

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.7
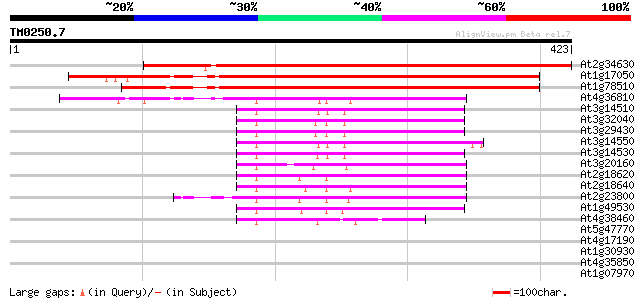
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g34630 trans-prenyltransferase like protein 479 e-136
At1g17050 prenyl transferase (prephytoene pyrophosphatase dehydr... 235 4e-62
At1g78510 polyprenyl diphosphate synthase (At-trans-PT) 230 1e-60
At4g36810 geranylgeranyl pyrophosphate synthase 92 7e-19
At3g14510 geranylgeranyl pyrophosphate synthetase, putative 87 1e-17
At3g32040 geranylgeranyl pyrophosphate synthase, putative 86 5e-17
At3g29430 geranylgeranyl pyrophosphate synthase, putative 86 5e-17
At3g14550 geranylgeranyl pyrophosphate synthase (GGPS3) 86 5e-17
At3g14530 putative geranylgeranyl pyrophosphate synthase GGPS3 85 9e-17
At3g20160 geranyl geranyl pyrophosphate synthase, putative 79 4e-15
At2g18620 putative geranylgeranyl pyrophosphate synthase 79 6e-15
At2g18640 putative geranylgeranyl pyrophosphate synthase 76 4e-14
At2g23800 pregeranylgeranyl pyrophosphate synthase 74 1e-13
At1g49530 geranyl geranyl pyrophosphate synthase, putative 71 1e-12
At4g38460 geranylgeranyl pyrophosphate synthase-related protein 56 4e-08
At5g47770 farnesyl diphosphate synthase precursor (gb|AAB49290.1) 35 0.078
At4g17190 farnesyl-pyrophosphate synthetase FPS2 35 0.10
At1g30930 F17F8.20 34 0.17
At4g35850 unknown protein 32 0.66
At1g07970 unknown protein 31 1.5
>At2g34630 trans-prenyltransferase like protein
Length = 321
Score = 479 bits (1234), Expect = e-136
Identities = 246/324 (75%), Positives = 291/324 (88%), Gaps = 5/324 (1%)
Query: 102 MVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKP--PPSNDLGGTFTAD 159
MV+AEVPKLAS AEYFFK GV+GK+FR T+LLLM+TALN+ +P+ S D+ T++
Sbjct: 1 MVLAEVPKLASAAEYFFKRGVQGKQFRSTILLLMATALNVRVPEALIGESTDI---VTSE 57
Query: 160 LRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACV 219
LR RQ+ IAEITEMIHVASLLHDDVLDDADTRRG+GSLN VMGNK++VLAGDFLLSRAC
Sbjct: 58 LRVRQRGIAEITEMIHVASLLHDDVLDDADTRRGVGSLNVVMGNKMSVLAGDFLLSRACG 117
Query: 220 ALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASLISNSCKA 279
ALA+LKNTEVV+LLA VEHLVTGETM+++++++QR SM+YYMQKTYYKTASLISNSCKA
Sbjct: 118 ALAALKNTEVVALLATAVEHLVTGETMEITSSTEQRYSMDYYMQKTYYKTASLISNSCKA 177
Query: 280 IAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPIL 339
+A+L GQTA+VA+LAF+YG+NLGLAFQLIDD+LDFTGTSASLGKGSLSDIRHG++TAPIL
Sbjct: 178 VAVLTGQTAEVAVLAFEYGRNLGLAFQLIDDILDFTGTSASLGKGSLSDIRHGVITAPIL 237
Query: 340 FAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAAIDSLPES 399
FAMEEFPQLR VVD+ +P NVDIALEYLGKS+GIQR RELA++HA+LAAAAI SLPE+
Sbjct: 238 FAMEEFPQLREVVDQVEKDPRNVDIALEYLGKSKGIQRARELAMEHANLAAAAIGSLPET 297
Query: 400 DDGEVRLSRRALVDLTQRVITRTK 423
D+ +V+ SRRAL+DLT RVITR K
Sbjct: 298 DNEDVKRSRRALIDLTHRVITRNK 321
>At1g17050 prenyl transferase (prephytoene pyrophosphatase
dehydrogenase) like protein
Length = 417
Score = 235 bits (599), Expect = 4e-62
Identities = 140/367 (38%), Positives = 223/367 (60%), Gaps = 26/367 (7%)
Query: 45 GSTAKVMRSLAFSKGLPALHSSRYQIH----DQSSSIS--EEELDPFSL------VADEL 92
G+ + R LA + +PA ++ DQ+ ++ +E P SL VAD+L
Sbjct: 47 GNLVFLRRDLATCRAVPAKSKENSLVNGIGQDQTVMLNLRQESRKPISLETLFEVVADDL 106
Query: 93 SFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIPKPPPSNDL 152
+ +NL ++V AE P L S AE F G GKR RP ++ L+S A + +L
Sbjct: 107 QRLNDNLLSIVGAENPVLISAAEQIFSAG--GKRMRPGLVFLVSRA----------TAEL 154
Query: 153 GGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDF 212
G +L +R+ EI EMIH ASL+HDDVLD++D RRG +++ + G ++AVLAGDF
Sbjct: 155 AGL--KELTVEHRRLGEIIEMIHTASLIHDDVLDESDMRRGRETVHELFGTRVAVLAGDF 212
Query: 213 LLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQKTYYKTASL 272
+ ++A LA+L+N EV+ L+++V++ +GE Q S+ D ++ YM K+YYKTASL
Sbjct: 213 MFAQASWYLANLENLEVIKLISQVIKDFASGEIKQASSLFDCDVKLDDYMLKSYYKTASL 272
Query: 273 ISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHG 332
++ S K AI + + VA + +GKNLGL+FQ++DD+LDFT ++ LGK + +D+ G
Sbjct: 273 VAASTKGAAIFSKVESKVAEQMYQFGKNLGLSFQVVDDILDFTQSTEQLGKPAANDLAKG 332
Query: 333 IVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIKHADLAAAA 392
+TAP++FA+E P+LR +++ F P +++ A+E + GI++ +ELA + A+LA
Sbjct: 333 NITAPVIFALENEPRLREIIESEFCEPGSLEEAIEIVRNRGGIKKAQELAKEKAELALKN 392
Query: 393 IDSLPES 399
++ LP S
Sbjct: 393 LNCLPRS 399
>At1g78510 polyprenyl diphosphate synthase (At-trans-PT)
Length = 406
Score = 230 bits (587), Expect = 1e-60
Identities = 130/315 (41%), Positives = 202/315 (63%), Gaps = 14/315 (4%)
Query: 85 FSLVADELSFIANNLRAMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALNLPIP 144
F LVA +L + +NL ++V AE P L S AE F G GKR RP ++ L+S A
Sbjct: 88 FELVAVDLQTLNDNLLSIVGAENPVLISAAEQIF--GAGGKRMRPGLVFLVSHA------ 139
Query: 145 KPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDVLDDADTRRGIGSLNFVMGNK 204
+ +L G +L + +R+AEI EMIH ASL+HDDVLD++D RRG +++ + G +
Sbjct: 140 ----TAELAGL--KELTTEHRRLAEIIEMIHTASLIHDDVLDESDMRRGKETVHELFGTR 193
Query: 205 LAVLAGDFLLSRACVALASLKNTEVVSLLAKVVEHLVTGETMQMSTTSDQRCSMEYYMQK 264
+AVLAGDF+ ++A LA+L+N EV+ L+++V++ +GE Q S+ D ++ Y+ K
Sbjct: 194 VAVLAGDFMFAQASWYLANLENLEVIKLISQVIKDFASGEIKQASSLFDCDTKLDEYLLK 253
Query: 265 TYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKG 324
++YKTASL++ S K AI + DV +++GKNLGL+FQ++DD+LDFT ++ LGK
Sbjct: 254 SFYKTASLVAASTKGAAIFSRVEPDVTEQMYEFGKNLGLSFQIVDDILDFTQSTEQLGKP 313
Query: 325 SLSDIRHGIVTAPILFAMEEFPQLRTVVDEGFDNPENVDIALEYLGKSRGIQRTRELAIK 384
+ SD+ G +TAP++FA+E P+LR +++ F +++ A+E + K GI+R +ELA +
Sbjct: 314 AGSDLAKGNLTAPVIFALEREPRLREIIESEFCEAGSLEEAIEAVTKGGGIKRAQELARE 373
Query: 385 HADLAAAAIDSLPES 399
AD A + LP S
Sbjct: 374 KADDAIKNLQCLPRS 388
>At4g36810 geranylgeranyl pyrophosphate synthase
Length = 371
Score = 91.7 bits (226), Expect = 7e-19
Identities = 99/339 (29%), Positives = 151/339 (44%), Gaps = 54/339 (15%)
Query: 38 HHYHTPTGSTAKVMRSLAFSKGLPALHSSRYQIHDQSSSISEE---------ELDPFSLV 88
+H+H + T RS + P S+ + SS +S + +P S
Sbjct: 16 NHHHPSSILTKSRSRSCPITLTKPISFRSKRTVSSSSSIVSSSVVTKEDNLRQSEPSSF- 74
Query: 89 ADELSFIANNLR--------AMVIAEVPKLASGAEYFFKMGVEGKRFRPTVLLLMSTALN 140
D +S+I A+ + E K+ Y G GKR RP VL + + L
Sbjct: 75 -DFMSYIITKAELVNKALDSAVPLREPLKIHEAMRYSLLAG--GKRVRP-VLCIAACEL- 129
Query: 141 LPIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDDV--LDDADTRRGIGSLN 198
+GG S A EMIH SL+HDD+ +D+ D RRG + +
Sbjct: 130 -----------VGGE-----ESTAMPAACAVEMIHTMSLIHDDLPCMDNDDLRRGKPTNH 173
Query: 199 FVMGNKLAVLAGDFLLSRACVALASLKNTEVVSL---------LAKVV--EHLVTGETMQ 247
V G +AVLAGD LLS A LAS +++VVS LAK + E LV G+ +
Sbjct: 174 KVFGEDVAVLAGDALLSFAFEHLASATSSDVVSPVRVVRAVGELAKAIGTEGLVAGQVVD 233
Query: 248 MSTTSDQR--CSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAMLAFDYGKNLGLAF 305
+S+ +E+ +KTA+L+ S AI+ G + D + + +GL F
Sbjct: 234 ISSEGLDLNDVGLEHLEFIHLHKTAALLEASAVLGAIVGGGSDDEIERLRKFARCIGLLF 293
Query: 306 QLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEE 344
Q++DD+LD T +S LGK + D+ +T P + +E+
Sbjct: 294 QVVDDILDVTKSSKELGKTAGKDLIADKLTYPKIMGLEK 332
>At3g14510 geranylgeranyl pyrophosphate synthetase, putative
Length = 284
Score = 87.4 bits (215), Expect = 1e-17
Identities = 63/186 (33%), Positives = 100/186 (52%), Gaps = 14/186 (7%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EMIH +SL+HDD+ +D+AD RRG + + V G +AVLAGD LL+ A + + + V
Sbjct: 59 EMIHTSSLIHDDLPCMDNADLRRGKPTNHKVFGEDMAVLAGDALLALAFEHMTVVSSGLV 118
Query: 230 VS--LLAKVVE--------HLVTGETMQMSTT--SDQRCSMEYYMQKTYYKTASLISNSC 277
S ++ VVE LV G+ + +S+ + +E +KTA+L+ +
Sbjct: 119 ASERMIRAVVELARAIGTKGLVAGQVVDLSSERLNPHDVGLERLEFIHLHKTAALLEAAA 178
Query: 278 KAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAP 337
AI+ G T + Y + +GL FQ++DD+LD T ++ LGK + D+ G +T P
Sbjct: 179 VIGAIMGGGTEEEIEKLRKYARCIGLLFQVVDDILDVTKSTEELGKTAGKDVMAGKLTYP 238
Query: 338 ILFAME 343
L +E
Sbjct: 239 RLIGLE 244
>At3g32040 geranylgeranyl pyrophosphate synthase, putative
Length = 360
Score = 85.5 bits (210), Expect = 5e-17
Identities = 62/186 (33%), Positives = 99/186 (52%), Gaps = 14/186 (7%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EMIH +SL+HDD+ +DDAD RRG + + V G +AVLAGD LL+ A + + + V
Sbjct: 135 EMIHTSSLIHDDLPCMDDADLRRGKPTNHKVFGEHMAVLAGDALLALAFEHMTVVSSGLV 194
Query: 230 --------VSLLAKVV--EHLVTGETMQMSTT--SDQRCSMEYYMQKTYYKTASLISNSC 277
V+ LAK + + LV G+ + + + +E +KTA+L+ +
Sbjct: 195 APERMIRSVTELAKAIGTKGLVAGQVSDLCSQGLNPYDVGLERLEFIHLHKTAALLEAAA 254
Query: 278 KAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAP 337
AI+ G T + YG+ +GL FQ++DD++D T ++ LGK + D+ +T P
Sbjct: 255 VLGAIIGGGTEEEIQKLRKYGRCIGLLFQVVDDIIDVTESTEELGKTAGKDVMARKLTYP 314
Query: 338 ILFAME 343
L +E
Sbjct: 315 RLIGLE 320
>At3g29430 geranylgeranyl pyrophosphate synthase, putative
Length = 357
Score = 85.5 bits (210), Expect = 5e-17
Identities = 63/186 (33%), Positives = 97/186 (51%), Gaps = 14/186 (7%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EMIH +SL+HDD+ +DDAD RRG + + G +AVLAGD LL+ A + + N V
Sbjct: 132 EMIHTSSLIHDDLPCMDDADLRRGKPTNHKEFGEDMAVLAGDALLALAFEHMTFVSNGLV 191
Query: 230 --------VSLLAKVV--EHLVTGETMQMSTT--SDQRCSMEYYMQKTYYKTASLISNSC 277
V LAK + + LV G+ + + + +E +KTA+L+ +
Sbjct: 192 APERMIRAVMELAKAIGTKGLVAGQVTDLCSQGLNPDDVGLERLEFIHLHKTAALLEAAA 251
Query: 278 KAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAP 337
AI+ G T + Y + +GL FQ++DD+LD T ++ LGK + D+ G +T P
Sbjct: 252 VLGAIMGGGTEEEIEKLRKYARCIGLLFQVVDDILDVTESTKELGKTAGKDVMAGKLTYP 311
Query: 338 ILFAME 343
L +E
Sbjct: 312 RLIGLE 317
>At3g14550 geranylgeranyl pyrophosphate synthase (GGPS3)
Length = 360
Score = 85.5 bits (210), Expect = 5e-17
Identities = 66/207 (31%), Positives = 108/207 (51%), Gaps = 21/207 (10%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EMIH +SL+HDD+ +D+AD RRG + + V G +AVLAGD LL+ A + + + V
Sbjct: 135 EMIHTSSLIHDDLPCMDNADLRRGKPTNHKVYGEDMAVLAGDALLALAFEHMTVVSSGLV 194
Query: 230 VS--LLAKVVE--------HLVTGETMQMSTT--SDQRCSMEYYMQKTYYKTASLISNSC 277
++ VVE LV G+ + +++ + + +E+ +KTA+L+ +
Sbjct: 195 APERMIRAVVELARAIGTTGLVAGQMIDLASERLNPDKVGLEHLEFIHLHKTAALLEAAA 254
Query: 278 KAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAP 337
I+ G T + Y + +GL FQ++DD+LD T ++ LGK + D+ G +T P
Sbjct: 255 VLGVIMGGGTEEEIEKLRKYARCIGLLFQVVDDILDVTKSTEELGKTAGKDVMAGKLTYP 314
Query: 338 ILFAMEEFPQ----LRTVVDE---GFD 357
L +E + LR +E GFD
Sbjct: 315 RLIGLERSKEVAEKLRREAEEQLLGFD 341
>At3g14530 putative geranylgeranyl pyrophosphate synthase GGPS3
Length = 360
Score = 84.7 bits (208), Expect = 9e-17
Identities = 60/186 (32%), Positives = 99/186 (52%), Gaps = 14/186 (7%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EMIH +SL+HDD+ +D+AD RRG + + V G +AVLAGD LL+ A + + + V
Sbjct: 135 EMIHTSSLIHDDLPCMDNADLRRGKPTNHKVYGEDMAVLAGDALLALAFEHMTVVSSGLV 194
Query: 230 V--SLLAKVVE--------HLVTGETMQMSTT--SDQRCSMEYYMQKTYYKTASLISNSC 277
++ VVE LV G+ + +++ + + +E+ +KTA+L+ +
Sbjct: 195 APEKMIRAVVELARAIGTTGLVAGQMIDLASERLNPDKVGLEHLEFIHLHKTAALLEAAA 254
Query: 278 KAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAP 337
I+ G T Y + +GL FQ++DD+LD T ++ LGK + D+ G +T P
Sbjct: 255 VLGVIMGGGTEQEIEKLRKYARCIGLLFQVVDDILDVTKSTEELGKTAGKDVMAGKLTYP 314
Query: 338 ILFAME 343
L +E
Sbjct: 315 RLIGLE 320
>At3g20160 geranyl geranyl pyrophosphate synthase, putative
Length = 344
Score = 79.3 bits (194), Expect = 4e-15
Identities = 60/191 (31%), Positives = 96/191 (49%), Gaps = 23/191 (12%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNT-- 227
EM+H +SL+ DD+ +D+ RRG + + V G +A+LA S+A +ALA K T
Sbjct: 120 EMLHASSLILDDLPCMDNDSLRRGKPTNHIVFGESIAILA-----SQALIALAVQKTTSS 174
Query: 228 -----------EVVSLLAKVVEHLVTGETMQMSTTS---DQRCSMEYYMQKTYYKTASLI 273
+ V + K VE LV G+ ++ D +E+ +KTA+L+
Sbjct: 175 TFADVPPERILKTVQEMVKAVEGLVAGQQADLAGEGMRFDSDTGLEHLEFIHIHKTAALL 234
Query: 274 SNSCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGI 333
+ AI+ G + + Y + +GL FQ++DDVLD T +S LGK + D+ G
Sbjct: 235 EAAAVMGAIMGGGSDEEIERLRSYARCIGLMFQVVDDVLDVTKSSEELGKTAGKDLIAGK 294
Query: 334 VTAPILFAMEE 344
+T P L +E+
Sbjct: 295 LTYPRLMGVEK 305
>At2g18620 putative geranylgeranyl pyrophosphate synthase
Length = 347
Score = 78.6 bits (192), Expect = 6e-15
Identities = 60/185 (32%), Positives = 98/185 (52%), Gaps = 12/185 (6%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRA------CVALAS 223
EMIH SL+HDD+ +D+ D RRG + + V G +AVLAGD L+S A A++
Sbjct: 124 EMIHTMSLIHDDLPCMDNDDLRRGKPTNHKVFGEDVAVLAGDALISFAFEHLATSTAVSP 183
Query: 224 LKNTEVVSLLAKVV--EHLVTGETMQMSTTS-DQR-CSMEYYMQKTYYKTASLISNSCKA 279
+ + LAK + + LV G+ + +++ DQ +E +KTA L+ +
Sbjct: 184 ARVVRAIGELAKAIGSKGLVAGQVVDLTSGGMDQNDVGLEVLEFIHVHKTAVLLEAATVL 243
Query: 280 IAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPIL 339
AI+ G + + + + +GL FQ++DD+LD T +S LGK + D+ +T P L
Sbjct: 244 GAIVGGGSDEEVEKLRRFARCIGLLFQVVDDILDVTKSSEELGKTAGKDLIADKLTYPKL 303
Query: 340 FAMEE 344
+E+
Sbjct: 304 MGLEK 308
>At2g18640 putative geranylgeranyl pyrophosphate synthase
Length = 372
Score = 75.9 bits (185), Expect = 4e-14
Identities = 59/184 (32%), Positives = 95/184 (51%), Gaps = 11/184 (5%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVAL-----ASL 224
EMIH SL+ DD+ +D+ D RRG + + V G +A+L+G LL+ A L +S
Sbjct: 150 EMIHTMSLIKDDLPCMDNDDLRRGKPTTHKVFGESVAILSGGALLALAFEHLTEADVSSK 209
Query: 225 KNTEVVSLLAKVV--EHLVTGETMQMSTTSDQR--CSMEYYMQKTYYKTASLISNSCKAI 280
K V LAK + + LV G+ +S+ ++ +E +KT SL+ S
Sbjct: 210 KMVRAVKELAKSIGTKGLVAGQAKDLSSEGLEQNDVGLEDLEYIHVHKTGSLLEASAVIG 269
Query: 281 AILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILF 340
A++ G T ++ + +GL FQ++DD+LD T +S LGK + D G +T P +
Sbjct: 270 AVIGGGTEKEIEKVRNFARCIGLLFQVVDDILDETKSSEELGKTAGKDKVAGKLTYPKVI 329
Query: 341 AMEE 344
+E+
Sbjct: 330 GVEK 333
>At2g23800 pregeranylgeranyl pyrophosphate synthase
Length = 376
Score = 74.3 bits (181), Expect = 1e-13
Identities = 71/232 (30%), Positives = 111/232 (47%), Gaps = 29/232 (12%)
Query: 124 GKRFRPTVLLLMSTALNLPIPKPPPSNDLGGTFTADLRSRQQRIAEITEMIHVASLLHDD 183
GKR RP +L L S L +GG A + + A EMIH SL+ DD
Sbjct: 124 GKRVRP-ILCLASCEL------------VGGQENAAMPA-----ACAVEMIHTMSLIKDD 165
Query: 184 V--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRA-----CVALASLKNTEVVSLLAKV 236
+ +D+ D RRG + + V G +A+L+G LLS A ++S + V LA+
Sbjct: 166 LPCMDNDDLRRGKPTTHKVYGEGVAILSGGALLSLAFEHMTTAEISSERMVWAVRELARS 225
Query: 237 V--EHLVTGETMQMSTTSDQ--RCSMEYYMQKTYYKTASLISNSCKAIAILAGQTADVAM 292
+ LV G+ M +S+ +E+ +KTA L+ + AI+ G + +
Sbjct: 226 IGTRGLVAGQAMDISSEGLDLNEVGLEHLEFIHVHKTAVLLETAAVLGAIIGGGSDEEIE 285
Query: 293 LAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPILFAMEE 344
+ + +GL FQ++DD+LD T +S LGK + D G +T P L +E+
Sbjct: 286 SVRKFARCIGLLFQVVDDILDETKSSEELGKTAGKDQLAGKLTYPKLIGLEK 337
>At1g49530 geranyl geranyl pyrophosphate synthase, putative
Length = 336
Score = 71.2 bits (173), Expect = 1e-12
Identities = 57/188 (30%), Positives = 94/188 (49%), Gaps = 16/188 (8%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRAC--------VAL 221
EMIH ASL+ DD+ +DD RRG + + V G K ++LA + L S A + +
Sbjct: 109 EMIHAASLILDDLPCMDDDSLRRGKPTNHKVFGEKTSILASNALRSLAVKQTLASTSLGV 168
Query: 222 ASLKNTEVVSLLAKVV--EHLVTGETMQMS----TTSDQRCSMEYYMQKTYYKTASLISN 275
S + V +A+ V E LV G+ ++ + ++ + Y +KTA L+
Sbjct: 169 TSERVLRAVQEMARAVGTEGLVAGQAADLAGERMSFKNEDDELRYLELMHVHKTAVLVEA 228
Query: 276 SCKAIAILAGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVT 335
+ AI+ G + + Y + +GL FQ++DDVLD T +S LGK + D+ G +T
Sbjct: 229 AAVVGAIMGGGSDEEIERLKSYARCVGLMFQVMDDVLDETKSSEELGKTAGKDLITGKLT 288
Query: 336 APILFAME 343
P + ++
Sbjct: 289 YPKVMGVD 296
>At4g38460 geranylgeranyl pyrophosphate synthase-related protein
Length = 326
Score = 55.8 bits (133), Expect = 4e-08
Identities = 45/153 (29%), Positives = 76/153 (49%), Gaps = 15/153 (9%)
Query: 172 EMIHVASLLHDDV--LDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKNTEV 229
EM+H ASL+HDD+ +DD RRG S + V G+ +A+LAGD L A + S ++
Sbjct: 117 EMVHAASLIHDDLPCMDDDPVRRGKPSNHTVYGSGMAILAGDALFPLAFQHIVSHTPPDL 176
Query: 230 V---SLLAKVVEHLVTGETMQMSTTSDQRCSME------YYMQKTYYKTASLISNSCKAI 280
V ++L + E T + M+ + Q +E ++Q+ + S C
Sbjct: 177 VPRATILRLITEIARTVGSTGMA--AGQYVDLEGGPFPLSFVQEKKFGAMGECSAVCG-- 232
Query: 281 AILAGQTADVAMLAFDYGKNLGLAFQLIDDVLD 313
+L G T D YG+ +G+ +Q++DD+ +
Sbjct: 233 GLLGGATEDELQSLRRYGRAVGMLYQVVDDITE 265
>At5g47770 farnesyl diphosphate synthase precursor (gb|AAB49290.1)
Length = 384
Score = 35.0 bits (79), Expect = 0.078
Identities = 49/208 (23%), Positives = 93/208 (44%), Gaps = 16/208 (7%)
Query: 172 EMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLAGDFLLSRACVALASLKN----- 226
E + L+ DD++D++ TRRG F + V D +L R + K+
Sbjct: 125 EWLQAYFLVLDDIMDNSVTRRGQPCW-FRVPQVGMVAINDGILLRNHIHRILKKHFRDKP 183
Query: 227 --TEVVSLLAKVVEHLVTGETMQMSTTSD-----QRCSMEYYMQKTYYKTASLISNSCKA 279
++V L +V G+ + + TT + + S+ + + YKTA A
Sbjct: 184 YYVDLVDLFNEVELQTACGQMIDLITTFEGEKDLAKYSLSIHRRIVQYKTAYYSFYLPVA 243
Query: 280 IAIL-AGQTADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGKGSLSDIRHGIVTAPI 338
A+L AG+ + + + ++G+ FQ+ DD LD +LGK +DI + +
Sbjct: 244 CALLMAGENLENHIDVKNVLVDMGIYFQVQDDYLDCFADPETLGKIG-TDIEDFKCSWLV 302
Query: 339 LFAMEEFPQLRT-VVDEGFDNPENVDIA 365
+ A+E + +T ++ E + P+ ++A
Sbjct: 303 VKALERCSEEQTKILYENYGKPDPSNVA 330
>At4g17190 farnesyl-pyrophosphate synthetase FPS2
Length = 342
Score = 34.7 bits (78), Expect = 0.10
Identities = 45/170 (26%), Positives = 76/170 (44%), Gaps = 24/170 (14%)
Query: 172 EMIHVASLLHDDVLDDADTRRGIGSLNFVMGNKLAVLA-GDFLLSRACVALASLKN---- 226
E + L+ DD++D++ TRRG + K+ ++A D +L R + K+
Sbjct: 83 EWLQAYFLVLDDIMDNSVTRRGQPC--WFRKPKVGMIAINDGILLRNHIHRILKKHFREM 140
Query: 227 ---TEVVSLLAKVVEHLVTGETMQMSTTSD-----QRCSMEYYMQKTYYKTASLISNSCK 278
++V L +V G+ + + TT D + S++ + + YKTA
Sbjct: 141 PYYVDLVDLFNEVEFQTACGQMIDLITTFDGEKDLSKYSLQIHRRIVEYKTAYYSFYLPV 200
Query: 279 AIAIL-AGQT----ADVAMLAFDYGKNLGLAFQLIDDVLDFTGTSASLGK 323
A A+L AG+ DV + D +G+ FQ+ DD LD +LGK
Sbjct: 201 ACALLMAGENLENHTDVKTVLVD----MGIYFQVQDDYLDCFADPETLGK 246
>At1g30930 F17F8.20
Length = 708
Score = 33.9 bits (76), Expect = 0.17
Identities = 31/150 (20%), Positives = 63/150 (41%), Gaps = 21/150 (14%)
Query: 17 SLNGTRRLLSIQEQNHQSLQSHHYHTPTGSTAKVMRSLAFSKGLPALHSSRYQ------I 70
+ N R L +Q Q L ++ H + + + S + L SS Q
Sbjct: 296 NFNPDRNTLQVQSVEIQGLGANRDHIACHAFVDYVEDFSVSDAVLQLKSSPLQQQGQDTS 355
Query: 71 HDQSSSISEEELDPFSLVADELS--------FIANNLRAMVIAEVPKLASGAEYFFKMGV 122
HD S S+ ++LD + +A++L+ F + R +++ + +YF ++ +
Sbjct: 356 HDLSKSVKNKQLDNYEFLANKLNHSVSLAILFSKSVARCHCVSKQWRAIFRRKYFIELFL 415
Query: 123 EGKRFRPTVLLLMS-------TALNLPIPK 145
+ RP +L ++ + L+LP P+
Sbjct: 416 TRSKARPRILFVVQDGEWSEWSFLSLPQPQ 445
>At4g35850 unknown protein
Length = 444
Score = 32.0 bits (71), Expect = 0.66
Identities = 22/65 (33%), Positives = 31/65 (46%), Gaps = 2/65 (3%)
Query: 43 PTGSTAKVMRSLAFSKGLPALHSSRYQIHDQSSSISEEEL--DPFSLVADELSFIANNLR 100
P + K++ + SKG A+ SSR D SISEEEL P + + F+ NL
Sbjct: 198 PDNLSTKILEFVEQSKGWSAIDSSRKSAEDVMFSISEEELYNIPTADYSHRTRFLQRNLT 257
Query: 101 AMVIA 105
+A
Sbjct: 258 VYHVA 262
>At1g07970 unknown protein
Length = 693
Score = 30.8 bits (68), Expect = 1.5
Identities = 18/57 (31%), Positives = 32/57 (55%), Gaps = 2/57 (3%)
Query: 54 LAFSKGLPALHSSRYQIHDQSSSISEEELDPFSLVADELSFIANNLRA-MVIAEVPK 109
+A SKG+ +LH +++ ++ S S+E D FSL +L + +A ++E PK
Sbjct: 107 MALSKGI-SLHRAKFAARGETKSFSKETKDQFSLSNRQLELLGIKKKADQSVSEFPK 162
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,579,802
Number of Sequences: 26719
Number of extensions: 350385
Number of successful extensions: 1108
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1055
Number of HSP's gapped (non-prelim): 28
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0250.7


