

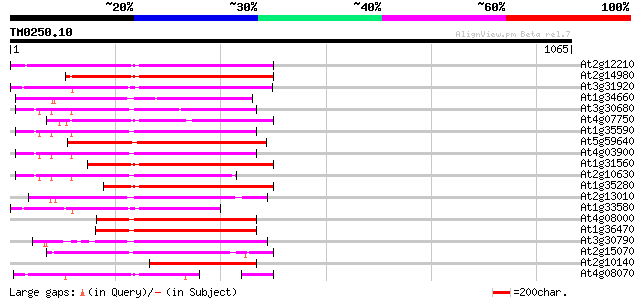
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.10
(1065 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g12210 putative TNP2-like transposon protein 387 e-107
At2g14980 pseudogene 353 3e-97
At3g31920 hypothetical protein 347 2e-95
At1g34660 hypothetical protein 337 2e-92
At3g30680 hypothetical protein 335 6e-92
At4g07750 putative transposon protein 334 1e-91
At1g35590 CACTA-element, putative 332 5e-91
At5g59640 serine/threonine-specific protein kinase - like 331 2e-90
At4g03900 putative transposon protein 325 6e-89
At1g31560 putative protein 322 5e-88
At2g10630 En/Spm transposon hypothetical protein 1 311 1e-84
At1g35280 hypothetical protein 309 5e-84
At2g13010 pseudogene 305 9e-83
At1g33580 En/Spm-like transposon protein, putative 288 9e-78
At4g08000 putative transposon protein 284 2e-76
At1g36470 hypothetical protein 281 1e-75
At3g30790 hypothetical protein 259 4e-69
At2g15070 En/Spm-like transposon protein 253 4e-67
At2g10140 putative protein 223 4e-58
At4g08070 209 7e-54
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 387 bits (994), Expect = e-107
Identities = 213/509 (41%), Positives = 291/509 (56%), Gaps = 16/509 (3%)
Query: 1 MDKEWTKQPRFSSEYINGVESFL-DFAYTKGRPQGDEILCPCAKCGNCCWEKRNVIYNHL 59
+DK W + R Y G + F+ D A G D I+CPC C N + + + +HL
Sbjct: 2 LDKAWVRLCRADPGYEKGAQKFVTDVAVALG--DIDMIVCPCKDCRNVERKSGSDVVDHL 59
Query: 60 ICTGFLEGYNV---WIHHGEQMMEIDEEME-EDYHDDIDGLLYETFKYVAGAEPEGPNED 115
+ G E Y + W HHG+ D E ++ +I L + A +ED
Sbjct: 60 VRRGMDEAYKLRADWYHHGDVDSVADCESNFSRWNAEILELYQAAQGFDADLGEIAEDED 119
Query: 116 ARK--FYNLIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTALLELLKEAMPD 173
R+ F + +++ LYP C N S LS I+ ++ +K +GWS+ SF LL L E +P
Sbjct: 120 IREDEFLAKLADAEIPLYPSCLNHSKLSAIVTLFRIKTKNGWSDKSFNELLGTLPEMLPA 179
Query: 174 LNIPE-SFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVA 232
N+ S + K +R + Y++IHAC NDC L+ K Y+N +C C ASRWK Q
Sbjct: 180 DNVLHTSLYEVKKFLRSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRWKSNMQT- 238
Query: 233 GEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQA 292
GE +K VP KVLR+FP+IPR +R+F + +RWH +S DGKLRHPVD
Sbjct: 239 GEAKKG-----VPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSTDGKLRHPVDSVT 293
Query: 293 WQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYM 352
W + + +FA + RN+RLGL++DGFNPF + +S WPVLLV YN PP LCMK E +
Sbjct: 294 WDKMNDKYPEFAIEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPPDLCMKKENI 353
Query: 353 MMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYP 412
M+SLLIPG Q PG +IDVYL+PLIE+L LW+ G TYDA K TF + A LLWTI+D+P
Sbjct: 354 MLSLLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFP 413
Query: 413 AYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGKM 472
AY L+G KGK+ C C T S +LK+S K +M HR L H++R + F+GK+
Sbjct: 414 AYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHVFMCHRKGLAPTHSYRNKKAWFDGKI 473
Query: 473 EFRTASPLLKGTEILEILKNFNNVFGKTK 501
E + +L G E+ + LKNF N FG K
Sbjct: 474 EHGRKARILTGREVSQTLKNFKNDFGNAK 502
>At2g14980 pseudogene
Length = 665
Score = 353 bits (906), Expect = 3e-97
Identities = 184/396 (46%), Positives = 246/396 (61%), Gaps = 9/396 (2%)
Query: 107 AEPEGPNEDARKFYNLIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTALLEL 166
AE + ED L+D ++ LYP C N S LS I+ ++ LK +GWS+ SF LLE
Sbjct: 8 AERDDKKED-ELLAKLVD-AETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLET 65
Query: 167 LKEAMPDLNIPE-SFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRW 225
L E +P+ N+ S + K ++ + Y++IHAC NDC L+ K+Y+ +C C ASRW
Sbjct: 66 LPEMLPEDNVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFRKKYKKLQNCLKCNASRW 125
Query: 226 KEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLR 285
K GE +K VP KVLR+F ++PRL+R+F + +RWH +S DGKL
Sbjct: 126 KTNMHT-GEVKKG-----VPQKVLRYFSIVPRLKRMFRSEEMARDLRWHFHNKSTDGKLC 179
Query: 286 HPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWL 345
HPVD W + + FAS+ RN+RLGL+++GFNPF + +S WPVLLV YN PP L
Sbjct: 180 HPVDSVTWDQMNAKYPLFASEERNLRLGLSTNGFNPFNMKNTRYSCWPVLLVNYNLPPDL 239
Query: 346 CMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALL 405
CMK E +M++LLIPG Q PG +IDVYL+PLI++L LW+ G TYDA K +F + A LL
Sbjct: 240 CMKKENIMLTLLIPGSQQPGNSIDVYLEPLIDDLCHLWDNGEWTYDAFSKSSFTLKAMLL 299
Query: 406 WTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTE 465
WTI+D+PAY L+G KGK+AC C +T S +LK S K YM HR+ LP H +R +
Sbjct: 300 WTISDFPAYGNLAGCKVKGKMACLLCGKHTESMWLKFSRKHVYMGHRMGLPPSHRFRNKK 359
Query: 466 NLFNGKMEFRTASPLLKGTEILEILKNFNNVFGKTK 501
+ F+GK E R S +L G EI LKNF N FG K
Sbjct: 360 SWFDGKAEHRRKSRILSGLEISHNLKNFQNNFGNFK 395
>At3g31920 hypothetical protein
Length = 725
Score = 347 bits (891), Expect = 2e-95
Identities = 194/516 (37%), Positives = 275/516 (52%), Gaps = 27/516 (5%)
Query: 1 MDKEWTKQPRFSSEYINGVESFLDFAYTKGRPQGD--EILCPCAKCGNCCWEKRNVIYNH 58
MDK W PR + EY G +F+ Y R GD E+ CPC C N C + I H
Sbjct: 1 MDKSWVWLPRNNIEYEKGATTFV---YASSRSLGDPAEMFCPCIDCRNLCHQPIETILEH 57
Query: 59 LICTGFLEGYN---VWIHHGE---QMMEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEGP 112
L+ G Y W HGE +E++ + + + F ++ +
Sbjct: 58 LVMKGMDHKYKRNRCWSKHGEIRADKLEVEPSSTTSECEAYELIRTAYFDGDVHSDSQNQ 117
Query: 113 NEDARK---------FYNLIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTAL 163
NED K F + E++ LY C ++ +S I+ +Y +K G S F L
Sbjct: 118 NEDDSKEPETKEEADFREKLKEAETPLYHGCPKYTKVSAIMGLYRIKVKSGMSENYFDQL 177
Query: 164 LELLKEAMPDLNI-PESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKA 222
L L+ + +P N+ P S N+ K ++ G Y IHAC NDC+LY KEYE ++C C A
Sbjct: 178 LSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACQNDCILYRKEYELRDTCPRCNA 237
Query: 223 SRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDG 282
SRW E + GE +K +PAKVL +FP+ R +R+F + ++ RWH S DG
Sbjct: 238 SRW-ERDKHTGEEKKG-----IPAKVLGYFPIKDRFKRMFRSKKMAEDQRWHFSNASDDG 291
Query: 283 KLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYP 342
+RH VD AW + +FA++PRN+RLGL++DG NPF + +STWP+LLV YN
Sbjct: 292 TMRHHVDSLAWAQVNDKWPEFAAEPRNLRLGLSADGMNPFSIQNTKYSTWPILLVNYNMA 351
Query: 343 PWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHA 402
P +CMK+E +M++LLI G +P NIDVYL PLI++LK+LW G+ YD+ +KE+F + A
Sbjct: 352 PTMCMKAENIMLTLLIHGPTAPSNNIDVYLAPLIDDLKDLWSEGIHVYDSFKKESFTLRA 411
Query: 403 ALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWR 462
LW+I+DYPA L+G KGK AC C +T +LK S K Y+ +R L H +R
Sbjct: 412 MFLWSISDYPALGTLAGCKVKGKQACNVCGKDTPFRWLKFSRKHVYLRNRRRLRPGHPYR 471
Query: 463 TTENLFNGKMEFRTASPLLKGTEILEILKNFNNVFG 498
F+ +E T S + G EI E LK + G
Sbjct: 472 RRRGWFDNTVEEGTTSRIQTGEEIFEALKALKMILG 507
>At1g34660 hypothetical protein
Length = 501
Score = 337 bits (864), Expect = 2e-92
Identities = 176/475 (37%), Positives = 273/475 (57%), Gaps = 36/475 (7%)
Query: 12 SSEYINGVESFLDFAYTKGRP-QGDEILCPCAKCGNCCWEKRNVIYNHLICTGFLEGYNV 70
+ E+ G+ F+ FA + P + ++ CPC+ CGN + + I+ HL GF+ GY +
Sbjct: 13 TKEFKEGLVDFMRFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKHLYSRGFMPGYYI 72
Query: 71 WIHHGEQMM------EIDE------------EMEEDYHDDIDGLLY---ETFKYVAGAEP 109
W HG+ E DE E EE+ D G++ F+ +
Sbjct: 73 WFSHGKDFETLANSNEDDEIHGLEPEDNNSNEEEENIIRDPTGVIQMVTNAFRETTESGI 132
Query: 110 EGPNEDARKFYNLIDESQKELYP*CKN-FSTLSFIIRMYLLKCTHGWSNVSFTALLELLK 168
E PN +A+KF++++D ++K +Y CK S LS R+ +K + S A+ + +K
Sbjct: 133 EEPNLEAKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKTEYNLSEDCVDAIADFVK 192
Query: 169 EAMPDLNI-PESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKE 227
+ +P+ N+ P S+ + + ++ LGL Y+ I C ++CM++W++ N ++C C+ R
Sbjct: 193 DLLPEPNLAPGSYYEIQKLVSSLGLPYQMIDVCVDNCMIFWRDTVNLDACTFCQKPR--- 249
Query: 228 FPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHP 287
F+ + + +VP K + + P+ RL+RL+ RT+ MRWH E S DG + HP
Sbjct: 250 -------FQVTSGKSRVPFKRMWYLPITDRLKRLYQSERTAGPMRWHAEH-SSDGNISHP 301
Query: 288 VDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCM 347
D +AW+ F+ ++ DFA++ RNV LGL +DGFNPF +S WPV+L YN PP LCM
Sbjct: 302 SDAEAWKHFNSMYKDFANEHRNVYLGLCTDGFNPFGKSGRKYSLWPVILTPYNLPPSLCM 361
Query: 348 KSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWT 407
K E++ +S+L+PG + P +++DV+LQPLI EL+ LW GV YD S K F M A L+WT
Sbjct: 362 KREFLFLSILVPGPEHPRRSLDVFLQPLIHELQLLWSEGVVAYDVSLKNNFVMRAVLMWT 421
Query: 408 INDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD-HRVFLPADHTW 461
I+D+PAY MLSGW+T G+L+C C + LKH K C+ D HR FLP +H++
Sbjct: 422 ISDFPAYGMLSGWTTHGRLSCPYCQDTIDAFQLKHGKKSCWFDCHRRFLPKNHSY 476
>At3g30680 hypothetical protein
Length = 1116
Score = 335 bits (860), Expect = 6e-92
Identities = 186/498 (37%), Positives = 272/498 (54%), Gaps = 53/498 (10%)
Query: 12 SSEYINGVESFLDFAYTKGRPQG--DEILCPCAKCGNCCWEKRNV----IYNHLICTGFL 65
S+EY+ GVE F+ FA ++ Q + CPC+ C N EK + + +HL GF+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKN---EKHIISGRRVSSHLFSQGFM 86
Query: 66 EGYNVWIHHGEQM--------------MEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEG 111
Y VW HGE++ E EE+ D ++ + F + G +
Sbjct: 87 PDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVNDAFNFNVGYDDNV 146
Query: 112 PNED------------------ARKFYNLIDESQKELYP*CKNF-STLSFIIRMYLLKCT 152
++D + KFY+L++ + LY C+ S LS R+ K
Sbjct: 147 GHDDCYHHDGSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCREGQSQLSLASRLMHNKAE 206
Query: 153 HGWSNVSFTALLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEY 211
+ S ++ E+ + +P+ N S +T+ ++R+LGL Y I C N+CML+WKE
Sbjct: 207 YNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCKNNCMLFWKED 266
Query: 212 ENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSM 271
E ++ CR C A RWK + KVP + + P+ +L+R++ +T+ +M
Sbjct: 267 EKEDQCRFCGAQRWKP--------KDDRRRTKVPYSRMWYLPIADQLKRMYQSHKTAAAM 318
Query: 272 RWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHST 331
RWH E +SK+G++ HP D W+ F LH FA +PRNV LGL +DGFNPF +MS +HS
Sbjct: 319 RWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLCTDGFNPF-SMSRNHSL 377
Query: 332 WPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYD 391
WPV+L YN PP +CM +EY+ +++L G P ++DV+LQPLIEELKELW GV+ YD
Sbjct: 378 WPVILTPYNLPPRMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKELWCTGVDAYD 437
Query: 392 ASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD- 450
S + F + A LLWTI+D+PAY+MLSGW+T GKL+C C +T S YL + K C+ D
Sbjct: 438 VSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDC 497
Query: 451 HRVFLPADHTWRTTENLF 468
HR FLP H R + F
Sbjct: 498 HRRFLPHGHPSRRNKKDF 515
>At4g07750 putative transposon protein
Length = 569
Score = 334 bits (857), Expect = 1e-91
Identities = 186/441 (42%), Positives = 249/441 (56%), Gaps = 28/441 (6%)
Query: 71 WIHHGEQMMEIDEEME-EDYHDDI----DGLLYETFKYVAG----AEPEGPNEDARKFYN 121
W HHGE + + + ++D++ L E F Y G AE + ED +F
Sbjct: 11 WYHHGESNSRAESDTKVTQWNDEVLYQAAEFLDEEFAYKVGLSEIAECDDKKED--EFLA 68
Query: 122 LIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTALLELLKEAMPDLNIPE-SF 180
+ +++ LYP C N S LS I+ ++ L +GWS+ SF LLE+L E +P N+ S
Sbjct: 69 KLADAETPLYPSCVNHSKLSAIVSLFRLNTKNGWSDKSFNDLLEILPEMLPKDNVLHTSL 128
Query: 181 NKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSES 240
K K ++ + Y++IHAC NDC L+ K Y+ +SC C ASRWK + GE +K
Sbjct: 129 YKVKKFLKSFDMGYQKIHACVNDCCLFRKNYKKLDSCPKCNASRWKTNLHI-GEIKKG-- 185
Query: 241 EYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLH 300
VP KVLR+FP+IPRL+ +F +RWH +S D KL HPVD W + +
Sbjct: 186 ---VPQKVLRYFPIIPRLKGMFRSKEMDKDLRWHFSNKSSDEKLCHPVDLVTWDQMNAKY 242
Query: 301 LDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPG 360
FA + RN+R GL++DGFNPF + +S WP L CMK E +M++LLIPG
Sbjct: 243 PSFAGEERNLRHGLSTDGFNPFNMKNTRYSCWPDL----------CMKKENIMLTLLIPG 292
Query: 361 QQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGW 420
Q PG NIDVYL+PLIE+L LW+ G TYDA K TF + A LLWTI+D+PAY L+G
Sbjct: 293 PQQPGNNIDVYLEPLIEDLCHLWDNGELTYDAYSKSTFDLKAMLLWTISDFPAYGNLAGC 352
Query: 421 STKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGKMEFRTASPL 480
KGK+ C C NT S +LK S K YM H LP H +R ++ F+GK E +
Sbjct: 353 KVKGKMGCPLCGKNTDSMWLKFSRKHVYMCHIKGLPPTHHFRMKKSWFDGKAENGRKRRI 412
Query: 481 LKGTEILEILKNFNNVFGKTK 501
L G EI + LKNF N FG K
Sbjct: 413 LSGQEIYQNLKNFKNNFGTLK 433
>At1g35590 CACTA-element, putative
Length = 1180
Score = 332 bits (852), Expect = 5e-91
Identities = 186/498 (37%), Positives = 269/498 (53%), Gaps = 53/498 (10%)
Query: 12 SSEYINGVESFLDFAYTKGRPQG--DEILCPCAKCGNCCWEKRNV----IYNHLICTGFL 65
S+EY+ GVE F+ FA ++ Q + CPC+ C N EK + + +HL GF+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKN---EKHIISGRRVSSHLFSQGFM 86
Query: 66 EGYNVWIHHGEQM--------------MEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEG 111
Y VW HGE++ E EE+ D ++ + F + G +
Sbjct: 87 PDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVNDAFNFNVGYDDNV 146
Query: 112 PNED------------------ARKFYNLIDESQKELYP*CKNF-STLSFIIRMYLLKCT 152
++D + KFY+L++ + LY C+ S LS R+ K
Sbjct: 147 GHDDCYHHDGSYQNVEEPVRNHSNKFYDLLEGANNLLYDGCREGQSQLSLASRLMHNKAE 206
Query: 153 HGWSNVSFTALLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEY 211
+ S ++ E+ + +P+ N S +T+ ++ +LGL Y I C N+CML+WKE
Sbjct: 207 YNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMHNLGLPYHTIDVCKNNCMLFWKED 266
Query: 212 ENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSM 271
E ++ CR C A RWK + KVP + + P+ RL+R++ C +T+ +M
Sbjct: 267 EKEDQCRFCGAQRWKP--------KDDRRRTKVPYSRMWYLPIADRLKRMYQCHKTAAAM 318
Query: 272 RWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHST 331
RWH E +SK+G++ HP D W+ F LH FA +PRNV LGL +DGFNPF MS +HS
Sbjct: 319 RWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLCTDGFNPFG-MSRNHSL 377
Query: 332 WPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYD 391
WPV+L YN PP +CM +EY+ +++L G P ++DV+LQPLIEELKELW GV+ Y+
Sbjct: 378 WPVILTPYNLPPGMCMNTEYLFLTILNYGPNHPRASLDVFLQPLIEELKELWCTGVDAYE 437
Query: 392 ASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD- 450
S + F + A LLWTI+D+PAY MLSGW+T GKL+C C +T S YL K C+ D
Sbjct: 438 VSLSQNFNLKAVLLWTISDFPAYNMLSGWTTHGKLSCPVCMESTKSFYLPIGRKTCWFDC 497
Query: 451 HRVFLPADHTWRTTENLF 468
HR FLP H R + F
Sbjct: 498 HRRFLPHGHPSRRNKKDF 515
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 331 bits (848), Expect = 2e-90
Identities = 156/381 (40%), Positives = 242/381 (62%), Gaps = 12/381 (3%)
Query: 110 EGPNEDARKFYNLIDESQKELYP*CKN-FSTLSFIIRMYLLKCTHGWSNVSFTALLELLK 168
E PN +A+KFY+++D +++ +Y CK S LS R+ LK + S ++ ++++
Sbjct: 48 EDPNFEAKKFYDILDAAKQPIYDGCKEGLSKLSLAARLMSLKTDNNLSQNCMDSIAQIMQ 107
Query: 169 EAMPD-LNIPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKE 227
E +P+ N P+S+ + K ++R LGL Y++I C ++CM++WKE E + C CK R++
Sbjct: 108 EYLPEGNNSPKSYYEIKKLMRSLGLPYQKIDVCQDNCMIFWKETEKEEYCLFCKKDRYRP 167
Query: 228 FPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHP 287
++ + +P + + + P+ RL+RL+ T+ MRWH E + DG++ HP
Sbjct: 168 TQKIG--------QKSIPYRQMFYLPIADRLKRLYQSHNTAKHMRWHAEHLASDGEMGHP 219
Query: 288 VDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCM 347
DG+AW+ F +H DFAS PRNV LGL +DGFNPF ++S WPV+L YN PP +CM
Sbjct: 220 SDGEAWKHFHKVHPDFASKPRNVYLGLCTDGFNPFGMSGHNYSLWPVILTPYNLPPEMCM 279
Query: 348 KSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWT 407
K E+M +++L+PG P +++D++LQPLIEELK+LW GVE YD S K+ F + L+WT
Sbjct: 280 KQEFMFLTILVPGPNHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVLMWT 339
Query: 408 INDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD-HRVFLPADHTWRTTEN 466
I+D+PAY MLSGW+T G+LAC C+ T + +LK+ K C+ D HR FLP +H++R +
Sbjct: 340 ISDFPAYGMLSGWTTHGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRGNKK 399
Query: 467 LF-NGKMEFRTASPLLKGTEI 486
F GK+ + +L G E+
Sbjct: 400 DFKKGKVVEDSKPEILTGEEL 420
>At4g03900 putative transposon protein
Length = 817
Score = 325 bits (834), Expect = 6e-89
Identities = 184/498 (36%), Positives = 267/498 (52%), Gaps = 53/498 (10%)
Query: 12 SSEYINGVESFLDFAYTKGRPQG--DEILCPCAKCGNCCWEKRNV----IYNHLICTGFL 65
S+EY+ GVE F+ FA ++ Q + CPC+ C N EK + + +HL F+
Sbjct: 30 SAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKN---EKHIISGRRVSSHLFSHEFM 86
Query: 66 EGYNVWIHHGEQM--------------MEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEG 111
Y VW HGE++ E EE+ D ++ + F + G +
Sbjct: 87 PDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVNDAFNFNVGYDDNV 146
Query: 112 PNED------------------ARKFYNLIDESQKELYP*CKNF-STLSFIIRMYLLKCT 152
++D + KFY+L++ + LY C+ S LS R+ K
Sbjct: 147 GHDDNYHHDGSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCREGQSQLSLASRLMHNKAE 206
Query: 153 HGWSNVSFTALLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEY 211
+ S ++ E+ +P+ N S +T+ ++R+LGL Y I N+CML+WKE
Sbjct: 207 YNMSEKLVDSVCEMFTAFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVYKNNCMLFWKED 266
Query: 212 ENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSM 271
E ++ CR C A RWK + KVP + + P+ RL+R++ +T+ +M
Sbjct: 267 EKEDQCRFCGAQRWKP--------KDDRRRTKVPYSRMWYLPIADRLKRMYQSHKTAAAM 318
Query: 272 RWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHST 331
RWH E +SK+G++ HP D W+ F LH FA +P NV LGL +DGFNPF MS +HS
Sbjct: 319 RWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPHNVYLGLYTDGFNPFG-MSRNHSL 377
Query: 332 WPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYD 391
WPV+L YN PP +CM +EY+ +++L G P ++DV+LQPLIEELKELW GV+ YD
Sbjct: 378 WPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKELWCTGVDAYD 437
Query: 392 ASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD- 450
S + F + A LLWTI+D+PAY+MLSGW+T GKL+C C +T S YL + K C+ D
Sbjct: 438 VSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDC 497
Query: 451 HRVFLPADHTWRTTENLF 468
HR FLP H R + F
Sbjct: 498 HRRFLPHGHPSRRNKKDF 515
>At1g31560 putative protein
Length = 1431
Score = 322 bits (826), Expect = 5e-88
Identities = 161/354 (45%), Positives = 218/354 (61%), Gaps = 7/354 (1%)
Query: 149 LKCTHGWSNVSFTALLELLKEAMPDLNIPE-SFNKTKGMIRDLGLDYKRIHACPNDCMLY 207
+K +GWS+ SF LL L E +P N+ S + K ++ + Y++IHAC NDC L+
Sbjct: 86 IKTKNGWSDKSFNELLGTLPEMLPADNVLRTSLYEVKKFLKSFDMGYEKIHACVNDCCLF 145
Query: 208 WKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRT 267
K Y+N +C C ASRWK + Q GE +K VP KVLR+F +IPR +++F
Sbjct: 146 RKRYKNLENCPKCNASRWKNYMQT-GEAKKG-----VPQKVLRYFLIIPRFKKMFRSKEM 199
Query: 268 SDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSI 327
+ +RWH +S DGK RHPVD W + +FA++ RN+RLGL++DGFNPF +
Sbjct: 200 AKDLRWHYSNKSTDGKHRHPVDSVTWDKMIDKYPEFAAEERNMRLGLSTDGFNPFNMKNT 259
Query: 328 SHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGV 387
+S WPVLLV YN PP CMK+E +M+SLLIPG Q PG +IDVYL+P IE+L LW+ G
Sbjct: 260 RYSCWPVLLVNYNLPPDQCMKNENIMLSLLIPGPQQPGNSIDVYLEPFIEDLNHLWKKGE 319
Query: 388 ETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMC 447
TYDA K TF + A L WTI+D+PAY L+G KGK+ C C T S +LK+S K
Sbjct: 320 LTYDAFSKTTFTLKAMLFWTISDFPAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHV 379
Query: 448 YMDHRVFLPADHTWRTTENLFNGKMEFRTASPLLKGTEILEILKNFNNVFGKTK 501
+M HR L H+++ + F+GK+E + +L G E+ LKNF N FG K
Sbjct: 380 FMCHRKGLAPTHSYKNKKAWFDGKVEHGRKARILTGREVSHNLKNFKNDFGNAK 433
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 311 bits (797), Expect = 1e-84
Identities = 171/453 (37%), Positives = 249/453 (54%), Gaps = 49/453 (10%)
Query: 12 SSEYINGVESFLDFAYTKGRPQGD--EILCPCAKCGNCCWEKRNV----IYNHLICTGFL 65
S+EY+ GVE F+ FA ++ Q + CPC+ C N EK + + +HL GF+
Sbjct: 30 SAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKN---EKHIISGRRVSSHLFSHGFM 86
Query: 66 EGYNVWIHHGEQM--------------MEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEG 111
Y VW HGE+M E EE+ D ++ + F + G +
Sbjct: 87 HDYYVWYKHGEEMNMDLGTSYTNRTYFSENHEEVGNIVEDPYVDMVNDAFNFNVGYDDNY 146
Query: 112 PNEDA------------RKFYNLIDESQKELYP*CKNF-STLSFIIRMYLLKCTHGWSNV 158
++D+ KFY+L++ + LY C+ S LS R+ K + +
Sbjct: 147 RHDDSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCRERQSQLSLASRLMHNKAEYNMNEK 206
Query: 159 SFTALLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSC 217
++ E + +P+ N S +T+ ++R+LGL Y I C N+CML+WKE E ++ C
Sbjct: 207 FVDSVCETFTDFLPEENQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLFWKEEEKEDQC 266
Query: 218 RTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEE 277
R C A RWK + KVP + + P+ RL+R++ +T+ +MRWH E
Sbjct: 267 RFCGAKRWKP--------KDDRRRTKVPYSCMWYLPIGDRLKRMYQSHKTAAAMRWHAEH 318
Query: 278 RSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLV 337
+SK+G++ HP D W+ F GLH FA DPRNV LGL +DGFNPF MS +HS WPV+L
Sbjct: 319 QSKEGEMNHPSDAAEWRYFQGLHPQFAEDPRNVYLGLCTDGFNPFG-MSRNHSLWPVILT 377
Query: 338 VYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKET 397
YN PP +CM +EY+ +++L G P ++DV+LQPLIEELKELW GV+ YD S +
Sbjct: 378 PYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQN 437
Query: 398 FQMHAALLWTINDYPAYAMLSGWSTKGKLACAC 430
F + A LLWTI+D+PAY+MLSGW+T+ C C
Sbjct: 438 FNLKAVLLWTISDFPAYSMLSGWTTQ---FCKC 467
>At1g35280 hypothetical protein
Length = 557
Score = 309 bits (792), Expect = 5e-84
Identities = 153/323 (47%), Positives = 203/323 (62%), Gaps = 6/323 (1%)
Query: 179 SFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKS 238
S + K ++ + Y++IHAC NDC L+ K Y+N +C C ASRWK Q GE +K
Sbjct: 11 SLYEVKKFLKSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRWKSNMQT-GEAKKG 69
Query: 239 ESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDG 298
VP KVLR+FP+IPR +R+F + +RWH +S DGKLRHPVD W +
Sbjct: 70 -----VPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSIDGKLRHPVDSVTWDKMND 124
Query: 299 LHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLI 358
+ +FA++ RN+RLGL++DGFNPF + +S WPVLLV YN P LCMK E +M+SLLI
Sbjct: 125 KYPEFAAEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPLDLCMKKENIMLSLLI 184
Query: 359 PGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLS 418
PG Q PG +IDVYL+PLIE+L LW+ G TYDA K TF + A LLWTI+D+ AY L+
Sbjct: 185 PGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFSAYGNLA 244
Query: 419 GWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGKMEFRTAS 478
G KGK+ C C T S +LK+S K +M HR L H++R + F+GK+E +
Sbjct: 245 GCKVKGKMGCPLCGKGTDSMWLKYSRKHVFMCHRKGLAPTHSYRNKKAWFDGKIEHGRKA 304
Query: 479 PLLKGTEILEILKNFNNVFGKTK 501
+L G E+ + LKNF N F K
Sbjct: 305 RILTGREVSQTLKNFKNDFRNAK 327
>At2g13010 pseudogene
Length = 1040
Score = 305 bits (781), Expect = 9e-83
Identities = 169/486 (34%), Positives = 256/486 (51%), Gaps = 55/486 (11%)
Query: 37 ILCPCAKCGNCCWEKRNVIYNHLICTGFLEGYNVWIHHGE-------------------- 76
ILCPC KC N + + NV+ HL GF+ Y VW+ HG+
Sbjct: 211 ILCPCRKCLNEKYLEFNVVKKHLYNRGFMPNYYVWLRHGDVVEIAHELVIQIMVSSNLVI 270
Query: 77 ----QMMEIDEEM------EEDYHDDIDGLLYETFKYVAGAEPEGPNEDARKFYNLIDES 126
++M++ ++ E +HD + +ET E PN DA+ FY+++D +
Sbjct: 271 LIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHETIASFPENISEEPNVDAQHFYDMLDAA 330
Query: 127 QKELYP*CKNF-STLSFIIRMYLLKCTHGWSNVSFTALLELLKEAMPDLNIP-ESFNKTK 184
+ +Y C+ S LS RM +K + S + EL+KE +P NI ES+ + +
Sbjct: 331 NQPIYEGCREGNSKLSLASRMMTIKADNNLSEKCMDSWAELIKEYLPPDNISAESYYEIQ 390
Query: 185 GMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKV 244
++ GL + I C + CM++W + N CR C R ++ + +V
Sbjct: 391 KLVSSHGLPSEMIDVCIDYCMIFWGDDVNLQECRFCGKPR----------YQTTGGRTRV 440
Query: 245 PAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFA 304
P K + + P+ RL+RL+ RT+ MRWH + + DG++ HP D +AW+ F ++ DFA
Sbjct: 441 PYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEITHPSDAKAWKHFQTVYPDFA 500
Query: 305 SDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSP 364
++ RNV LGL +DGF+PF +S WPV+L Y PP +CM+SE+M +S+L+PG + P
Sbjct: 501 NECRNVYLGLCTDGFSPFGLCGRQYSLWPVILTPYKLPPDMCMQSEFMFLSILVPGPKHP 560
Query: 365 GKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKG 424
+ +DV+LQPLI ELK+LW GV T+D S K+ F M A L+WTI+D+PAY MLSGW+ G
Sbjct: 561 KRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAMLMWTISDFPAYRMLSGWTMHG 620
Query: 425 KLACACCNYNTSSTYLKHSHKMCYMD-HRVFLPADHTWRTTENLFNGKMEFRTASP-LLK 482
+L K C+ + HR FLP H +R + LF R P +
Sbjct: 621 RLYVP-----------TKDRKPCWFNCHRRFLPLHHPYRRNKKLFRKNKIVRLLPPSYVS 669
Query: 483 GTEILE 488
GT++ E
Sbjct: 670 GTDLFE 675
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 288 bits (738), Expect = 9e-78
Identities = 160/418 (38%), Positives = 231/418 (54%), Gaps = 30/418 (7%)
Query: 1 MDKEWTKQPRFSSEYINGVESFLDFAYTKGRPQGD--EILCPCAKCGNCCWEKRNVIYNH 58
MDK W PR S EY G F+ Y R GD E+ CPC C N C + + + H
Sbjct: 11 MDKSWVWLPRNSIEYEKGATKFV---YASSRRLGDPSEMFCPCTDCRNLCHQPIDTVLEH 67
Query: 59 LICTGFLEGYN---VWIHHGE---QMMEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEGP 112
L+ G Y W HGE +E + E ++ I ++ + ++ +
Sbjct: 68 LVIKGMNHKYKRNGCWSKHGEIRADKLEAESSSEFGAYELIRTAYFDGEDH---SDSQNQ 124
Query: 113 NEDARK---------FYNLIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTAL 163
NE+ K F + +++ LY C ++ +S I+R+Y +K G S F L
Sbjct: 125 NENDSKEPSTKEESDFREKLKDAETPLYYGCPKYTKVSAIMRLYRIKVKSGMSENYFDQL 184
Query: 164 LELLKEAMPDLNIP-ESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKA 222
L L+ + +P N+ S N+ K ++ G Y IHACPNDC+LY KEYE ++C C A
Sbjct: 185 LSLVHDILPGENVLLTSTNEIKKFLKMFGFGYDIIHACPNDCILYRKEYELRDTCPRCSA 244
Query: 223 SRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDG 282
RW E + GE +K +PAKVLR+FP+ R +R+F + ++ +RWH S DG
Sbjct: 245 FRW-ERDKHTGEEKKG-----IPAKVLRYFPIKDRFKRMFRSKKMAEDLRWHFSNASDDG 298
Query: 283 KLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYP 342
+RHPVD AW + FA++PRN+RLGL++DG NPF + +STWPVLLV YN
Sbjct: 299 TMRHPVDSLAWAQVNYKWPQFAAEPRNLRLGLSTDGMNPFSIQNTKYSTWPVLLVNYNMA 358
Query: 343 PWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQM 400
P +CMK+E +M++LLIPG +P NIDVYL PLI++LK+LW G++ YD+ KE+F +
Sbjct: 359 PTMCMKAENIMLTLLIPGPTTPNNNIDVYLAPLIDDLKDLWSDGIQVYDSFLKESFTL 416
>At4g08000 putative transposon protein
Length = 609
Score = 284 bits (726), Expect = 2e-76
Identities = 139/306 (45%), Positives = 192/306 (62%), Gaps = 11/306 (3%)
Query: 165 ELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKAS 223
+LL+E +P+ N S +T+ ++R+LGL Y I C N+CML+WKE E ++ CR C A
Sbjct: 73 DLLEEFLPEGNQATSSHYQTEKLMRNLGLPYHTIDVCSNNCMLFWKEDEQEDHCRFCGAQ 132
Query: 224 RWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGK 283
RWK + KVP + + P+ RL+R++ +T+ +MRWH E +SK+G+
Sbjct: 133 RWKP--------KDDRRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGE 184
Query: 284 LRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPP 343
+ HP D W+ F LH FA +PRNV LGL +DGFNPF MS +HS WPV+L YN PP
Sbjct: 185 MNHPSDAAEWRYFQELHPRFAEEPRNVYLGLCTDGFNPFG-MSRNHSLWPVILTPYNLPP 243
Query: 344 WLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAA 403
+CM ++Y+ +++L P ++DV+LQPLIEELKELW GV+ YD S + F + A
Sbjct: 244 SMCMNTKYLFLTILNVEPNHPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQNFNLKAV 303
Query: 404 LLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD-HRVFLPADHTWR 462
LLWTI+D+PAY+MLSGW T GK +C C +T S YL + K C+ D HR FLP H R
Sbjct: 304 LLWTISDFPAYSMLSGWITHGKFSCPICMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSR 363
Query: 463 TTENLF 468
+ F
Sbjct: 364 RNKKDF 369
>At1g36470 hypothetical protein
Length = 753
Score = 281 bits (719), Expect = 1e-75
Identities = 137/308 (44%), Positives = 191/308 (61%), Gaps = 11/308 (3%)
Query: 163 LLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCK 221
+++ ++ +P+ N S +T+ ++R+LGL Y I C N+CML WKE E ++ CR C
Sbjct: 49 IVQSSRDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLSWKEDEKEDQCRFCG 108
Query: 222 ASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKD 281
A RWK + KVP + + P+ RL+R++ +T+ +MRWH E +SK+
Sbjct: 109 AKRWKP--------KDDRRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKE 160
Query: 282 GKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNY 341
G++ H D W+ F GLH +FA +PRNV LGL +DGFNPF MS +HS WPV+L YN
Sbjct: 161 GEMNHLSDAAEWRYFQGLHPEFAEEPRNVYLGLCTDGFNPFG-MSRNHSLWPVILTPYNL 219
Query: 342 PPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMH 401
PP +CM +EY+ +++L G P ++DV+LQPLIEELKE W GV+ YD S + F +
Sbjct: 220 PPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKEFWSTGVDAYDVSLSQNFNLK 279
Query: 402 AALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD-HRVFLPADHT 460
A LLWTI+D+PAY ML GW+T GKL+C C +T S YL + K C+ D HR FLP H
Sbjct: 280 AVLLWTISDFPAYNMLLGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHP 339
Query: 461 WRTTENLF 468
R F
Sbjct: 340 SRRNRKDF 347
>At3g30790 hypothetical protein
Length = 953
Score = 259 bits (663), Expect = 4e-69
Identities = 154/467 (32%), Positives = 239/467 (50%), Gaps = 57/467 (12%)
Query: 44 CGNCCWEKRNVIYNHLICTGF------------LEGY----NVWIHHGE--QMMEIDEEM 85
C W+ + + IC GF L G +W E ++ D ++
Sbjct: 5 CEESAWKASFLANSSQICEGFTANQPPSQIATSLRGMIILSQLWPRSSESWRVRSNDRKV 64
Query: 86 EEDYHDDIDGLLYETFKYVAGAEPEGPNEDARKFYNLIDESQKELYP*CKNFSTLSF-II 144
+Y+D + G +Y+ EP AR+ + +K C N L F ++
Sbjct: 65 TREYYDGLRGFMYQAIN-----EPF-----ARENGTIFCPCRK-----CLNEKYLEFNVV 109
Query: 145 RMYLLKCTHGWSNVSFTALLELLKEAMPDLNIP-ESFNKTKGMIRDLGLDYKRIHACPND 203
+ +L + E +P NI ES+ + + ++ LGL + I C +
Sbjct: 110 KKHLYN----------RGFMPNYYEYLPPDNISAESYFEIQKLVLSLGLPSEMIDVCIDY 159
Query: 204 CMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFM 263
CM++W + N CR C R ++ + +VP K + + P+ RL+RL+
Sbjct: 160 CMIFWGDDVNLQECRFCGKPR----------YQTTSGRTRVPYKRMWYLPITDRLKRLYQ 209
Query: 264 CSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFR 323
RT+ MRWH + + DG+++HP D +AW+ F ++ DFAS+ RNV LGL +DGF+PF
Sbjct: 210 SERTAAKMRWHAQHTTADGEIKHPSDTKAWKHFQTVYPDFASECRNVYLGLCTDGFSPFV 269
Query: 324 TMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELW 383
+S W ++L YN PP + M+SE++ +S+L+PG + P + +DV+LQPLI ELK+LW
Sbjct: 270 LSGRQYSLWHIILTPYNLPPDMYMQSEFLFLSILVPGPKHPKRALDVFLQPLIHELKKLW 329
Query: 384 EVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHS 443
GV T+D S K+ F M A L+WTI+D+PAY MLSGW+T G+L+C C T + LK
Sbjct: 330 YEGVHTFDYSSKQNFNMRAVLMWTISDFPAYGMLSGWTTHGRLSCPYCMGRTDAFQLKKG 389
Query: 444 HKMCYMD-HRVFLPADHTWRTTENLFNGKMEFRTASP-LLKGTEILE 488
K C+ D HR FLP H +R + LF R P + GT++ +
Sbjct: 390 RKPCWFDYHRRFLPLHHPYRRNKKLFRKNKIVRLPPPSYVSGTDLFK 436
Score = 36.2 bits (82), Expect = 0.10
Identities = 21/61 (34%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Query: 10 RFSSEYINGVESFLDFAYTK--GRPQGDEILCPCAKCGNCCWEKRNVIYNHLICTGFLEG 67
+ + EY +G+ F+ A + R G I CPC KC N + + NV+ HL GF+
Sbjct: 63 KVTREYYDGLRGFMYQAINEPFARENGT-IFCPCRKCLNEKYLEFNVVKKHLYNRGFMPN 121
Query: 68 Y 68
Y
Sbjct: 122 Y 122
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 253 bits (646), Expect = 4e-67
Identities = 148/439 (33%), Positives = 229/439 (51%), Gaps = 30/439 (6%)
Query: 71 WIHHGEQMMEIDEEMEEDYHDDIDGLLYETFKYVAGAEP--EGPNEDARKFYNLIDESQK 128
W HGE + ++E + + Y G E E ++ F + + +
Sbjct: 11 WSKHGEIWAD---KLEAEPSSEFGAYELIRTAYFDGEEDSKEPVTKEESYFREKLKDVET 67
Query: 129 ELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTALLELLKEAMPDLNI-PESFNKTKGMI 187
LY C ++ +S I+ +Y +K G S F LL L+ + +P N+ P S N+ K +
Sbjct: 68 PLYYGCPKYTKVSAIMGLYRIKVKSGMSENYFDQLLSLVHDILPGENVLPTSTNEIKKFL 127
Query: 188 RDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAK 247
+ G Y IHACPNDC+LY KEYE ++C C ASRWK + E + +PAK
Sbjct: 128 KMFGFGYDIIHACPNDCILYKKEYELRDTCLGCSASRWKR------DKHTGEEKKGIPAK 181
Query: 248 VLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDP 307
VL +F + R +R+F + + ++WH S DG +RHP+D AW + FA++
Sbjct: 182 VLWYFSIKDRFKRMFRSKKMVEDLQWHFSNASVDGTMRHPIDSLAWAQVNDKWPQFAAES 241
Query: 308 RNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKN 367
RN+RLGL++D NPF + +STWPVLLV YN P +CMK++ +M++LLI G ++P N
Sbjct: 242 RNLRLGLSTDKMNPFSIQNTKYSTWPVLLVNYNMAPTMCMKAKNIMLTLLIHGPKAPSNN 301
Query: 368 IDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLA 427
IDVYL PLI +LK+L G++ YD+ KE F + A LLW+INDYP L+G
Sbjct: 302 IDVYLAPLINDLKDLSSDGIQVYDSFFKENFTLRAMLLWSINDYPTLGTLAG-------- 353
Query: 428 CACCNY-NTSSTYLKHSHK-----MCYMDHRVFLPADHTWRTTENLFNGKMEFRTASPLL 481
C ++ N L+ ++K +C D + H WR + ++ + P+
Sbjct: 354 --CKDFKNDFGRPLERNNKRKRSDLCEDDE--YGEDTHQWRWKKRSILFELPYWKDLPVR 409
Query: 482 KGTEILEILKNFNNVFGKT 500
+++ + KN ++V T
Sbjct: 410 HNIDVMHVEKNVSDVIVST 428
>At2g10140 putative protein
Length = 663
Score = 223 bits (568), Expect = 4e-58
Identities = 104/204 (50%), Positives = 138/204 (66%), Gaps = 2/204 (0%)
Query: 266 RTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTM 325
+T+ +MRWH E +SK+G++ HP D W+ F LH FA +PRNV LGL DGFNPF +
Sbjct: 244 KTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHTWFAKEPRNVYLGLCIDGFNPFG-I 302
Query: 326 SISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEV 385
S +HS WPV+L YN PP +CM ++Y+ +++L G P ++DV+LQPLIEELKELW
Sbjct: 303 SRNHSLWPVILTSYNLPPGMCMNTKYLFLTILNSGPNHPRASLDVFLQPLIEELKELWCT 362
Query: 386 GVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHK 445
GV+TYD S + F + A LLW I+D+PAY+MLSGW+T GKL+C C +T S YL + K
Sbjct: 363 GVDTYDVSLSQNFNLKAVLLWAISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRK 422
Query: 446 MCYMD-HRVFLPADHTWRTTENLF 468
C+ D HR FLP H R + F
Sbjct: 423 TCWFDCHRRFLPHSHPSRRNKKDF 446
>At4g08070
Length = 767
Score = 209 bits (532), Expect = 7e-54
Identities = 130/374 (34%), Positives = 187/374 (49%), Gaps = 30/374 (8%)
Query: 8 QPRFSSEYINGVESFL-DFAYTKGRPQGDEILCPCAKCGNCCWEKRNVIYNHLICTGFLE 66
Q R Y G F+ D A G D I+CPC C N V+ +HL+ G E
Sbjct: 25 QQRADPTYERGAIKFVRDVAAALG---DDMIVCPCIDCRNIDRHSGCVVVDHLVTRGMDE 81
Query: 67 GYNV---WIHHGEQMMEIDEEMEEDYHDDIDGLLYETFKYV---------AGAEPEGPNE 114
Y W HHGE + + E + +D LY+ +Y+ G E ++
Sbjct: 82 AYKRRCDWYHHGELVSGGESESKVSQWNDEVVELYQAAEYLDEEFDAMVQLGEIAERDDK 141
Query: 115 DARKFYNLIDESQKELYP*CKNFSTLSFIIRMYLLKCTHGWSNVSFTALLELLKEAMPDL 174
+F + +++ LYP C N S LS I+ ++ LK +GWS+ SF LLE L E +P+
Sbjct: 142 KEDEFLAKLADAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLETLSEMLPED 201
Query: 175 NIPE-SFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEYENDNSCRTCKASRWKEFPQVAG 233
N+ S + K ++ + Y++IHAC NDC L+ K+Y+ +C C ASRWK G
Sbjct: 202 NVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFRKKYKKLQNCPKCNASRWKTNMHT-G 260
Query: 234 EFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAW 293
E +K VP KVLR+FP+IPRL+R+F + +RWH +S DGKLRHPVD W
Sbjct: 261 EVKKG-----VPQKVLRYFPIIPRLKRMFRSEEMARDLRWHFHNKSTDGKLRHPVDSVTW 315
Query: 294 QDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTW-------PVLLVVYNYPPWLC 346
+ + FAS+ RN+RLGL++DGFNPF + + P N W
Sbjct: 316 DQMNAKYPLFASEERNLRLGLSTDGFNPFNMKNTRKHVYMGHRMGLPPSHRFRNKKSWFD 375
Query: 347 MKSEYMMMSLLIPG 360
K+E+ S ++ G
Sbjct: 376 GKAEHRRKSRILSG 389
Score = 56.6 bits (135), Expect = 7e-08
Identities = 28/62 (45%), Positives = 37/62 (59%)
Query: 440 LKHSHKMCYMDHRVFLPADHTWRTTENLFNGKMEFRTASPLLKGTEILEILKNFNNVFGK 499
+K++ K YM HR+ LP H +R ++ F+GK E R S +L G EI LKNF N FG
Sbjct: 346 MKNTRKHVYMGHRMGLPPSHRFRNKKSWFDGKAEHRRKSRILSGLEISHNLKNFQNNFGN 405
Query: 500 TK 501
K
Sbjct: 406 FK 407
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.153 0.544
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,229,573
Number of Sequences: 26719
Number of extensions: 917932
Number of successful extensions: 3958
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 3832
Number of HSP's gapped (non-prelim): 49
length of query: 1065
length of database: 11,318,596
effective HSP length: 110
effective length of query: 955
effective length of database: 8,379,506
effective search space: 8002428230
effective search space used: 8002428230
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0250.10


