

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.3
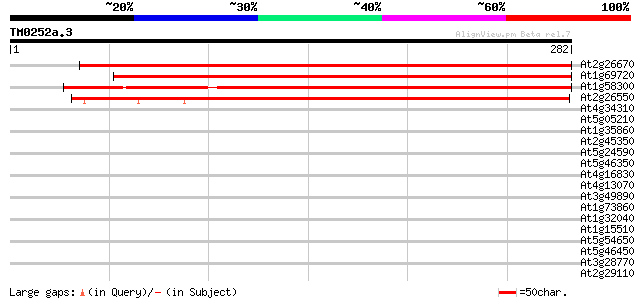
(282 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g26670 heme oxygenase 1 (HO1) 384 e-107
At1g69720 putative heme oxygenase 364 e-101
At1g58300 hypothetical protein 301 2e-82
At2g26550 heme oxygenase 2 (HO2) 231 4e-61
At4g34310 hypothetical protein 37 0.012
At5g05210 unknown protein 31 0.65
At1g35860 hypothetical protein 31 0.86
At2g45350 hypothetical protein 30 1.5
At5g24590 NAC2-like protein 30 1.9
At5g46350 putative protein 29 2.5
At4g16830 nuclear antigen homolog 29 2.5
At4g13070 putative protein 29 3.2
At3g49890 unknown protein 28 4.2
At1g73860 kinesin-related protein 28 4.2
At1g32040 hypothetical protein 28 4.2
At1g15510 hypothetical protein 28 4.2
At5g54650 unknown protein 28 5.5
At5g46450 disease resistance protein-like 28 5.5
At3g28770 hypothetical protein 28 5.5
At2g29110 putative ligand-gated ion channel protein (AtGLR2.8) 28 5.5
>At2g26670 heme oxygenase 1 (HO1)
Length = 282
Score = 384 bits (987), Expect = e-107
Identities = 180/247 (72%), Positives = 213/247 (85%)
Query: 36 PTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGE 95
P ++L + VV+ + TAAE +KRYPGESKGFVEEMRFVAM+LHTKDQAKEGE
Sbjct: 36 PRIQILSMTMNKSPSLVVVAATTAAEKQKKRYPGESKGFVEEMRFVAMRLHTKDQAKEGE 95
Query: 96 KEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSAS 155
KE EE+ V KWEPTV+GYL+FLVDSK+VYDTLE I+QD+ P+YAEF+NTGLER+
Sbjct: 96 KETKSIEERPVAKWEPTVEGYLRFLVDSKLVYDTLELIIQDSNFPTYAEFKNTGLERAEK 155
Query: 156 LAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGK 215
L+ DLEWFKEQGY IP+P++PG Y+QYL++L++ DPQAFICHFYNIYFAHSAGGRMIG+
Sbjct: 156 LSTDLEWFKEQGYEIPEPTAPGKTYSQYLKELAEKDPQAFICHFYNIYFAHSAGGRMIGR 215
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGE 275
K+A +L+NK LEFYKWDG+LSQLLQNVR+KLNKVAE+WTREEKNHCLEETEKSFK+SGE
Sbjct: 216 KVAERILDNKELEFYKWDGELSQLLQNVREKLNKVAEEWTREEKNHCLEETEKSFKYSGE 275
Query: 276 ILRLILS 282
ILRLILS
Sbjct: 276 ILRLILS 282
>At1g69720 putative heme oxygenase
Length = 285
Score = 364 bits (934), Expect = e-101
Identities = 171/230 (74%), Positives = 199/230 (86%)
Query: 53 VIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPT 112
V+ +A E +K+YPGESKGFVEEMRFVAM+LHTKDQA+EGEKE PEE V KWEPT
Sbjct: 56 VVTAAAITEKQQKKYPGESKGFVEEMRFVAMRLHTKDQAREGEKESRSPEEGPVAKWEPT 115
Query: 113 VDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQ 172
V+GYL FLVDSK+VYDTLE I+ + P+YA F+NTGLER+ SL KDLEWFKEQGY IP+
Sbjct: 116 VEGYLHFLVDSKLVYDTLEGIIDGSNFPTYAGFKNTGLERAESLRKDLEWFKEQGYEIPE 175
Query: 173 PSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKW 232
P +PG Y++YL+DL++NDPQAFICHFYNIYFAHSAGG+MIG K++ ++L+NK LEFYKW
Sbjct: 176 PMAPGKTYSEYLKDLAENDPQAFICHFYNIYFAHSAGGQMIGTKVSKKILDNKELEFYKW 235
Query: 233 DGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
DG LSQLLQNVR KLNKVAE WTREEK+HCLEETEKSFK+SGEILRLILS
Sbjct: 236 DGQLSQLLQNVRQKLNKVAEWWTREEKSHCLEETEKSFKFSGEILRLILS 285
>At1g58300 hypothetical protein
Length = 283
Score = 301 bits (772), Expect = 2e-82
Identities = 148/255 (58%), Positives = 192/255 (75%), Gaps = 5/255 (1%)
Query: 28 RQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHT 87
R R+I P L+++ Q V +V+A A E +RYP E GFVEEMRFV MK+H
Sbjct: 34 RYVRTIAAPRRHLVRRANEDQTLVVNVVAA-AGEKPERRYPREPNGFVEEMRFVVMKIHP 92
Query: 88 KDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRN 147
+DQ KEG+ + + V+ W T++GYLKFLVDSK+V++TLE+I+ ++ +YA +N
Sbjct: 93 RDQVKEGKSDSND----LVSTWNFTIEGYLKFLVDSKLVFETLERIINESAIQAYAGLKN 148
Query: 148 TGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHS 207
TGLER+ +L++DLEWFKEQGY IP+ PG Y+QYL+++++ DP AFICHFYNI FAHS
Sbjct: 149 TGLERAENLSRDLEWFKEQGYEIPESMVPGKAYSQYLKNIAEKDPPAFICHFYNINFAHS 208
Query: 208 AGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETE 267
AGGRMIG K+A ++L+NK LEFYKWDG LS+LLQNV ++LNKVAE WTREEKNHCLEETE
Sbjct: 209 AGGRMIGTKVAEKILDNKELEFYKWDGQLSELLQNVSEELNKVAELWTREEKNHCLEETE 268
Query: 268 KSFKWSGEILRLILS 282
KSFK+ EI R +LS
Sbjct: 269 KSFKFYWEIFRYLLS 283
>At2g26550 heme oxygenase 2 (HO2)
Length = 299
Score = 231 bits (588), Expect = 4e-61
Identities = 118/274 (43%), Positives = 176/274 (64%), Gaps = 24/274 (8%)
Query: 32 SIYFP----TTRLLQQHRFRQMKSVVIVSATAAETA-----RKRYPGESKGFVEEMRFVA 82
SI FP T R Q+H +S S A RK+YPGE+ G EEMRFVA
Sbjct: 25 SISFPFQISTQRKPQKHLLNLCRSTPTPSQQKASQRKRTRYRKQYPGENIGITEEMRFVA 84
Query: 83 MKLH---------------TKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVY 127
M+L T+ + +E E++ + +E W+P+ +G+LK+LVDSK+V+
Sbjct: 85 MRLRNVNGKKLDLSEDKTDTEKEEEEEEEDDDDDDEVKEETWKPSKEGFLKYLVDSKLVF 144
Query: 128 DTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDL 187
DT+E+IV ++ + SYA FR TGLER S+ KDL+W +EQ IP+PS+ G++YA+YL +
Sbjct: 145 DTIERIVDESENVSYAYFRRTGLERCESIEKDLQWLREQDLVIPEPSNVGVSYAKYLEEQ 204
Query: 188 SQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKL 247
+ F+ HFY+IYF+H AGG+++ ++++ +LL K LEF +W+GD LL+ VR+KL
Sbjct: 205 AGESAPLFLSHFYSIYFSHIAGGQVLVRQVSEKLLEGKELEFNRWEGDAQDLLKGVREKL 264
Query: 248 NKVAEQWTREEKNHCLEETEKSFKWSGEILRLIL 281
N + E W+R+EKN CL+ET K+FK+ G+I+RLI+
Sbjct: 265 NVLGEHWSRDEKNKCLKETAKAFKYMGQIVRLII 298
>At4g34310 hypothetical protein
Length = 1074
Score = 37.0 bits (84), Expect = 0.012
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 8/55 (14%)
Query: 12 SLYIIKPRLSPPPPPHRQ---FRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETA 63
SL +IKP+L+P PPPH+ RSIY + KSV ++SA A TA
Sbjct: 38 SLNLIKPKLTPSPPPHQSRGLLRSIY-----TVSPSSSFSKKSVFVLSAAALSTA 87
>At5g05210 unknown protein
Length = 386
Score = 31.2 bits (69), Expect = 0.65
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 15/95 (15%)
Query: 61 ETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFL 120
ET RK YPG SK + K T D K+ +++ +PE+ A+T T+D LK
Sbjct: 47 ETERKWYPGLSKA-----QKARAKKKTNDNLKKAKRDKLDPEKSALT----TLD-LLKEK 96
Query: 121 VDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSAS 155
++ + + +K+ + +A+ R LE+ S
Sbjct: 97 IEKERLASQKQKLKK-----KHADLREQKLEQEKS 126
>At1g35860 hypothetical protein
Length = 399
Score = 30.8 bits (68), Expect = 0.86
Identities = 15/31 (48%), Positives = 21/31 (67%)
Query: 222 LNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE 252
LN+K L F +GD++QL+ +DKL VAE
Sbjct: 66 LNSKELIFEVMEGDITQLVIQFQDKLGNVAE 96
>At2g45350 hypothetical protein
Length = 606
Score = 30.0 bits (66), Expect = 1.5
Identities = 16/67 (23%), Positives = 29/67 (42%), Gaps = 2/67 (2%)
Query: 68 PGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKF--LVDSKV 125
P E K + ++ T D K + EK + W +DGY+K + D+K
Sbjct: 207 PMEMKNLISWNSMISGYAQTSDGVDIASKLFADMPEKDLISWNSMIDGYVKHGRIEDAKG 266
Query: 126 VYDTLEK 132
++D + +
Sbjct: 267 LFDVMPR 273
>At5g24590 NAC2-like protein
Length = 451
Score = 29.6 bits (65), Expect = 1.9
Identities = 31/116 (26%), Positives = 44/116 (37%), Gaps = 14/116 (12%)
Query: 80 FVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATH 139
FV KL K G EPEE + EP V S V D +E
Sbjct: 156 FVVCKLFKKQDIVNG---AAEPEESKSCEVEPAVS--------SPTVVDEVEMSEVSPVF 204
Query: 140 PSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAF 195
P E + S SL E GY++P+ ++ GL+ +L + + P+ F
Sbjct: 205 PKTEETNPCDVAES-SLVIPSEC--RSGYSVPEVTTTGLDDIDWLSFMEFDSPKLF 257
>At5g46350 putative protein
Length = 326
Score = 29.3 bits (64), Expect = 2.5
Identities = 20/74 (27%), Positives = 36/74 (48%), Gaps = 4/74 (5%)
Query: 49 MKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTK 108
+KS V VSA+ + + +PGE G + + R ++ + + ++V + ++K K
Sbjct: 111 IKSEVRVSASPSSSEADHHPGEDSGKIRKKR----EVRDGGEDDQRSQKVVKTKKKEEKK 166
Query: 109 WEPTVDGYLKFLVD 122
EP V K VD
Sbjct: 167 KEPRVSFMTKTEVD 180
>At4g16830 nuclear antigen homolog
Length = 355
Score = 29.3 bits (64), Expect = 2.5
Identities = 21/61 (34%), Positives = 27/61 (43%)
Query: 70 ESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDT 129
E+ VE + V K D A + KE T EEK E T+D Y K L + K +
Sbjct: 185 EAVAGVETEKDVGEKPAVDDVAADANKEDTVVEEKEPEDKEMTLDEYEKILEEKKKALQS 244
Query: 130 L 130
L
Sbjct: 245 L 245
>At4g13070 putative protein
Length = 332
Score = 28.9 bits (63), Expect = 3.2
Identities = 50/232 (21%), Positives = 86/232 (36%), Gaps = 17/232 (7%)
Query: 55 VSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVD 114
V T E KR K V E+RF +K K + E + EKA K E ++
Sbjct: 92 VVPTRKEKKWKRAKLSRKAKVNELRFYRLKAKKKMNSPNPEVRIRYKLEKAKRKEEWLIE 151
Query: 115 GYLKFLVDSKVV--YDTLEKIVQDATH---------PSYAEFRNTGLERSASLAKDLEWF 163
K+ V YD E + ++ H ++ G+ L L W
Sbjct: 152 KLRKYDVPKSPAEPYDP-ESLTEEEQHYLKRTGEKRKNFVLVGRRGVFGGVVLNLHLHWK 210
Query: 164 K-EQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKI---AG 219
K E I +P + +Y +L++ I N G + ++
Sbjct: 211 KHETVKVICKPCNKPGQVHEYAEELARLSKGIVIDVKPNNTIVLYRGKNYVRPEVMSPVD 270
Query: 220 ELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFK 271
L +K LE Y+++ L + + +KL K E++ + H ++ E++ K
Sbjct: 271 TLSKDKALEKYRYEQSLEHTSEFI-EKLEKELEEYHKYVARHKKKKDEEAEK 321
>At3g49890 unknown protein
Length = 220
Score = 28.5 bits (62), Expect = 4.2
Identities = 18/76 (23%), Positives = 37/76 (48%), Gaps = 14/76 (18%)
Query: 47 RQMKSVVI----VSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPE 102
+++KS+ + + A E A+K R A ++ KD+AK+ E+ + E +
Sbjct: 139 KRVKSIAVDGSDILTAAVEAAKKA----------SARLDAKEVAAKDKAKKEEERIAELK 188
Query: 103 EKAVTKWEPTVDGYLK 118
+ KW P+++ +K
Sbjct: 189 KVRGEKWLPSIERAMK 204
>At1g73860 kinesin-related protein
Length = 1050
Score = 28.5 bits (62), Expect = 4.2
Identities = 23/103 (22%), Positives = 45/103 (43%), Gaps = 10/103 (9%)
Query: 37 TTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGF----------VEEMRFVAMKLH 86
TTR + + QM+S +V+ T E+ K E K V+E+ + + H
Sbjct: 331 TTRKAYEQQCSQMESQTMVATTGLESRLKELEQEGKVVNTAKNALEERVKELEQMGKEAH 390
Query: 87 TKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDT 129
+ A E + + + EK +++G ++ L + V++ T
Sbjct: 391 SAKNALEEKIKQLQQMEKETKTANTSLEGKIQELEQNLVMWKT 433
>At1g32040 hypothetical protein
Length = 408
Score = 28.5 bits (62), Expect = 4.2
Identities = 19/86 (22%), Positives = 36/86 (41%), Gaps = 4/86 (4%)
Query: 24 PPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAM 83
PPP + S + F M +V + ++ + G ++ E + +A
Sbjct: 195 PPPEKLLESEQYAEASSFFLRAFSSMNKMV----RSYDSTTRACLGANEKLAEAEKKLAA 250
Query: 84 KLHTKDQAKEGEKEVTEPEEKAVTKW 109
+ H + +A+ E E T +EKA +W
Sbjct: 251 EGHARAEAESKEVEATLAKEKAKEEW 276
>At1g15510 hypothetical protein
Length = 866
Score = 28.5 bits (62), Expect = 4.2
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 3/50 (6%)
Query: 92 KEGEKEVTEPEEKAVTKWEPTVDGY-LKFLVDSKVVYDTLEKIVQDATHP 140
+E EK + E K + W + GY FL D + DT + QD+ P
Sbjct: 349 REAEKLFSRMERKDIVSWTTMISGYEYNFLPDKAI--DTYRMMDQDSVKP 396
>At5g54650 unknown protein
Length = 900
Score = 28.1 bits (61), Expect = 5.5
Identities = 12/37 (32%), Positives = 23/37 (61%)
Query: 129 TLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKE 165
T + +V++ + S +RN GLE+ + L+ +LE K+
Sbjct: 704 TEDLLVEETSEESEENYRNLGLEKVSGLSSELEHVKK 740
>At5g46450 disease resistance protein-like
Length = 1123
Score = 28.1 bits (61), Expect = 5.5
Identities = 21/81 (25%), Positives = 35/81 (42%), Gaps = 16/81 (19%)
Query: 104 KAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWF 163
K + + + G +FLVDSK +YD LE NTG ++ +A D+
Sbjct: 496 KEIVRAQSNEPGEREFLVDSKHIYDVLED--------------NTGTKKVLGIALDIN-- 539
Query: 164 KEQGYTIPQPSSPGLNYAQYL 184
+ G I + + G+ +L
Sbjct: 540 ETDGLYIHESAFKGMRNLLFL 560
>At3g28770 hypothetical protein
Length = 2081
Score = 28.1 bits (61), Expect = 5.5
Identities = 14/27 (51%), Positives = 18/27 (65%)
Query: 84 KLHTKDQAKEGEKEVTEPEEKAVTKWE 110
K +KDQ K+ EKE+ E EEK + K E
Sbjct: 1183 KKSSKDQQKKKEKEMKESEEKKLKKNE 1209
>At2g29110 putative ligand-gated ion channel protein (AtGLR2.8)
Length = 958
Score = 28.1 bits (61), Expect = 5.5
Identities = 19/76 (25%), Positives = 32/76 (42%), Gaps = 5/76 (6%)
Query: 25 PPHRQF-RSIYFPTTRLLQQHRFRQMKS----VVIVSATAAETARKRYPGESKGFVEEMR 79
PPH Q S +F + ++ HR + + + VV+V + Y F+ R
Sbjct: 620 PPHHQIGTSFWFSFSTMVFAHREKVVSNLARFVVVVWCFVVLVLTQSYTANLTSFLTVQR 679
Query: 80 FVAMKLHTKDQAKEGE 95
F ++ KD K G+
Sbjct: 680 FQPAAINVKDLIKNGD 695
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,639,334
Number of Sequences: 26719
Number of extensions: 289335
Number of successful extensions: 1496
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1480
Number of HSP's gapped (non-prelim): 28
length of query: 282
length of database: 11,318,596
effective HSP length: 98
effective length of query: 184
effective length of database: 8,700,134
effective search space: 1600824656
effective search space used: 1600824656
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0252a.3


