

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252c.1
(325 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
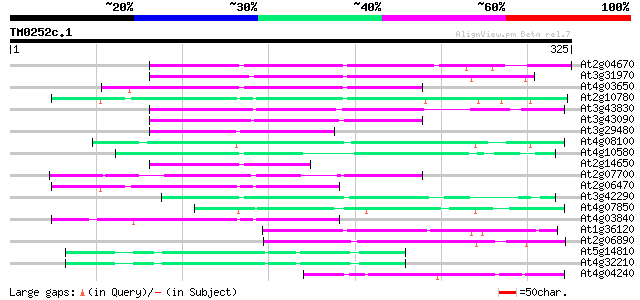
Score E
Sequences producing significant alignments: (bits) Value
At2g04670 putative retroelement pol polyprotein 104 6e-23
At3g31970 hypothetical protein 100 1e-21
At4g03650 putative reverse transcriptase 97 9e-21
At2g10780 pseudogene 92 5e-19
At3g43830 putative protein 90 1e-18
At3g43090 putative protein 82 3e-16
At3g29480 hypothetical protein 77 2e-14
At4g08100 putative polyprotein 76 3e-14
At4g10580 putative reverse-transcriptase -like protein 74 8e-14
At2g14650 putative retroelement pol polyprotein 72 5e-13
At2g07700 putative retroelement pol polyprotein 66 2e-11
At2g06470 putative retroelement pol polyprotein 63 2e-10
At3g42290 putative protein 62 4e-10
At4g07850 putative polyprotein 61 7e-10
At4g03840 putative transposon protein 60 1e-09
At1g36120 putative reverse transcriptase gb|AAD22339.1 54 1e-07
At2g06890 putative retroelement integrase 53 2e-07
At5g14810 putative protein 48 6e-06
At4g32210 putative protein 48 6e-06
At4g04240 putative protein 48 8e-06
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 104 bits (260), Expect = 6e-23
Identities = 76/252 (30%), Positives = 110/252 (43%), Gaps = 26/252 (10%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
F+ GT P+EAD W +++ F + +V LA + L GDA WWR +
Sbjct: 138 FSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTA--RRRQAD 195
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDE 201
++W F A F KYFP A D E++FL L QG ++ EY + L + R D+
Sbjct: 196 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRLLAYAG--RGMEDD 253
Query: 202 PYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRS 261
+RF+RGLR D+ R ALVE A EVE R++ G ++T +
Sbjct: 254 QAQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQRQV--VGVSPAVQTKNTQ 311
Query: 262 YQ------GKGKLQRKKPYQRPT--GEGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGE 313
Q GK +K+ + P+ G+G G + GS + T C E
Sbjct: 312 QQVTPSKGGKPAQGQKRKWDHPSRAGQGGRAGYF------------SCGSLDHTVADCTE 359
Query: 314 IGHYSTKCPKGK 325
GH K G+
Sbjct: 360 RGHLHVKVAGGQ 371
>At3g31970 hypothetical protein
Length = 1329
Score = 100 bits (249), Expect = 1e-21
Identities = 76/229 (33%), Positives = 101/229 (43%), Gaps = 10/229 (4%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
F GGT P+EAD W +E F + +V LA + L GDA WWR H
Sbjct: 158 FFGGTSPEEADSWKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAH-- 215
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDE 201
++W F A F KYFP A D E++FL L QG ++ EY + L + R D+
Sbjct: 216 MSWADFVAEFNAKYFPQEALDRMEARFLELTQGERSVREYEREFNRLLVYAG--RGMQDD 273
Query: 202 PYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTG-GPMRTSSR 260
+RF+RGLR D+ R L ALVE A EVE R++ P +T +
Sbjct: 274 QAQMRRFLRGLRPDLRVRCRVLQYATKAALVETAAEVEEDLQRQVVGVSPAVKPKKTQQQ 333
Query: 261 SYQGKG--KLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA---GSQAGSR 304
KG Q +K +R P L R +A A G+Q R
Sbjct: 334 VAPSKGGKPAQGQKRKERGHLRPNCPKLQRMAVAVVQPAVQHGAQVQQR 382
>At4g03650 putative reverse transcriptase
Length = 839
Score = 97.4 bits (241), Expect = 9e-21
Identities = 63/190 (33%), Positives = 90/190 (47%), Gaps = 8/190 (4%)
Query: 54 RELHLRQREASLDQN----RGLNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTSE 109
R + ++QR A ++ R + R F+GGT P+EAD W ++E+ F +
Sbjct: 124 RVVEVQQRAAVAEEVPSYLRMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPA 183
Query: 110 GAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERESQFL 169
+V L + L GDA WWR +++W F A F KYFP A D E++FL
Sbjct: 184 EYRVDLTVHFLEGDAHLWWRSVTA--RRRQADMSWADFMAEFNAKYFPREALDRMEARFL 241
Query: 170 TLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQ 229
L QG ++ EY K L + R D+ +RF+RGLR ++ R
Sbjct: 242 ELTQGVRSVREYDRKFNRLLVYAG--RGMEDDQAQMRRFLRGLRPNLRVRCRVSQYATKA 299
Query: 230 ALVEKATEVE 239
ALVE A EVE
Sbjct: 300 ALVETAAEVE 309
>At2g10780 pseudogene
Length = 1611
Score = 91.7 bits (226), Expect = 5e-19
Identities = 88/329 (26%), Positives = 133/329 (39%), Gaps = 37/329 (11%)
Query: 25 QLAEMVATLVQAMTVQTNDNAQRRAAE----DARELHLRQREASLDQNRGLNNFIRQNPP 80
QLA + ++ V DN + +E + + H R+R L L + R
Sbjct: 124 QLATLTKAVIADPVVLKVDNQKDDDSEVVEVNPQPNHGRKRGDYLSL---LEHVSRLGTR 180
Query: 81 KFTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHV 140
F G TDP AD W +++ F+ + E + +A + L GDA WW + V
Sbjct: 181 HFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVE-KRRGDEV 239
Query: 141 EVNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVD 200
++ F F +KYFP A D E +L L QG+ T+ EY + L ++ R+ +
Sbjct: 240 R-SFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVG--RELEE 296
Query: 201 EPYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVE--LMKNRRMDRAGTGGPMRTS 258
E +RF+RGLR +I + LVE+A +E + + R ++R S
Sbjct: 297 EQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMIEEGIEEERYLNREKAPIRNNQS 356
Query: 259 SRSYQGKGKLQR--KKPYQRPTGEGFT-----PGLYRPTIAAAGGAGSQ----------- 300
++ K K + TGE T G I A G GS+
Sbjct: 357 TKPADKKRKFDKVDNTKSDAKTGECVTCGKNHSGTCWKAIGACGRCGSKDHAIQSCPKME 416
Query: 301 ------AGSREITCFKCGEIGHYSTKCPK 323
G TCF CG+ GH +CPK
Sbjct: 417 PGQSKVLGEETRTCFYCGKTGHLKRECPK 445
>At3g43830 putative protein
Length = 329
Score = 90.1 bits (222), Expect = 1e-18
Identities = 66/240 (27%), Positives = 103/240 (42%), Gaps = 36/240 (15%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
F GGTDP +A W +E F+ ++ LA+ L G+A+ WW + + V+
Sbjct: 116 FNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWWERVKQREQVGCVD 175
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDE 201
W+ F+ F +Y P+ D+ E +FL L+QG+ T+ +Y + SL RF R + E
Sbjct: 176 -QWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLE---RFERRKRGE 231
Query: 202 PYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRS 261
+ +F+ GLR DI LVEKA +E+
Sbjct: 232 HELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLEI--------------------- 270
Query: 262 YQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKC 321
L+ + Y+R E G Y T S+ +E+TC++CG GH + C
Sbjct: 271 -----GLEEEARYKRIAQEKEAMGSYGQT------GHSKRRCQEVTCYRCGVAGHIARDC 319
>At3g43090 putative protein
Length = 357
Score = 82.4 bits (202), Expect = 3e-16
Identities = 53/158 (33%), Positives = 72/158 (45%), Gaps = 3/158 (1%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
FTGG DP +AD W + +E FE + + + + L +A +WW G G HV
Sbjct: 131 FTGGIDPVKADDWRKLLENNFENARCPVEFQKDIIVHFLREEASHWWDGVLGNTPVQHV- 189
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDE 201
+ W FR F K+FP A D E F LRQ + + E +L L++ R E
Sbjct: 190 IFWEDFREEFNRKFFPQEAMDSLEDDFEELRQDTKKVRENERELSHLSRF--SVRAGRGE 247
Query: 202 PYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVE 239
M +R +RGLR DI LVEKA +E
Sbjct: 248 QSMIRRLMRGLRPDILTRCASREFFSMIDLVEKAARIE 285
>At3g29480 hypothetical protein
Length = 718
Score = 76.6 bits (187), Expect = 2e-14
Identities = 39/107 (36%), Positives = 53/107 (49%), Gaps = 2/107 (1%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
F+GGT P+EAD W +E+ F + +V LA + L GDA WW+
Sbjct: 51 FSGGTSPEEADSWRSRVERNFGSSRCPVEYRVDLAVHFLEGDAHLWWKSV--TTRRRQAN 108
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESL 188
++W F A F KYFP A D E+ FL L QG ++ EY + L
Sbjct: 109 MSWADFVAEFNAKYFPQEALDRMEAHFLELTQGERSVREYDREFNRL 155
>At4g08100 putative polyprotein
Length = 1054
Score = 75.9 bits (185), Expect = 3e-14
Identities = 69/281 (24%), Positives = 113/281 (39%), Gaps = 25/281 (8%)
Query: 49 AAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTS 108
++ ++ H R+R D GL I P F G +PD W Q+IE +F +
Sbjct: 70 SSHSSQRRHRRERPPPRDPLGGLKLKI----PAFHGTDNPDTYLEWEQKIELVFLCQECL 125
Query: 109 EGAKVGLATYLLLGDAEYWWRG--ARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERES 166
+ KV +A A WW ++ WN + +++ P E
Sbjct: 126 QSNKVKIAATKFYNYALSWWDQLVTSRRRTRDYPIKTWNQLKFVMRKRFVPSYYHRELHQ 185
Query: 167 QFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIM 226
+ L QGS T+ EY ++E+L Q D + RF+ GL +I+D + +
Sbjct: 186 RLRNLVQGSKTVEEYFLEMETLMLRADL---QEDGEAVMSRFMGGLNREIQDRLETQHYV 242
Query: 227 RFQALVEKATEVELMKNRRMDRAG-TGGPMRTSSRSYQGKGKL---QRKKPYQRPTGEGF 282
+ ++ KA E R+ R+ T + SYQ + K KP+ +P
Sbjct: 243 ELEEMLHKAVMFEQQIKRKNARSSHTKTNYSSGKPSYQKEEKFGYQNDSKPFVKP----- 297
Query: 283 TPGLYRPTIAAAGGAGSQ--AGSREITCFKCGEIGHYSTKC 321
+P G G + +R+I CFK + HY++KC
Sbjct: 298 -----KPVDLDPKGKGKEVITRARDIRCFKSQGLRHYASKC 333
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 74.3 bits (181), Expect = 8e-14
Identities = 70/256 (27%), Positives = 94/256 (36%), Gaps = 49/256 (19%)
Query: 62 EASLDQNRGLNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLL 121
E L R + R + F+GGT P+EAD W + + F + +V LA + L
Sbjct: 132 EEVLSYLRMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRCPAEYRVDLAVHFLE 191
Query: 122 GDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEY 181
GDA WWR +++W F A F KYFP A D Q +
Sbjct: 192 GDAHLWWRSVTA--RRRQTDMSWADFVAEFKAKYFPQEALDPYAGQGME----------- 238
Query: 182 AAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVELM 241
D+ +RF+RGLR D+ R ALVE A EVE
Sbjct: 239 ------------------DDQAQMRRFLRGLRPDLRVRCRVSQYATKAALVETAAEVEED 280
Query: 242 KNRR-MDRAGTGGPMRTSSRSYQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGAGSQ 300
R+ + + P +T + KG KP Q G P+ A GG
Sbjct: 281 FQRQVVGVSPVVQPKKTQQQVTPSKG----GKPAQ-----GQKRKWDHPSRAGQGG---- 327
Query: 301 AGSREITCFKCGEIGH 316
CF CG + H
Sbjct: 328 ----RARCFSCGSLDH 339
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 71.6 bits (174), Expect = 5e-13
Identities = 35/93 (37%), Positives = 47/93 (49%), Gaps = 2/93 (2%)
Query: 82 FTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVE 141
F+GGT P+ AD W +E+ F + ++ LA + L GDA WWR +
Sbjct: 156 FSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLWWRSVTA--RRRQAD 213
Query: 142 VNWNSFRAAFLEKYFPDSARDERESQFLTLRQG 174
++W F A F KYFP A D E+ FL L QG
Sbjct: 214 ISWADFVAEFNAKYFPQEALDRMEAHFLELTQG 246
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 66.2 bits (160), Expect = 2e-11
Identities = 53/216 (24%), Positives = 90/216 (41%), Gaps = 38/216 (17%)
Query: 24 NQLAEMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFT 83
+QL M+ T +Q + + +A+ + L + R+ + +G +N +
Sbjct: 10 SQLGHMIETQQHQQQIQESQREESTSAKFLK-LVMMMRDLGIRVFKGEHNTV-------- 60
Query: 84 GGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVN 143
AD W++++E FE+ + + K +A L DA W + + +
Sbjct: 61 ------VADKWLRDLEMNFEISRCPDNFKRQIAVNFLDKDARIWRESVTARNQGQMI--S 112
Query: 144 WNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPY 203
W +FR F +KYFP ARD E QF+ K + + Q DE
Sbjct: 113 WEAFRREFEKKYFPPEARDRLEQQFM--------------------KRY-LYNGQDDEAL 151
Query: 204 MCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVE 239
M +RF+RGLR +I ++ + L E+A VE
Sbjct: 152 MVRRFLRGLRLEIWGRLQAVKYDSVNELTERADNVE 187
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 63.2 bits (152), Expect = 2e-10
Identities = 50/171 (29%), Positives = 76/171 (44%), Gaps = 9/171 (5%)
Query: 25 QLAEMVATLVQAMTVQTNDNAQRRAAE----DARELHLRQREASLDQNRGLNNFIRQNPP 80
QLA + +V VQ DN + +E + + H R+R L L + R
Sbjct: 89 QLATLTKAVVADPVVQRVDNQKDDDSEVVEVNPQPNHGRKRGDYLSL---LEHVSRLGTR 145
Query: 81 KFTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHV 140
F G TDP AD W +++ F+ + E + +A + L GDA WW + V
Sbjct: 146 HFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVE-KRRGDEV 204
Query: 141 EVNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKH 191
++ F F +KYFP A D E +L L QG+ T+ EY + L ++
Sbjct: 205 R-SFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At3g42290 putative protein
Length = 462
Score = 62.0 bits (149), Expect = 4e-10
Identities = 56/228 (24%), Positives = 83/228 (35%), Gaps = 44/228 (19%)
Query: 89 DEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFR 148
++AD W +++ F + +V LA + L DA WW+ +++W F
Sbjct: 161 EKADSWRSRVKRNFSSSRCLMEYRVDLAVHFLEEDAHLWWKSVTA--RRRQADMSWADFV 218
Query: 149 AAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRF 208
F KYF D E +FL L ++ EY + + + + D +RF
Sbjct: 219 VEFNAKYFLQDELDRMEVRFLELTLVERSVWEYDREFNRVLVYAGW--GMKDGQAELRRF 276
Query: 209 VRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKGKL 268
+RGLR D+ R ALVE A EV K G PM+ RS+ +
Sbjct: 277 MRGLRPDLRVRCRMSQYALKAALVETAAEVAPRKG--------GKPMQGKKRSWNHLSR- 327
Query: 269 QRKKPYQRPTGEGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGH 316
G G +AG CF CG + H
Sbjct: 328 --------------------------AGQGGRAG-----CFSCGNLAH 344
>At4g07850 putative polyprotein
Length = 1138
Score = 61.2 bits (147), Expect = 7e-10
Identities = 56/222 (25%), Positives = 89/222 (39%), Gaps = 27/222 (12%)
Query: 108 SEGAKVGLATYLLLGDAEYWWRGA---RGMMEANHVEVNWNSFRAAFLEKYFPDSARDER 164
+E KV +A A WW + + + +E WN + ++ P E
Sbjct: 91 TEANKVKVAATEFYDYALSWWDQVVTTKRRLGDDSIET-WNQLKNIMKRRFVPSHYHREL 149
Query: 165 ESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMC--KRFVRGLRADIEDSVRP 222
+ L QG+ T+ EY ++E+L R V E RF+ GL DI D
Sbjct: 150 HQRLRNLVQGNRTVEEYFKEMETL-----MLRADVQEECEATMSRFMGGLNRDILDRFEV 204
Query: 223 LGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKGKL---QRKKPYQRPTG 279
+ + L KA E RR + +S SYQ + K + KP+ +P
Sbjct: 205 IHYENLEELFHKAVMFEKQIKRRSAKPS----YNSSKPSYQREEKSGFQKEYKPFVKPKV 260
Query: 280 EGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKC 321
E I++ G +R++ CFKC +GHY+++C
Sbjct: 261 E---------EISSKGKEKEVTRTRDLKCFKCHGLGHYASEC 293
>At4g03840 putative transposon protein
Length = 973
Score = 60.5 bits (145), Expect = 1e-09
Identities = 48/172 (27%), Positives = 75/172 (42%), Gaps = 11/172 (6%)
Query: 25 QLAEMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRG-----LNNFIRQNP 79
QLA + +V VQ DN + +D E+ + + + RG L + R
Sbjct: 89 QLATLTKAIVANPVVQRVDNQK----DDELEVVEVNPQPNHGRKRGDYLSLLEHVSRLGT 144
Query: 80 PKFTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANH 139
F G TDP AD W +++ F+ + E + +A + L DA WW +
Sbjct: 145 RHFMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLERDAHNWWLTVE-KRRGDE 203
Query: 140 VEVNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKH 191
V ++ F F +KYFP A D E +L L QG+ T+ EY + L ++
Sbjct: 204 VR-SFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRY 254
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 53.9 bits (128), Expect = 1e-07
Identities = 46/178 (25%), Positives = 79/178 (43%), Gaps = 11/178 (6%)
Query: 147 FRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCK 206
F A F KYFP A D E++FL L +G +++ EY + + L + R D+ +
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAG--RGMEDDQAQMR 214
Query: 207 RFVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKG 266
RF+RGLR D+ R AL E +L + + + + + P + + KG
Sbjct: 215 RFLRGLRPDLRVRCRVSQYATKAALTAAEVEEDLQR-QVVAVSPSVQPKKIQQQVAPSKG 273
Query: 267 ----KLQRKK---PYQRPTGEGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGHY 317
++Q++K P++ G + + AGGA AG+ + +C + Y
Sbjct: 274 GKPAQVQKRKWDHPFRAGQGGRAGSAADDSSTSTAGGAAQSAGAAR-SAAECAAVSTY 330
>At2g06890 putative retroelement integrase
Length = 1215
Score = 53.1 bits (126), Expect = 2e-07
Identities = 41/181 (22%), Positives = 74/181 (40%), Gaps = 15/181 (8%)
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+A +++ P E + L QG+ ++ EY ++E+L D+ R
Sbjct: 2 KAIMRKRFVPSHYHRELHLKLRNLTQGNRSVEEYYKEMETLMLRADISEDR---EATLSR 58
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKGK 267
F+ L DI+D + ++ + ++ KA E R+ + G + +YQ + +
Sbjct: 59 FLGDLNRDIQDRLETQYYVQIEEMLHKAILFEQQVKRKSSSRSSYGSGTIAKPTYQREER 118
Query: 268 LQ--RKKPYQRPTGEGFTPGLYRPTIAAAGGAG----SQAGSREITCFKCGEIGHYSTKC 321
KP P E +P A G S + R++ C+KC GHY+ +C
Sbjct: 119 TSSYHNKPIVSPRAES------KPYAAVQDHKGKAEISTSRVRDVRCYKCQGKGHYANEC 172
Query: 322 P 322
P
Sbjct: 173 P 173
>At5g14810 putative protein
Length = 716
Score = 48.1 bits (113), Expect = 6e-06
Identities = 48/197 (24%), Positives = 79/197 (39%), Gaps = 22/197 (11%)
Query: 33 LVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDEAD 92
L+ + T D ++ R + L +R ++F R +F G D
Sbjct: 72 LISTAPIVTPDTSKERVGMSNKYFPLEER----------SDFARLEHLRFNG----DRIT 117
Query: 93 LWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFL 152
W+ +IE+ F + +T E KVG A+ A + M HV +W S++ L
Sbjct: 118 EWLFQIEQFFLIDRTPEELKVGFASLHFDDTAATLHQSIVQSMWWKHVRHDWWSYK-LLL 176
Query: 153 EKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGL 212
+ + + D LT Q + I EY A+ E ++ F D Y+ ++ GL
Sbjct: 177 QVRYDEHVNDSIAK--LTQLQETEGIEEYHARFELISTRLNFAED-----YLVSVYLAGL 229
Query: 213 RADIEDSVRPLGIMRFQ 229
R D + +VR G Q
Sbjct: 230 RTDTQLNVRMFGPQTIQ 246
>At4g32210 putative protein
Length = 1086
Score = 48.1 bits (113), Expect = 6e-06
Identities = 48/197 (24%), Positives = 79/197 (39%), Gaps = 22/197 (11%)
Query: 33 LVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDEAD 92
L+ + T D ++ R + L +R ++F R +F G D
Sbjct: 72 LISTAPIVTPDTSKERVGMSNKYFPLEER----------SDFARLEHLRFNG----DRIT 117
Query: 93 LWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFL 152
W+ +IE+ F + +T E KVG A+ A + M HV +W S++ L
Sbjct: 118 EWLFQIEQFFLIDRTPEELKVGFASLHFDDTAATLHQSIVQSMWWKHVRHDWWSYK-LLL 176
Query: 153 EKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGL 212
+ + + D LT Q + I EY A+ E ++ F D Y+ ++ GL
Sbjct: 177 QVRYDEHVNDSIAK--LTQLQETEGIEEYHARFELISTRLNFAED-----YLVSVYLAGL 229
Query: 213 RADIEDSVRPLGIMRFQ 229
R D + +VR G Q
Sbjct: 230 RTDTQLNVRMFGPQTIQ 246
>At4g04240 putative protein
Length = 334
Score = 47.8 bits (112), Expect = 8e-06
Identities = 42/155 (27%), Positives = 67/155 (43%), Gaps = 10/155 (6%)
Query: 171 LRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQA 230
L QG+ T+ EY + E L ++ DE M + F+ GL+ I+ V
Sbjct: 135 LSQGTRTVDEYFEEFEKLMNALEL--EESDEALMAQ-FIDGLQERIQRKVERAQYNGLHE 191
Query: 231 LVEKATEVELMKNRRM---DRAGTGGPMRTSSRSYQGKGKL-QRKKPYQRPTGEGFTPGL 286
L+ A +VE R+ +R+ P S+ K K+ + ++ + E P
Sbjct: 192 LLHLAVQVEQQIRRKASLSNRSRNNTPWNASNNRAMDKSKVVESDHRFKNKSNEA--PKT 249
Query: 287 YRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKC 321
RP + +Q+ SR ITCFKC GH + +C
Sbjct: 250 SRPKLGKFPST-NQSRSRYITCFKCQGRGHMAREC 283
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,123,695
Number of Sequences: 26719
Number of extensions: 299534
Number of successful extensions: 1021
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 894
Number of HSP's gapped (non-prelim): 121
length of query: 325
length of database: 11,318,596
effective HSP length: 100
effective length of query: 225
effective length of database: 8,646,696
effective search space: 1945506600
effective search space used: 1945506600
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0252c.1


