAKSLVEKFEEEGIPTGKVAAIFN------DGDSTFTNRDCWNYIRNVRRK 207
+ ++K+ + + + + G P ++ + G F +D N++R ++
Sbjct: 183 LPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRACKKS 242
Query: 208 NLD---------VGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQ 258
+ D + + CK + +F Y DE+ ++ N+ W S Y
Sbjct: 243 VQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRGYS 302
Query: 259 LFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAM 318
LFGDV+ FDT+Y++ Y + F G++N+ +++L GC LLQDES SF W +T+++ M
Sbjct: 303 LFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVRFM 362
Query: 319 GGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHIIKK----FPEKLAHIYHKQSIFKR 374
G+ P +I+TD D +K A+ + P + H + + HI+ K F + L Y + F+
Sbjct: 363 RGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFSQTLGSHYEE---FRA 419
Query: 375 DMKRCIRGSHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTT 434
R ++ FE++W ++ + L + LY R SW+P R F A T+
Sbjct: 420 GFDMLCRAG-NVDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQTMTS 478
Query: 435 QRSESINAFFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEE 494
+ + SI++F VD +T +Q +L E A++ A + R L T +E+
Sbjct: 479 EFNLSIDSFLKRVVDGATCMQ--LLLEESALQVSAAASLAKQILPRFTYPSLKTCMPMED 536
Query: 495 HAGHLYTPEIF 505
HA + TP F
Sbjct: 537 HARGILTPYAF 547
>At3g45340 putative protein
Length = 693
Score = 65.5 bits (158), Expect = 9e-11
Identities = 74/330 (22%), Positives = 121/330 (36%), Gaps = 43/330 (13%)
Query: 44 IGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVCCNEGQHKVKISRTEEIKDST 103
+G EF S E V + N K+ FGV+ IK T
Sbjct: 190 VGQEFRSKEAVWDLINRASKEEVFGVKT---------------------------IKSDT 222
Query: 104 NQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVA 163
+ +CS GC+ L V++ + W +K H P++ Y + +
Sbjct: 223 GRLMLECSQASKGCDWYLRVTKTSETDFWCVKKHTQIHK-CSRCPEATRYEKQKGTPRLV 281
Query: 164 AKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDVGDA--------Q 215
A L E + + T K I + DC +Y +R K V D +
Sbjct: 282 ADVLHEDYPGL-LNTPKPKQIMSIVKGRL-GLDC-SYSTTLRGKKKHVSDVSGSPETSYK 338
Query: 216 AVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKY 275
+F Y + N ++ D D R LF S ++ VI D T+ Y
Sbjct: 339 LLFAYLHILRSVNHGTITEVELDADDRFKFLFIALGASIEGFKAMRKVIVIDATFLKTIY 398
Query: 276 S--MPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLA 333
+ A N+H I +G ++ E+ AS+ W F+ + + I D +
Sbjct: 399 GGMLVIATAQDPNHHHYPIAFG--VIDSENHASWNWFFRKLNSIIPDDPELVFIRDIHGS 456
Query: 334 MKAALAKVFPESRHRLCLWHIIKKFPEKLA 363
+ +A VFP++ H C+WH+ + + LA
Sbjct: 457 IIKGVADVFPKASHGHCVWHLSQNIKKMLA 486
>At3g59470 unknown protein
Length = 251
Score = 56.6 bits (135), Expect = 4e-08
Identities = 38/145 (26%), Positives = 64/145 (43%), Gaps = 17/145 (11%)
Query: 9 HQLMDDNGIGSSSKDFTTSILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFG 68
++L +++G D + L +D+ +P +G EFES FYN++ K GF
Sbjct: 37 NELGENSGEQDEKVDLDSIPLAVADMTEAQGDEPYVGQEFESEAAAHGFYNAYATKVGFV 96
Query: 69 VRV-------RSSKPKRAVLVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASL 121
+RV P LVC EG + ++ R+ + R GC+A +
Sbjct: 97 IRVSKLSRSRHDGSPIGRQLVCNKEGY---------RLPSKRDKVIRQRAETRVGCKAMI 147
Query: 122 IVSRGTTKSKWMIKSFNNDHNHVMV 146
++ R KW+I F +HNH ++
Sbjct: 148 LI-RKENSGKWVITKFVKEHNHSLM 171
>At3g07500 hypothetical protein
Length = 214
Score = 56.6 bits (135), Expect = 4e-08
Identities = 40/142 (28%), Positives = 69/142 (48%), Gaps = 22/142 (15%)
Query: 18 GSSSKDFTTSILDTSDIGGELNSK------PEIGMEFESIEKVREFYNSFDKKNGFGVRV 71
G+S + +++ SD+ + + P IGMEFES E + FY+++ GF +RV
Sbjct: 11 GNSLPEEDVEMMENSDVMKTVTDEASPMVEPFIGMEFESEEAAKSFYDNYATCMGFVMRV 70
Query: 72 ----RSSKPKRAV---LVCCNEGQHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVS 124
RS + V LVC EG + + R+E +++ + R GC+A ++V
Sbjct: 71 DAFRRSMRDGTVVWRRLVCNKEGFRRSRPRRSE--------SRKPRAITREGCKALIVVK 122
Query: 125 RGTTKSKWMIKSFNNDHNHVMV 146
R W++ F +HNH ++
Sbjct: 123 R-EKSGTWLVTKFEKEHNHPLL 143
>At2g23500 Mutator-like transposase
Length = 784
Score = 54.7 bits (130), Expect = 2e-07
Identities = 78/386 (20%), Positives = 145/386 (37%), Gaps = 64/386 (16%)
Query: 9 HQL---MDDNGIGSSSKDFTTSILDTSDIGGELNSKP---EIGM---------------E 47
HQL ++D+G + S +D SD G + P E GM E
Sbjct: 146 HQLEAIVEDHGTQEDETRYDES-MDNSDRGEQYVESPPAVEPGMFKKEWEDGIGLTLRQE 204
Query: 48 FESIEKVREFYNSFDKKNGFGVRVRSSKPKRAVLVC----CNEGQHKVKISRTEEIKDST 103
F + + E + N FG ++ S +R VL C C+ IS T+
Sbjct: 205 FPNKAALHEVVDRAAFANSFGYVIKKSDKERYVLKCAKESCSWRLRASNISTTDIFSIRR 264
Query: 104 NQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHKKMSVA 163
C+ + G + L +G + ++ + +DH + S + M +
Sbjct: 265 YNKMHSCTRLSKG-SSRLRKRKGNPQ---LVAALLHDHFPGQLETPVPSII-----MELV 315
Query: 164 AKSLVEKFEEEGIPTGKVAAIFN---DGDSTFTNRDCWNYIRNVRRKNLDVGDAQAVFDY 220
L K GK AI++ + ++ + +C+ Y+
Sbjct: 316 QTKLGVKVSYSTALRGKYHAIYDLKGSPEESYKDINCYLYM------------------- 356
Query: 221 CKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYS--MP 278
K+V + Y ++ DE+ + +F S +++ V+ D T+ N Y +
Sbjct: 357 --LKKVNDGTVTY-LKLDENDKFQYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLV 413
Query: 279 FAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAAL 338
FA N H I + A+L E++AS+ W F+ + + +TD++ ++ A+
Sbjct: 414 FASAQDPNRHHYIIAF--AVLDGENDASWEWFFEKLKTVVPDTSELVFMTDRNASLIKAI 471
Query: 339 AKVFPESRHRLCLWHIIKKFPEKLAH 364
V+ + H C+WH+ + H
Sbjct: 472 RNVYTAAHHGYCIWHLSQNVKGHATH 497
>At3g32060 hypothetical protein
Length = 487
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/143 (23%), Positives = 65/143 (45%), Gaps = 5/143 (3%)
Query: 224 KQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYS--MPFAP 281
K+V + Y ++ DE+ + +F S +++ V+ D T+ N Y + FA
Sbjct: 228 KKVNDGTVTY-LKLDENDKFQYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFAS 286
Query: 282 FVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAALAKV 341
N H I + A+L E++AS+ W F+ + + +TD++ ++ A+ V
Sbjct: 287 AQDPNRHHYIIAF--AVLDGENDASWEWFFEKLKTVVPDTSELVFMTDRNASLIKAIRNV 344
Query: 342 FPESRHRLCLWHIIKKFPEKLAH 364
+ + H C+WH+ + H
Sbjct: 345 YTAAHHGYCIWHLSQNVKGHATH 367
>At1g44840 hypothetical protein
Length = 926
Score = 51.6 bits (122), Expect = 1e-06
Identities = 26/127 (20%), Positives = 59/127 (45%)
Query: 235 IQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILY 294
++ + R + +F S ++ ++ D T+ KY G + + Q
Sbjct: 546 VEDESKERFLYMFLAFGASIAGFRHLRRILVVDGTHLKGKYKGVLLTSSGQDANFQVYPL 605
Query: 295 GCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKAALAKVFPESRHRLCLWHI 354
G A++ E++ S+ W F + + K ++I++D+ ++ A+ +VFP++ H C+ H+
Sbjct: 606 GFAVVDSENDESWTWFFTKLERIIADSKTLTILSDRHSSILVAVKRVFPQANHGACIIHL 665
Query: 355 IKKFPEK 361
+ K
Sbjct: 666 CRNIQTK 672
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,948,127
Number of Sequences: 26719
Number of extensions: 567526
Number of successful extensions: 2591
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 2436
Number of HSP's gapped (non-prelim): 127
length of query: 674
length of database: 11,318,596
effective HSP length: 106
effective length of query: 568
effective length of database: 8,486,382
effective search space: 4820264976
effective search space used: 4820264976
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0253.10


