

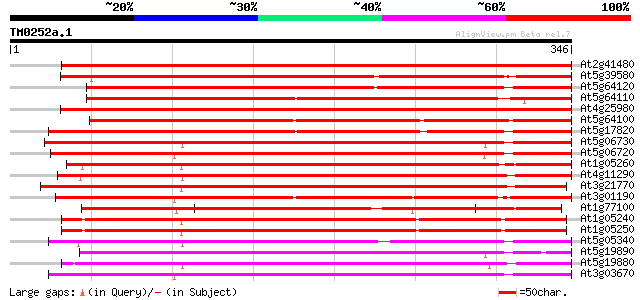
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.1
(346 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g41480 peroxidase like protein 456 e-129
At5g39580 peroxidase ATP24a 333 1e-91
At5g64120 peroxidase (emb|CAA67551.1) 328 3e-90
At5g64110 peroxidase ATP3a homolog 301 5e-82
At4g25980 putative peroxidase 299 2e-81
At5g64100 peroxidase ATP3a 292 2e-79
At5g17820 peroxidase ATP13a 274 5e-74
At5g06730 peroxidase 271 4e-73
At5g06720 peroxidase (emb|CAA68212.1) 266 1e-71
At1g05260 putative peroxidase 263 1e-70
At4g11290 peroxidase ATP19a 259 2e-69
At3g21770 putative peroxidase 251 3e-67
At3g01190 peroxidase like protein 251 4e-67
At1g77100 putative peroxidase 251 4e-67
At1g05240 putative peroxidase ATP12a 248 3e-66
At1g05250 putative peroxidase ATP12a 248 3e-66
At5g05340 peroxidase 247 6e-66
At5g19890 peroxidase ATP N 246 1e-65
At5g19880 peroxidase 244 7e-65
At3g03670 putative peroxidase 243 1e-64
>At2g41480 peroxidase like protein
Length = 328
Score = 456 bits (1173), Expect = e-129
Identities = 215/314 (68%), Positives = 264/314 (83%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
+++L++ + LK G+YS+SCP AE+ VRST+ES+F+ DPTI+P L+RLHFHDCFVQ
Sbjct: 15 MLVLVLGKEVRSQLLKNGYYSTSCPKAESIVRSTVESHFDSDPTISPGLLRLHFHDCFVQ 74
Query: 93 GCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYL 152
GCDGSVLI G SAE+ AL NL LRG EVI+DAK++LEA+CPGVVSCADILA+AARD+V L
Sbjct: 75 GCDGSVLIKGKSAEQAALPNLGLRGLEVIDDAKARLEAVCPGVVSCADILALAARDSVDL 134
Query: 153 SNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIG 212
S+GPSW VPTGR+DGRIS + +ASNLPSPLD V+VQ+QKF KGLD HDLVTL+GAHTIG
Sbjct: 135 SDGPSWRVPTGRKDGRISLATEASNLPSPLDSVAVQKQKFQDKGLDTHDLVTLLGAHTIG 194
Query: 213 QTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSF 272
QT+C FF YRLYNFT TGNSDPTI+ + L Q + LCP NG+G ++V+LD GSP+KFD SF
Sbjct: 195 QTDCLFFRYRLYNFTVTGNSDPTISPSFLTQLKTLCPPNGDGSKRVALDIGSPSKFDESF 254
Query: 273 FKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKT 332
FKN+RDGN +LESDQRLW D+ T +VV+ YA +RGLLG +F YEF KAMIK+S+I+VKT
Sbjct: 255 FKNLRDGNAILESDQRLWSDAETNAVVKKYASRLRGLLGFRFDYEFGKAMIKMSSIDVKT 314
Query: 333 DTEGEIRKVCSKFN 346
D +GE+RKVCSK N
Sbjct: 315 DVDGEVRKVCSKVN 328
>At5g39580 peroxidase ATP24a
Length = 319
Score = 333 bits (853), Expect = 1e-91
Identities = 172/319 (53%), Positives = 231/319 (71%), Gaps = 11/319 (3%)
Query: 32 SLMILMISFIIAQAQLKT--GFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDC 89
+L+I+ +S +IA T GFYS++CP AE VR+T+ S+F DP +AP L+R+H HDC
Sbjct: 8 ALVIVFLSCLIAVYGQGTRIGFYSTTCPNAETIVRTTVASHFGSDPKVAPGLLRMHNHDC 67
Query: 90 FVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDA 149
FVQGCDGSVL+ G ++ER A AN++L GFEVI+DAK QLEA CPGVVSCADILA+AARD+
Sbjct: 68 FVQGCDGSVLLSGPNSERTAGANVNLHGFEVIDDAKRQLEAACPGVVSCADILALAARDS 127
Query: 150 VYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLV-GA 208
V L+NG SW VPTGRRDGR+S ++ +NLPSP D +++Q++KF+A L+ DLVTLV G
Sbjct: 128 VSLTNGQSWQVPTGRRDGRVSLASNVNNLPSPSDSLAIQQRKFSAFRLNTRDLVTLVGGG 187
Query: 209 HTIGQTECRFFSYRLYNFTTTGN-SDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAK 267
HTIG C F + R++N ++GN +DPT++Q + Q Q LCP+NG+G +V LD+GS
Sbjct: 188 HTIGTAACGFITNRIFN--SSGNTADPTMDQTFVPQLQRLCPQNGDGSARVDLDTGSGNT 245
Query: 268 FDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSN 327
FD S+F N+ G+L+SD LW AT+S+VQ + RG F +F ++M+K+SN
Sbjct: 246 FDTSYFINLSRNRGILQSDHVLWTSPATRSIVQEFMAP-RG----NFNVQFARSMVKMSN 300
Query: 328 IEVKTDTEGEIRKVCSKFN 346
I VKT T GEIR+VCS N
Sbjct: 301 IGVKTGTNGEIRRVCSAVN 319
>At5g64120 peroxidase (emb|CAA67551.1)
Length = 328
Score = 328 bits (840), Expect = 3e-90
Identities = 163/299 (54%), Positives = 212/299 (70%), Gaps = 6/299 (2%)
Query: 48 KTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAER 107
+ GFY ++CP AE VR+ + + F+ DP IAP ++R+HFHDCFVQGCDGS+LI G++ ER
Sbjct: 36 RIGFYLTTCPRAETIVRNAVNAGFSSDPRIAPGILRMHFHDCFVQGCDGSILISGANTER 95
Query: 108 NALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDG 167
A NL+L+GFEVI++AK+QLEA CPGVVSCADILA+AARD V L+ G W VPTGRRDG
Sbjct: 96 TAGPNLNLQGFEVIDNAKTQLEAACPGVVSCADILALAARDTVILTQGTGWQVPTGRRDG 155
Query: 168 RISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFT 227
R+S ++ A+NLP P D V+VQ+QKF+A GL+ DLV LVG HTIG C F RL+N T
Sbjct: 156 RVSLASNANNLPGPRDSVAVQQQKFSALGLNTRDLVVLVGGHTIGTAGCGVFRNRLFN-T 214
Query: 228 TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
T +DPTI+ LAQ Q CP+NG+G +V LD+GS + +D S++ N+ G GVL+SDQ
Sbjct: 215 TGQTADPTIDPTFLAQLQTQCPQNGDGSVRVDLDTGSGSTWDTSYYNNLSRGRGVLQSDQ 274
Query: 288 RLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
LW D AT+ +VQ F EF ++M+++SNI V T GEIR+VCS N
Sbjct: 275 VLWTDPATRPIVQQLMAP-----RSTFNVEFARSMVRMSNIGVVTGANGEIRRVCSAVN 328
>At5g64110 peroxidase ATP3a homolog
Length = 330
Score = 301 bits (770), Expect = 5e-82
Identities = 153/304 (50%), Positives = 204/304 (66%), Gaps = 13/304 (4%)
Query: 48 KTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAER 107
+TG+Y S+C E+ VRS +ES + +P AP ++R+HFHDCFVQGCD SVL+ G ++ER
Sbjct: 35 RTGYYGSACWNVESIVRSVVESNYLANPANAPGILRMHFHDCFVQGCDASVLLAGPNSER 94
Query: 108 NALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDG 167
A+ NLSLRGF VIE+AK+QLE CP VSCADILA+AARD V+L+ GP W VP GR DG
Sbjct: 95 TAIPNLSLRGFNVIEEAKTQLEIACPRTVSCADILALAARDFVHLAGGPWWPVPLGRLDG 154
Query: 168 RISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFT 227
RIS ++ LP P D V+VQ+ +FA K L+ DLV L HTIG C F R +N+
Sbjct: 155 RISLASNVI-LPGPTDSVAVQKLRFAEKNLNTQDLVVLAAGHTIGTAGCIVFRDRFFNYD 213
Query: 228 TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
TG+ DPTI + + QA CP NG+ +V LD+GS +FD S+ N+++G G+LESDQ
Sbjct: 214 NTGSPDPTIAPSFVPLIQAQCPLNGDPATRVVLDTGSGDQFDTSYLNNLKNGRGLLESDQ 273
Query: 288 RLWGDSATQSVVQSYAGNIRGLLGLKFAY-----EFPKAMIKLSNIEVKTDTEGEIRKVC 342
LW + T+ +V+ LLGL+F + EF ++M K+S IE+KT +GEIR+VC
Sbjct: 274 VLWTNLETRPIVER-------LLGLRFPFLIFGLEFARSMTKMSQIEIKTGLDGEIRRVC 326
Query: 343 SKFN 346
S N
Sbjct: 327 SAVN 330
>At4g25980 putative peroxidase
Length = 371
Score = 299 bits (765), Expect = 2e-81
Identities = 147/316 (46%), Positives = 209/316 (65%), Gaps = 1/316 (0%)
Query: 32 SLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFV 91
+L ++ + F I+ A L+ GFYS++CP AE+ V+ + DP + L+RLHFHDCFV
Sbjct: 56 ALSLVTVFFGISLANLEVGFYSNTCPQAESIVKRVVSGAALSDPNLPAILLRLHFHDCFV 115
Query: 92 QGCDGSVLIV-GSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAV 150
+GCDGS+L+ G+ +E+NA + +RGFE++E K++LEA CPGVVSC+DI+A+AARDA+
Sbjct: 116 EGCDGSILVNNGAISEKNAFGHEGVRGFEIVEAVKAELEAACPGVVSCSDIVALAARDAI 175
Query: 151 YLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHT 210
L+NGP++ VPTGRRDGR+S+ + A ++P D + + + KF KGL+ DLV L AHT
Sbjct: 176 SLANGPAYEVPTGRRDGRVSNMSLAKDMPEVSDSIEILKAKFMQKGLNAKDLVLLSAAHT 235
Query: 211 IGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDV 270
IG T C F S RLY+F G DPTIN L + CP+NG+ ++ +D S FD
Sbjct: 236 IGTTACFFMSKRLYDFLPGGQPDPTINPTFLPELTTQCPQNGDINVRLPIDRFSERLFDK 295
Query: 271 SFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEV 330
+N++DG VL++D L+ D T+ VV SY G + G F +F KA++K+ I V
Sbjct: 296 QILQNIKDGFAVLQTDAGLYEDVTTRQVVDSYLGMLNPFFGPTFESDFVKAIVKMGKIGV 355
Query: 331 KTDTEGEIRKVCSKFN 346
KT +GEIR+VCS FN
Sbjct: 356 KTGFKGEIRRVCSAFN 371
>At5g64100 peroxidase ATP3a
Length = 331
Score = 292 bits (748), Expect = 2e-79
Identities = 152/297 (51%), Positives = 195/297 (65%), Gaps = 5/297 (1%)
Query: 50 GFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGSSAERNA 109
GFY + C E+ VRS ++S+ P AP ++R+HFHDCFV GCDGSVL+ G+++ER A
Sbjct: 40 GFYGNRCRNVESIVRSVVQSHVRSIPANAPGILRMHFHDCFVHGCDGSVLLAGNTSERTA 99
Query: 110 LANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRI 169
+ N SLRGFEVIE+AK++LE CP VSCADIL +AARDAV L+ G W VP GR DGRI
Sbjct: 100 VPNRSLRGFEVIEEAKARLEKACPRTVSCADILTLAARDAVVLTGGQRWEVPLGRLDGRI 159
Query: 170 SSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTT 229
S ++ NLP P D V+ Q+Q FAAK L+ DLVTLVG HTIG C R NF T
Sbjct: 160 SQASDV-NLPGPSDSVAKQKQDFAAKTLNTLDLVTLVGGHTIGTAGCGLVRGRFVNFNGT 218
Query: 230 GNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRL 289
G DP+I+ + + A CP+NG +V LD GS KFD SF + V VL+SD L
Sbjct: 219 GQPDPSIDPSFVPLILAQCPQNGG--TRVELDEGSVDKFDTSFLRKVTSSRVVLQSDLVL 276
Query: 290 WGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
W D T+++++ G R L+F EF K+M+K+S IEVKT ++GEIR+VCS N
Sbjct: 277 WKDPETRAIIERLLGLRRP--SLRFGTEFGKSMVKMSLIEVKTGSDGEIRRVCSAIN 331
>At5g17820 peroxidase ATP13a
Length = 313
Score = 274 bits (701), Expect = 5e-74
Identities = 142/322 (44%), Positives = 199/322 (61%), Gaps = 10/322 (3%)
Query: 25 MEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
M+ SL++L F IA AQL+ GFYS SCP AE VR+ + F PT+ +L+R+
Sbjct: 2 MKGAKFSSLLVLFFIFPIAFAQLRVGFYSQSCPQAETIVRNLVRQRFGVTPTVTAALLRM 61
Query: 85 HFHDCFVQGCDGSVLIVGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAV 144
HFHDCFV+GCD S+LI +++E+ A N S+R F++I+ K+QLEA CP VSCADI+ +
Sbjct: 62 HFHDCFVKGCDASLLIDSTNSEKTAGPNGSVREFDLIDRIKAQLEAACPSTVSCADIVTL 121
Query: 145 AARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVT 204
A RD+V L+ GPS+S+PTGRRDGR+S++ + LP P VS F KG++ D V
Sbjct: 122 ATRDSVALAGGPSYSIPTGRRDGRVSNNLDVT-LPGPTISVSGAVSLFTNKGMNTFDAVA 180
Query: 205 LVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGS 264
L+GAHT+GQ C FS R+ +F TG DP+++ A + + C + +LD S
Sbjct: 181 LLGAHTVGQGNCGLFSDRITSFQGTGRPDPSMDPALVTSLRNTCRNSAT----AALDQSS 236
Query: 265 PAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIK 324
P +FD FFK +R GVL+ DQRL D T+ +V YA N F +F +AM+K
Sbjct: 237 PLRFDNQFFKQIRKRRGVLQVDQRLASDPQTRGIVARYANN-----NAFFKRQFVRAMVK 291
Query: 325 LSNIEVKTDTEGEIRKVCSKFN 346
+ ++V T GEIR+ C +FN
Sbjct: 292 MGAVDVLTGRNGEIRRNCRRFN 313
>At5g06730 peroxidase
Length = 358
Score = 271 bits (693), Expect = 4e-73
Identities = 152/333 (45%), Positives = 210/333 (62%), Gaps = 13/333 (3%)
Query: 22 SSQMEAMLLGSLMILMIS-FIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPS 80
SS + + SL++++ S F + AQL FYS +CP A A VRSTI+ D I S
Sbjct: 7 SSTCDGFFIISLIVIVSSLFGTSSAQLNATFYSGTCPNASAIVRSTIQQALQSDARIGGS 66
Query: 81 LIRLHFHDCFVQGCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQLEAICPGVV 136
LIRLHFHDCFV GCDGS+L+ +S+ E+NA AN S RGF V++ K+ LE CPG+V
Sbjct: 67 LIRLHFHDCFVNGCDGSLLLDDTSSIQSEKNAPANANSTRGFNVVDSIKTALENACPGIV 126
Query: 137 SCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAK 195
SC+DILA+A+ +V L+ GPSW+V GRRDG ++ + A S+LPSP + ++ KF A
Sbjct: 127 SCSDILALASEASVSLAGGPSWTVLLGRRDGLTANLSGANSSLPSPFEGLNNITSKFVAV 186
Query: 196 GLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGL 255
GL D+V+L GAHT G+ +C F+ RL+NF TGN DPT+N L+ Q LCP+NG+
Sbjct: 187 GLKTTDVVSLSGAHTFGRGQCVTFNNRLFNFNGTGNPDPTLNSTLLSSLQQLCPQNGSNT 246
Query: 256 RKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGD--SATQSVVQSYAGNIRGLLGLK 313
+LD +P FD ++F N++ NG+L+SDQ L+ + SAT +V S+A N
Sbjct: 247 GITNLDLSTPDAFDNNYFTNLQSNNGLLQSDQELFSNTGSATVPIVNSFASN-----QTL 301
Query: 314 FAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
F F ++MIK+ NI T + GEIR+ C N
Sbjct: 302 FFEAFVQSMIKMGNISPLTGSSGEIRQDCKVVN 334
>At5g06720 peroxidase (emb|CAA68212.1)
Length = 335
Score = 266 bits (680), Expect = 1e-71
Identities = 148/329 (44%), Positives = 210/329 (62%), Gaps = 13/329 (3%)
Query: 26 EAMLLGSLMILMIS-FIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
+ + + SL++++ S F + AQL FYS +CP A A VRSTI+ D I SLIRL
Sbjct: 10 DGLFIISLIVIVSSIFGTSSAQLNATFYSGTCPNASAIVRSTIQQALQSDTRIGASLIRL 69
Query: 85 HFHDCFVQGCDGSVLI--VGS-SAERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCAD 140
HFHDCFV GCD S+L+ GS +E+NA N+ S RGF V+++ K+ LE CPGVVSC+D
Sbjct: 70 HFHDCFVNGCDASILLDDTGSIQSEKNAGPNVNSARGFNVVDNIKTALENACPGVVSCSD 129
Query: 141 ILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDD 199
+LA+A+ +V L+ GPSW+V GRRD ++ A A S++PSP++ +S KF+A GL+
Sbjct: 130 VLALASEASVSLAGGPSWTVLLGRRDSLTANLAGANSSIPSPIESLSNITFKFSAVGLNT 189
Query: 200 HDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVS 259
+DLV L GAHT G+ C F+ RL+NF+ TGN DPT+N L+ Q LCP+NG+ +
Sbjct: 190 NDLVALSGAHTFGRARCGVFNNRLFNFSGTGNPDPTLNSTLLSTLQQLCPQNGSASTITN 249
Query: 260 LDSGSPAKFDVSFFKNVRDGNGVLESDQRLWG--DSATQSVVQSYAGNIRGLLGLKFAYE 317
LD +P FD ++F N++ +G+L+SDQ L+ S+T ++V S+A N F
Sbjct: 250 LDLSTPDAFDNNYFANLQSNDGLLQSDQELFSTTGSSTIAIVTSFASN-----QTLFFQA 304
Query: 318 FPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
F ++MI + NI T + GEIR C K N
Sbjct: 305 FAQSMINMGNISPLTGSNGEIRLDCKKVN 333
>At1g05260 putative peroxidase
Length = 326
Score = 263 bits (672), Expect = 1e-70
Identities = 146/321 (45%), Positives = 204/321 (63%), Gaps = 14/321 (4%)
Query: 36 LMISFIIA------QAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDC 89
L +SF + QAQL+ FY++SCP AE V+ + ++ + P++A +LIR+HFHDC
Sbjct: 9 LSVSFFLVGIVGPIQAQLQMNFYANSCPNAEKIVQDFVSNHVSNAPSLAAALIRMHFHDC 68
Query: 90 FVQGCDGSVLIVGSS--AERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAAR 147
FV+GCDGSVLI +S AER+A NL++RGF I+ KS LEA CPG+VSCADI+A+A+R
Sbjct: 69 FVRGCDGSVLINSTSGNAERDATPNLTVRGFGFIDAIKSVLEAQCPGIVSCADIIALASR 128
Query: 148 DAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDLVTLV 206
DAV + GP+WSVPTGRRDGRIS++A+A +N+P P ++ + FA +GLD DLV L
Sbjct: 129 DAVVFTGGPNWSVPTGRRDGRISNAAEALANIPPPTSNITNLQTLFANQGLDLKDLVLLS 188
Query: 207 GAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQA-LCPKNGNGLRKVSLDSGSP 265
GAHTIG + C F+ RLYNFT G DP ++ A ++ CP + V +D GS
Sbjct: 189 GAHTIGVSHCSSFTNRLYNFTGRGGQDPALDSEYAANLKSRKCPSLNDNKTIVEMDPGSR 248
Query: 266 AKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKL 325
FD+S+++ V G+ +SD L + T S + + G +G F EF K+M K+
Sbjct: 249 KTFDLSYYQLVLKRRGLFQSDSALTTNPTTLSNINRI---LTGSVG-SFFSEFAKSMEKM 304
Query: 326 SNIEVKTDTEGEIRKVCSKFN 346
I VKT + G +R+ CS N
Sbjct: 305 GRINVKTGSAGVVRRQCSVAN 325
>At4g11290 peroxidase ATP19a
Length = 326
Score = 259 bits (661), Expect = 2e-69
Identities = 141/325 (43%), Positives = 202/325 (61%), Gaps = 12/325 (3%)
Query: 30 LGSLMILMISFII--AQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFH 87
L LMIL+I ++ ++AQLK GFY +CP AE V+ + + N P++A LIR+HFH
Sbjct: 6 LALLMILVIQGLVTFSEAQLKMGFYDQTCPYAEKIVQDVVNQHINNAPSLAAGLIRMHFH 65
Query: 88 DCFVQGCDGSVLIVGSSA----ERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILA 143
DCFV+GCDGS+LI +S+ E+ A NL++RGF+ I+ KS LE+ CPG+VSCADI+
Sbjct: 66 DCFVRGCDGSILINATSSNQQVEKLAPPNLTVRGFDFIDKVKSALESKCPGIVSCADIIT 125
Query: 144 VAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDL 202
+A RD++ GP+W+VPTGRRDGRIS+ A+A +N+P P + F +GLD DL
Sbjct: 126 LATRDSIVAIGGPTWNVPTGRRDGRISNFAEAMNNIPPPFGNFTTLITLFGNQGLDVKDL 185
Query: 203 VTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQA-LCPKNGNGLRKVSLD 261
V L GAHTIG + C FS RL+NFT G+ DP+++ ++ C + KV +D
Sbjct: 186 VLLSGAHTIGVSHCSSFSNRLFNFTGVGDQDPSLDSEYADNLKSRRCLSIADNTTKVEMD 245
Query: 262 SGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKA 321
GS FD+S+++ V G+ ESD L + A + V+ +AG +F EF +
Sbjct: 246 PGSRNTFDLSYYRLVLKRRGLFESDAALTMNPAALAQVKRFAGGSE----QEFFAEFSNS 301
Query: 322 MIKLSNIEVKTDTEGEIRKVCSKFN 346
M K+ I VKT ++GEIR+ C+ N
Sbjct: 302 MEKMGRIGVKTGSDGEIRRTCAFVN 326
>At3g21770 putative peroxidase
Length = 329
Score = 251 bits (642), Expect = 3e-67
Identities = 138/328 (42%), Positives = 199/328 (60%), Gaps = 8/328 (2%)
Query: 20 KVSSQMEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAP 79
K +Q+ ++ + +L+ ++AQL+ FY+ SCP AE + I+++ + P++A
Sbjct: 2 KTMTQLNIAVVVVVTVLIGMLRSSEAQLQMNFYAKSCPNAEKIISDHIQNHIHNGPSLAA 61
Query: 80 SLIRLHFHDCFVQGCDGSVLIVGSS--AERNALANLSLRGFEVIEDAKSQLEAICPGVVS 137
LIR+HFHDCFV+GCDGSVLI +S AER+A NL+LRGF +E K+ LE +CP VS
Sbjct: 62 PLIRMHFHDCFVRGCDGSVLINSTSGNAERDAPPNLTLRGFGFVERIKALLEKVCPKTVS 121
Query: 138 CADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKG 196
CADI+A+ ARDAV + GPSWSVPTGRRDGRIS+ +A +N+P P + ++ F +G
Sbjct: 122 CADIIALTARDAVVATGGPSWSVPTGRRDGRISNKTEATNNIPPPTSNFTTLQRLFKNQG 181
Query: 197 LDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQA-LCPKNGNGL 255
L+ DLV L GAHTIG + C + RLYNF+TT DP+++ A +A C +
Sbjct: 182 LNLKDLVLLSGAHTIGVSHCSSMNTRLYNFSTTVKQDPSLDSQYAANLKANKCKSLNDNS 241
Query: 256 RKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFA 315
+ +D GS FD+S+++ V G+ +SD L +SAT V+ KF
Sbjct: 242 TILEMDPGSSRSFDLSYYRLVLKRRGLFQSDSALTTNSATLKVINDLVNGSE----KKFF 297
Query: 316 YEFPKAMIKLSNIEVKTDTEGEIRKVCS 343
F K+M K+ ++VKT + G IR CS
Sbjct: 298 KAFAKSMEKMGRVKVKTGSAGVIRTRCS 325
>At3g01190 peroxidase like protein
Length = 321
Score = 251 bits (641), Expect = 4e-67
Identities = 140/321 (43%), Positives = 195/321 (60%), Gaps = 9/321 (2%)
Query: 29 LLGSLMILMISFIIAQAQ-LKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFH 87
L+ S + L++ F A +Q LK GFYS +CP E V+ + NK PT+ L+R+ FH
Sbjct: 7 LVVSCLFLVLLFAQANSQGLKVGFYSKTCPQLEGIVKKVVFDAMNKAPTLGAPLLRMFFH 66
Query: 88 DCFVQGCDGSVLI--VGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVA 145
DCFV+GCDGSVL+ + E++A+ NLSLRGF +I+D+K+ LE +CPG+VSC+DILA+
Sbjct: 67 DCFVRGCDGSVLLDKPNNQGEKSAVPNLSLRGFGIIDDSKAALEKVCPGIVSCSDILALV 126
Query: 146 ARDAVYLSNGPSWSVPTGRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTL 205
ARDA+ GPSW V TGRRDGR+S+ + NLPSP D ++ F +KGL++ DLV L
Sbjct: 127 ARDAMVALEGPSWEVETGRRDGRVSNINEV-NLPSPFDNITKLISDFRSKGLNEKDLVIL 185
Query: 206 VGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSP 265
G HTIG C + RLYNFT G+SDP+++ A+ + C K + + +D GS
Sbjct: 186 SGGHTIGMGHCPLLTNRLYNFTGKGDSDPSLDSEYAAKLRKKC-KPTDTTTALEMDPGSF 244
Query: 266 AKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKL 325
FD+S+F V G+ +SD L +S T++ V IR G F +F +M+K+
Sbjct: 245 KTFDLSYFTLVAKRRGLFQSDAALLDNSKTRAYVLQ---QIR-THGSMFFNDFGVSMVKM 300
Query: 326 SNIEVKTDTEGEIRKVCSKFN 346
V T GEIRK C N
Sbjct: 301 GRTGVLTGKAGEIRKTCRSAN 321
>At1g77100 putative peroxidase
Length = 660
Score = 251 bits (641), Expect = 4e-67
Identities = 139/299 (46%), Positives = 184/299 (61%), Gaps = 10/299 (3%)
Query: 45 AQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIV--G 102
AQL+ GFYS +CP+AE+ VR ++ DP A L+RL FHDCFV+GCDGS+LI G
Sbjct: 39 AQLQFGFYSETCPSAESIVRDVVQQAVTNDPGKAAVLLRLQFHDCFVEGCDGSILIKHGG 98
Query: 103 SSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPT 162
+ ER A N + GF+VI++AKS+LE CPGVVSCADI+A+AARDA+ + GP + VPT
Sbjct: 99 NDDERFAAGNAGVAGFDVIDEAKSELERFCPGVVSCADIVALAARDAIAEAKGPFYEVPT 158
Query: 163 GRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTL-VGAHTIGQTECRFFSY 221
GRRDG I++ A NLP D ++ + KF KGL D DLV L GAHTIG T C F
Sbjct: 159 GRRDGLIANVDHAKNLPDVQDSINTLKSKFREKGLSDQDLVLLSAGAHTIGTTACFFVIP 218
Query: 222 RLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNG 281
RL DPTIN ++ CP+ G+ ++ LD S FD F+N+++G G
Sbjct: 219 RL------DAQDPTINPEFFQILRSKCPQGGDVNVRIPLDWDSQFVFDNQIFQNIKNGRG 272
Query: 282 VLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEIRK 340
V+ SD L+ D+ + ++ SY + FA +F KAMIK+ I VK EGEIR+
Sbjct: 273 VILSDSVLYQDNNMKKIIDSYLETNQSSKA-NFAADFTKAMIKMGAIGVKIGAEGEIRR 330
Score = 94.0 bits (232), Expect = 1e-19
Identities = 64/176 (36%), Positives = 88/176 (49%), Gaps = 31/176 (17%)
Query: 115 LRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQ 174
+R F +I KS +E CP VVSCADI+A+AARDA+ + G + VPTGR DGRIS+
Sbjct: 328 IRRFNIIGKVKSAVERSCPDVVSCADIIAIAARDAIITAKGSFYEVPTGRNDGRISNVKN 387
Query: 175 ASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDP 234
A LP D ++ + KF +GL + DLV L + C + +R+
Sbjct: 388 AEKLPDAQDSIATLKAKFREQGLTNRDLVLL--------SRCAY--HRI--------DGM 429
Query: 235 TINQATLAQFQAL---CPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGVLESDQ 287
+ AT+ FQ L CP+ G K+ LD +FD N VL S+Q
Sbjct: 430 LLCHATVDFFQVLRLKCPRGGYVCFKIPLDWDGKFQFD----------NQVLRSNQ 475
>At1g05240 putative peroxidase ATP12a
Length = 325
Score = 248 bits (633), Expect = 3e-66
Identities = 136/314 (43%), Positives = 191/314 (60%), Gaps = 7/314 (2%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
L ++ +S I Q L +Y S CP AE VR Y ++ T+A L+R+HFHDCFV+
Sbjct: 13 LSVVGVSVAIPQL-LDLDYYRSKCPKAEEIVRGVTVQYVSRQKTLAAKLLRMHFHDCFVR 71
Query: 93 GCDGSVLIVGSS--AERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAV 150
GCDGSVL+ + AER+A+ NL+L+G+EV++ AK+ LE CP ++SCAD+LA+ ARDAV
Sbjct: 72 GCDGSVLLKSAKNDAERDAVPNLTLKGYEVVDAAKTALERKCPNLISCADVLALVARDAV 131
Query: 151 YLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAH 209
+ GP W VP GRRDGRIS A NLPSP + ++ FA KGL+ DLV L G H
Sbjct: 132 AVIGGPWWPVPLGRRDGRISKLNDALLNLPSPFADIKTLKKNFANKGLNAKDLVVLSGGH 191
Query: 210 TIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFD 269
TIG + C + RLYNFT G+SDP++N + + + + CP + +++D GS FD
Sbjct: 192 TIGISSCALVNSRLYNFTGKGDSDPSMNPSYVRELKRKCPPT-DFRTSLNMDPGSALTFD 250
Query: 270 VSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIE 329
+FK V G+ SD L D T++ VQ+ A I + F +F +M+KL ++
Sbjct: 251 THYFKVVAQKKGLFTSDSTLLDDIETKNYVQTQA--ILPPVFSSFNKDFSDSMVKLGFVQ 308
Query: 330 VKTDTEGEIRKVCS 343
+ T GEIRK C+
Sbjct: 309 ILTGKNGEIRKRCA 322
>At1g05250 putative peroxidase ATP12a
Length = 325
Score = 248 bits (633), Expect = 3e-66
Identities = 136/314 (43%), Positives = 191/314 (60%), Gaps = 7/314 (2%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
L ++ +S I Q L +Y S CP AE VR Y ++ T+A L+R+HFHDCFV+
Sbjct: 13 LSVVGVSVAIPQL-LDLDYYRSKCPKAEEIVRGVTVQYVSRQKTLAAKLLRMHFHDCFVR 71
Query: 93 GCDGSVLIVGSS--AERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAV 150
GCDGSVL+ + AER+A+ NL+L+G+EV++ AK+ LE CP ++SCAD+LA+ ARDAV
Sbjct: 72 GCDGSVLLKSAKNDAERDAVPNLTLKGYEVVDAAKTALERKCPNLISCADVLALVARDAV 131
Query: 151 YLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAH 209
+ GP W VP GRRDGRIS A NLPSP + ++ FA KGL+ DLV L G H
Sbjct: 132 AVIGGPWWPVPLGRRDGRISKLNDALLNLPSPFADIKTLKKNFANKGLNAKDLVVLSGGH 191
Query: 210 TIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFD 269
TIG + C + RLYNFT G+SDP++N + + + + CP + +++D GS FD
Sbjct: 192 TIGISSCALVNSRLYNFTGKGDSDPSMNPSYVRELKRKCPPT-DFRTSLNMDPGSALTFD 250
Query: 270 VSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIE 329
+FK V G+ SD L D T++ VQ+ A I + F +F +M+KL ++
Sbjct: 251 THYFKVVAQKKGLFTSDSTLLDDIETKNYVQTQA--ILPPVFSSFNKDFSDSMVKLGFVQ 308
Query: 330 VKTDTEGEIRKVCS 343
+ T GEIRK C+
Sbjct: 309 ILTGKNGEIRKRCA 322
>At5g05340 peroxidase
Length = 324
Score = 247 bits (631), Expect = 6e-66
Identities = 144/336 (42%), Positives = 204/336 (59%), Gaps = 26/336 (7%)
Query: 25 MEAMLLGSLMILMISFI-------IAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTI 77
M + L S+++L+++ + + +AQL T FYS+SCP +TV++ ++S N + +
Sbjct: 1 MASNKLISILVLVVTLLLQGDNNYVVEAQLTTNFYSTSCPNLLSTVQTAVKSAVNSEARM 60
Query: 78 APSLIRLHFHDCFVQGCDGSVLIVGSSA---ERNALANL-SLRGFEVIEDAKSQLEAICP 133
S++RL FHDCFV GCDGS+L+ +S+ E+NA N S RGF VI++ KS +E CP
Sbjct: 61 GASILRLFFHDCFVNGCDGSILLDDTSSFTGEQNAAPNRNSARGFNVIDNIKSAVEKACP 120
Query: 134 GVVSCADILAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKF 192
GVVSCADILA+AARD+V GP+W+V GRRD R +S A A SN+P+P +S F
Sbjct: 121 GVVSCADILAIAARDSVVALGGPNWNVKVGRRDARTASQAAANSNIPAPTSSLSQLISSF 180
Query: 193 AAKGLDDHDLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPK-N 251
+A GL D+V L GAHTIGQ+ C F R+YN T IN A Q CP+ +
Sbjct: 181 SAVGLSTRDMVALSGAHTIGQSRCTNFRARIYNET-------NINAAFATTRQRTCPRAS 233
Query: 252 GNGLRKVS-LDSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLL 310
G+G ++ LD + A FD ++FKN+ G+L SDQ L+ +T S+V+ Y+ N
Sbjct: 234 GSGDGNLAPLDVTTAASFDNNYFKNLMTQRGLLHSDQVLFNGGSTDSIVRGYSNN----- 288
Query: 311 GLKFAYEFPKAMIKLSNIEVKTDTEGEIRKVCSKFN 346
F +F AMIK+ +I T + GEIRKVC + N
Sbjct: 289 PSSFNSDFTAAMIKMGDISPLTGSSGEIRKVCGRTN 324
>At5g19890 peroxidase ATP N
Length = 328
Score = 246 bits (628), Expect = 1e-65
Identities = 133/308 (43%), Positives = 183/308 (59%), Gaps = 12/308 (3%)
Query: 44 QAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQGCDGSVLIVGS 103
+AQL Y+ SCP VR + + +A SLIRLHFHDCFV GCD S+L+ G+
Sbjct: 27 RAQLSPDIYAKSCPNLVQIVRKQVAIALKAEIRMAASLIRLHFHDCFVNGCDASLLLDGA 86
Query: 104 SAERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCADILAVAARDAVYLSNGPSWSVPT 162
+E+ A+ N+ S RGFEVI+ K+ +E CPGVVSCADIL +AARD+V LS GP W V
Sbjct: 87 DSEKLAIPNINSARGFEVIDTIKAAVENACPGVVSCADILTLAARDSVVLSGGPGWRVAL 146
Query: 163 GRRDGRISSSAQASNLPSPLDPVSVQRQKFAAKGLDDHDLVTLVGAHTIGQTECRFFSYR 222
GR+DG +++ A+NLPSP +P+ KF A L+ D+V L GAHT GQ +C FS R
Sbjct: 147 GRKDGLVANQNSANNLPSPFEPLDAIIAKFVAVNLNITDVVALSGAHTFGQAKCAVFSNR 206
Query: 223 LYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGSPAKFDVSFFKNVRDGNGV 282
L+NFT GN D T+ + L+ Q +CP GN LD + FD ++FKN+ +G G+
Sbjct: 207 LFNFTGLGNPDATLETSLLSNLQTVCPLGGNSNITAPLDRSTTDTFDNNYFKNLLEGKGL 266
Query: 283 LESDQRLWGD----SATQSVVQSYAGNIRGLLGLKFAYEFPKAMIKLSNIEVKTDTEGEI 338
L SDQ L+ + T+ +V++Y+ + F +F AMI++ NI GE+
Sbjct: 267 LSSDQILFSSDLAVNTTKKLVEAYSRS-----QSLFFRDFTCAMIRMGNI--SNGASGEV 319
Query: 339 RKVCSKFN 346
R C N
Sbjct: 320 RTNCRVIN 327
>At5g19880 peroxidase
Length = 329
Score = 244 bits (622), Expect = 7e-65
Identities = 141/324 (43%), Positives = 193/324 (59%), Gaps = 16/324 (4%)
Query: 33 LMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRLHFHDCFVQ 92
L ILM +++ AQL + FYS++CP A R IE D + ++RLHFHDCFV
Sbjct: 12 LPILMFG-VLSNAQLTSDFYSTTCPNVTAIARGLIERASRNDVRLTAKVMRLHFHDCFVN 70
Query: 93 GCDGSVLIVGSSA-----ERNALANL-SLRGFEVIEDAKSQLEAICPGVVSCADILAVAA 146
GCDGSVL+ + A E+ A N SL GFEVI+D K+ LE +CPGVVSCADILA+AA
Sbjct: 71 GCDGSVLLDAAPADGVEGEKEAFQNAGSLDGFEVIDDIKTALENVCPGVVSCADILAIAA 130
Query: 147 RDAVYLSNGPSWSVPTGRRDGRISSSAQA-SNLPSPLDPVSVQRQKFAAKGLDDHDLVTL 205
+V L+ GPS V GRRDGR + A A + LP D + + KF+ LD DLV L
Sbjct: 131 EISVALAGGPSLDVLLGRRDGRTAIRADAVAALPLGPDSLEILTSKFSVHNLDTTDLVAL 190
Query: 206 VGAHTIGQTECRFFSYRLYNFT-TTGNSDPTINQATLAQFQALCPKNGNGLRKVSLDSGS 264
GAHT G+ +C + RL+NF+ +G SDP+I L + CP+ G+ + +LD S
Sbjct: 191 SGAHTFGRVQCGVINNRLHNFSGNSGQSDPSIEPEFLQTLRRQCPQGGDLTARANLDPTS 250
Query: 265 PAKFDVSFFKNVRDGNGVLESDQRLWGDSA--TQSVVQSYAGNIRGLLGLKFAYEFPKAM 322
P FD +FKN+++ GV+ESDQ L+ + T S+V +A N +F F ++M
Sbjct: 251 PDSFDNDYFKNLQNNRGVIESDQILFSSTGAPTVSLVNRFAENQN-----EFFTNFARSM 305
Query: 323 IKLSNIEVKTDTEGEIRKVCSKFN 346
IK+ N+ + T EGEIR+ C + N
Sbjct: 306 IKMGNVRILTGREGEIRRDCRRVN 329
>At3g03670 putative peroxidase
Length = 321
Score = 243 bits (620), Expect = 1e-64
Identities = 136/326 (41%), Positives = 184/326 (55%), Gaps = 9/326 (2%)
Query: 25 MEAMLLGSLMILMISFIIAQAQLKTGFYSSSCPTAEATVRSTIESYFNKDPTIAPSLIRL 84
M+ L++L+ F +A AQLK FYS SCP AE V + + F +DP+I +L R+
Sbjct: 1 MKIATFSVLLLLLFIFPVALAQLKFKFYSESCPNAETIVENLVRQQFARDPSITAALTRM 60
Query: 85 HFHDCFVQGCDGSVLI---VGSSAERNALANLSLRGFEVIEDAKSQLEAICPGVVSCADI 141
HFHDCFVQGCD S+LI +E+NA N S+RGFE+I++ K+ LEA CP VSC+DI
Sbjct: 61 HFHDCFVQGCDASLLIDPTTSQLSEKNAGPNFSVRGFELIDEIKTALEAQCPSTVSCSDI 120
Query: 142 LAVAARDAVYLSNGPSWSVPTGRRDGRISSSAQASN-LPSPLDPVSVQRQKFAAKGLDDH 200
+ +A RDAV+L GPS+ VPTGRRDG +S+ A+ LP P V F KG++
Sbjct: 121 VTLATRDAVFLGGGPSYVVPTGRRDGFVSNPEDANEILPPPFISVEGMLSFFGNKGMNVF 180
Query: 201 DLVTLVGAHTIGQTECRFFSYRLYNFTTTGNSDPTINQATLAQFQALCPKNGNGLRKVSL 260
D V L+GAHT+G C F R+ NF TG DP+++ + + C G
Sbjct: 181 DSVALLGAHTVGIASCGNFVDRVTNFQGTGLPDPSMDPTLAGRLRNTCAVPGGFAALDQS 240
Query: 261 DSGSPAKFDVSFFKNVRDGNGVLESDQRLWGDSATQSVVQSYAGNIRGLLGLKFAYEFPK 320
+P FD FF +R+ G+L DQ + D AT VV YA N F +F
Sbjct: 241 MPVTPVSFDNLFFGQIRERKGILLIDQLIASDPATSGVVLQYASN-----NELFKRQFAI 295
Query: 321 AMIKLSNIEVKTDTEGEIRKVCSKFN 346
AM+K+ ++V T + GEIR C FN
Sbjct: 296 AMVKMGAVDVLTGSAGEIRTNCRAFN 321
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,280,414
Number of Sequences: 26719
Number of extensions: 296698
Number of successful extensions: 1051
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 723
Number of HSP's gapped (non-prelim): 96
length of query: 346
length of database: 11,318,596
effective HSP length: 100
effective length of query: 246
effective length of database: 8,646,696
effective search space: 2127087216
effective search space used: 2127087216
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0252a.1


