

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0248.4
(573 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
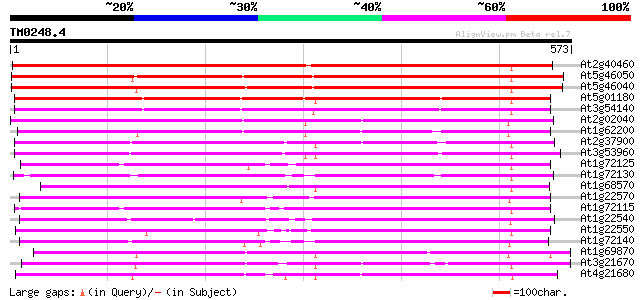
At2g40460 putative PTR2 family peptide transporter 668 0.0
At5g46050 peptide transporter 464 e-131
At5g46040 peptide transporter 451 e-127
At5g01180 oligopeptide transporter - like protein 433 e-121
At3g54140 peptide transport - like protein 421 e-118
At2g02040 histidine transport protein (PTR2-B) 402 e-112
At1g62200 unknown protein (At1g62200) 395 e-110
At2g37900 putative peptide/amino acid transporter 387 e-107
At3g53960 transporter like protein 383 e-106
At1g72125 oligopeptide like transporter 357 8e-99
At1g72130 peptide transporter PTR2-B -like protein 354 8e-98
At1g68570 peptide transporter like 353 1e-97
At1g22570 Similar to LeOPT1 [Lycopersicon esculentum 352 2e-97
At1g72115 oligopeptide like transporter 351 5e-97
At1g22540 Similar to peptide transporter 348 6e-96
At1g22550 Similar to LeOPT1 [Lycopersicon esculentum 341 6e-94
At1g72140 peptide transporter like protein (PTR2-B) 336 2e-92
At1g69870 unknown protein 323 2e-88
At3g21670 nitrate transporter 317 1e-86
At4g21680 peptide transporter - like protein 312 4e-85
>At2g40460 putative PTR2 family peptide transporter
Length = 583
Score = 668 bits (1723), Expect = 0.0
Identities = 313/558 (56%), Positives = 418/558 (74%), Gaps = 11/558 (1%)
Query: 4 KGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDV 63
K YT DGTVDL GRPVL+S TG+ +AC+++L Y ER A+YG+ +NLVN++T +L++D
Sbjct: 5 KVYTQDGTVDLQGRPVLASKTGRWRACSFLLGYEAFERMAFYGIASNLVNYLTKRLHEDT 64
Query: 64 VSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRP 123
+SS+ + NNWSG +TPI GAYIAD+Y GRFWT T S LIY +G++LL + T+KSLRP
Sbjct: 65 ISSVRNVNNWSGAVWITPIAGAYIADSYIGRFWTFTASSLIYVLGMILLTMAVTVKSLRP 124
Query: 124 ACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFN 183
CENG+C +AS+LQV FY SLYTIA+G+G KPN+STFGADQFD + EEK+QKVSFFN
Sbjct: 125 TCENGVCNKASSLQVTFFYISLYTIAIGAGGTKPNISTFGADQFDSYSIEEKKQKVSFFN 184
Query: 184 WWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARS 243
WW F+ G+L ATL +VYIQE GWGL Y + +G L+S ++F+ G+P YRHK + +
Sbjct: 185 WWMFSSFLGALFATLGLVYIQENLGWGLGYGIPTVGLLVSLVVFYIGTPFYRHKVIKTDN 244
Query: 244 PSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRES 303
+ + ++VP+ AF+NRKLQ P + EL+E +YY S+G ++HHT F FLD+AAI+ S
Sbjct: 245 LAKDLVQVPIAAFKNRKLQCPDDHLELYELDSHYYKSNGKHQVHHTPVFRFLDKAAIKTS 304
Query: 304 NTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFR 363
S PCTVT+VE K VLG+ IWL+ LIP+ WA +T+FV+QGTT+DR +G F+
Sbjct: 305 ----SRVPCTVTKVEVAKRVLGLIFIWLVTLIPSTLWAQVNTLFVKQGTTLDRKIGSNFQ 360
Query: 364 LPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTY 423
+PAASL F+ L+ L+ +P+YD F+PFMR++TGN RGI LLQR+G+G AIQ++A+A+
Sbjct: 361 IPAASLGSFVTLSMLLSVPMYDQSFVPFMRKKTGNPRGITLLQRLGVGFAIQIVAIAIAS 420
Query: 424 AVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMK 483
AVE +RM VIK+ HI P + VPMSIFWLLPQ +LG+ + F A G+LEFFYDQSPEEM+
Sbjct: 421 AVEVKRMRVIKEFHITSPTQVVPMSIFWLLPQYSLLGIGDVFNAIGLLEFFYDQSPEEMQ 480
Query: 484 GLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFVISAL 536
LGTT TS + G+++N+FLVTMIDK+ +WIGNNLNDS LDYYY FL VIS +
Sbjct: 481 SLGTTFFTSGIGLGNFLNSFLVTMIDKITSKGGGKSWIGNNLNDSRLDYYYGFLVVISIV 540
Query: 537 NFGVFLWVSSGYIYKKEN 554
N G+F+W +S Y+YK ++
Sbjct: 541 NMGLFVWAASKYVYKSDD 558
>At5g46050 peptide transporter
Length = 582
Score = 464 bits (1193), Expect = e-131
Identities = 243/576 (42%), Positives = 353/576 (61%), Gaps = 17/576 (2%)
Query: 3 GKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKD 62
G YT DGTVDL G PV S+ G+ KAC++++VY V ER AYYG+ +NL +MTT+L++
Sbjct: 7 GDDYTKDGTVDLQGNPVRRSIRGRWKACSFVVVYEVFERMAYYGISSNLFIYMTTKLHQG 66
Query: 63 VVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLR 122
V S + NW G + LTPILGAY+ D GR+ T S IY G+++L L+ T+ ++
Sbjct: 67 TVKSSNNVTNWVGTSWLTPILGAYVGDALLGRYITFVISCAIYFSGMMVLTLSVTIPGIK 126
Query: 123 PA------CENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKE 176
P EN C +AS LQ+A+F+ +LYT+A+G+G KPN+ST GADQFD F +EK
Sbjct: 127 PPECSTTNVEN--CEKASVLQLAVFFGALYTLAIGTGGTKPNISTIGADQFDVFDPKEKT 184
Query: 177 QKVSFFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRH 236
QK+SFFNWW F+ G+L A +VY+Q+ GW L Y L +G +S IF G+P YRH
Sbjct: 185 QKLSFFNWWMFSIFFGTLFANTVLVYVQDNVGWTLGYGLPTLGLAISITIFLLGTPFYRH 244
Query: 237 KSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLD 296
K SP RV + +FR + + + HE Y GA IH T FLD
Sbjct: 245 K-LPTGSPFTKMARVIVASFRKANAPMTHDITSFHELPSLEYERKGAFPIHPTPSLRFLD 303
Query: 297 RAAIRESNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDR 356
RA+++ N CT T+VE TK +L M + + +P+ A +T+FV+QGTT+DR
Sbjct: 304 RASLKTGTNHKWN-LCTTTEVEETKQMLRMLPVLFITFVPSMMLAQINTLFVKQGTTLDR 362
Query: 357 TLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQV 416
+ F +P ASL F+ L+ LI + +YD F+ R+ TGN RGI LLQR+GIG+ +
Sbjct: 363 KVTGSFSIPPASLSGFVTLSMLISIVLYDRVFVKITRKFTGNPRGITLLQRMGIGLIFHI 422
Query: 417 IAMAVTYAVETQRMSVIKKHHIVGPEET-VPMSIFWLLPQNIILGVSNAFLATGMLEFFY 475
+ M V E R+ V H ++ +P++IF LLPQ +++G++++FL LEFFY
Sbjct: 423 LIMIVASVTERYRLKVAADHGLIHQTGVKLPLTIFALLPQFVLMGMADSFLEVAKLEFFY 482
Query: 476 DQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL------NWIGNNLNDSHLDYYYAF 529
DQ+PE MK LGT+ T+ +A G+++++FL++ + ++ WI NNLN+S LDYYY F
Sbjct: 483 DQAPESMKSLGTSYSTTSLAIGNFMSSFLLSTVSEITKKRGRGWILNNLNESRLDYYYLF 542
Query: 530 LFVISALNFGVFLWVSSGYIYKKENTSTTEVHDIEM 565
V++ +NF +FL V Y+Y+ E T + +V ++EM
Sbjct: 543 FAVLNLVNFVLFLVVVKFYVYRAEVTDSVDVKEVEM 578
>At5g46040 peptide transporter
Length = 586
Score = 451 bits (1161), Expect = e-127
Identities = 238/573 (41%), Positives = 348/573 (60%), Gaps = 13/573 (2%)
Query: 3 GKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKD 62
G YT DGTVDL G V S TG+ KAC++++VY V ER AYYG+ +NLV +MTT+L++
Sbjct: 7 GDDYTKDGTVDLRGNRVRRSQTGRWKACSFVVVYEVFERMAYYGISSNLVIYMTTKLHQG 66
Query: 63 VVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLR 122
V S + NW G + LTPILGAY+AD + GR+ T S IY +G+ LL L+ +L L+
Sbjct: 67 TVKSSNNVTNWVGTSWLTPILGAYVADAHFGRYITFVISSAIYLLGMALLTLSVSLPGLK 126
Query: 123 -PACENG---ICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQK 178
P C C +AS +Q+A+F+ +LYT+A+G+G KPN+ST GADQFD+F ++K K
Sbjct: 127 PPKCSTANVENCEKASVIQLAVFFGALYTLAIGTGGTKPNISTIGADQFDEFDPKDKIHK 186
Query: 179 VSFFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKS 238
SFFNWW F+ G+ AT +VY+Q+ GW + Y LS +G S IF G+ +YRHK
Sbjct: 187 HSFFNWWMFSIFFGTFFATTVLVYVQDNVGWAIGYGLSTLGLAFSIFIFLLGTRLYRHKL 246
Query: 239 RQARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRA 298
SP RV + + R + + + + +E Y S A IH TS FL+RA
Sbjct: 247 PMG-SPFTKMARVIVASLRKAREPMSSDSTRFYELPPMEYASKRAFPIHSTSSLRFLNRA 305
Query: 299 AIRESNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL 358
+++ +T CT+T+VE TK +L M + + +P+ A T+F++QGTT+DR L
Sbjct: 306 SLKTGSTHKWR-LCTITEVEETKQMLKMLPVLFVTFVPSMMLAQIMTLFIKQGTTLDRRL 364
Query: 359 GPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIA 418
F +P ASL F + L+ + IYD F+ FMR+ TGN RGI LLQR+GIGM + ++
Sbjct: 365 TNNFSIPPASLLGFTTFSMLVSIVIYDRVFVKFMRKLTGNPRGITLLQRMGIGMILHILI 424
Query: 419 MAVTYAVETQRMSVIKKHHIVGPEET-VPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQ 477
M + E R+ V +H + +P+SIF LLPQ +++G+++AF+ LEFFYDQ
Sbjct: 425 MIIASITERYRLKVAAEHGLTHQTAVPIPLSIFTLLPQYVLMGLADAFIEIAKLEFFYDQ 484
Query: 478 SPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL------NWIGNNLNDSHLDYYYAFLF 531
+PE MK LGT+ ++ +A G ++++ L++ + ++ WI NNLN+S LD YY F
Sbjct: 485 APESMKSLGTSYTSTSMAVGYFMSSILLSSVSQITKKQGRGWIQNNLNESRLDNYYMFFA 544
Query: 532 VISALNFGVFLWVSSGYIYKKENTSTTEVHDIE 564
V++ LNF +FL V Y Y+ + T + V E
Sbjct: 545 VLNLLNFILFLVVIRFYEYRADVTQSANVEQKE 577
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 433 bits (1114), Expect = e-121
Identities = 220/561 (39%), Positives = 339/561 (60%), Gaps = 18/561 (3%)
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
YT DGT+D+ +P + TG KAC +IL ER AYYG+ NL+N++ Q+N + VS
Sbjct: 8 YTKDGTLDIHKKPANKNKTGTWKACRFILGTECCERLAYYGMSTNLINYLEKQMNMENVS 67
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPAC 125
+ S +NWSG TP++GA+IAD Y GR+WTI ++IY G+ LL ++ ++ L P C
Sbjct: 68 ASKSVSNWSGTCYATPLIGAFIADAYLGRYWTIASFVVIYIAGMTLLTISASVPGLTPTC 127
Query: 126 ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWW 185
C + Q A+ + +LY IA+G+G +KP +S+FGADQFDD +EKE K SFFNW+
Sbjct: 128 SGETCHATAG-QTAITFIALYLIALGTGGIKPCVSSFGADQFDDTDEKEKESKSSFFNWF 186
Query: 186 AFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPS 245
F G+++A+ +V+IQ GWG + + ++ + FF GS YR + SP
Sbjct: 187 YFVINVGAMIASSVLVWIQMNVGWGWGLGVPTVAMAIAVVFFFAGSNFYR-LQKPGGSPL 245
Query: 246 MNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNT 305
++V + + R K+++P + S L+E Q G+RK+ HT +F D+AA+ E+ +
Sbjct: 246 TRMLQVIVASCRKSKVKIPEDESLLYENQDAESSIIGSRKLEHTKILTFFDKAAV-ETES 304
Query: 306 DLSNPP-------CTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL 358
D CTVTQVE K ++ + IW ++ + ++ T+FV QG T+D+ +
Sbjct: 305 DNKGAAKSSSWKLCTVTQVEELKALIRLLPIWATGIVFASVYSQMGTVFVLQGNTLDQHM 364
Query: 359 GPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIA 418
GP F++P+ASL F L+ L P+YD +PF R+ TG+ RG LQR+GIG+ I + +
Sbjct: 365 GPNFKIPSASLSLFDTLSVLFWAPVYDKLIVPFARKYTGHERGFTQLQRIGIGLVISIFS 424
Query: 419 MAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQS 478
M +E R++ ++ H++ EET+PM+IFW +PQ ++G + F G LEFFYDQ+
Sbjct: 425 MVSAGILEVARLNYVQTHNLYN-EETIPMTIFWQVPQYFLVGCAEVFTFIGQLEFFYDQA 483
Query: 479 PEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLF 531
P+ M+ L + L + +A G+Y++TFLVT++ K+ WI NLN+ HLDY++ L
Sbjct: 484 PDAMRSLCSALSLTAIAFGNYLSTFLVTLVTKVTRSGGRPGWIAKNLNNGHLDYFFWLLA 543
Query: 532 VISALNFGVFLWVSSGYIYKK 552
+S LNF V+LW++ Y YKK
Sbjct: 544 GLSFLNFLVYLWIAKWYTYKK 564
>At3g54140 peptide transport - like protein
Length = 570
Score = 421 bits (1082), Expect = e-118
Identities = 220/561 (39%), Positives = 332/561 (58%), Gaps = 17/561 (3%)
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
YT DGTVD+ P TG KAC +IL ER AYYG+G NLVN++ ++LN+ +
Sbjct: 7 YTQDGTVDIHKNPANKEKTGNWKACRFILGNECCERLAYYGMGTNLVNYLESRLNQGNAT 66
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPA- 124
+ + NWSG +TP++GA+IAD Y GR+WTI + IY G+ LL L+ ++ L+P
Sbjct: 67 AANNVTNWSGTCYITPLIGAFIADAYLGRYWTIATFVFIYVSGMTLLTLSASVPGLKPGN 126
Query: 125 CENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNW 184
C C S+ Q A+F+ +LY IA+G+G +KP +S+FGADQFD+ EK +K SFFNW
Sbjct: 127 CNADTCHPNSS-QTAVFFVALYMIALGTGGIKPCVSSFGADQFDENDENEKIKKSSFFNW 185
Query: 185 WAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSP 244
+ F+ G+L+A +V+IQ GWG + + + +++ FF+GS YR R SP
Sbjct: 186 FYFSINVGALIAATVLVWIQMNVGWGWGFGVPTVAMVIAVCFFFFGSRFYR-LQRPGGSP 244
Query: 245 SMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESN 304
+V + AFR +++P + S L E + G+RK+ HT + F D+AA+ +
Sbjct: 245 LTRIFQVIVAAFRKISVKVPEDKSLLFETADDESNIKGSRKLVHTDNLKFFDKAAVESQS 304
Query: 305 TDLS----NP--PCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL 358
+ NP C+VTQVE K ++ + +W ++ ++ ST+FV QG TMD+ +
Sbjct: 305 DSIKDGEVNPWRLCSVTQVEELKSIITLLPVWATGIVFATVYSQMSTMFVLQGNTMDQHM 364
Query: 359 GPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIA 418
G F +P+ASL F ++ L P+YD + IP R+ T N RG LQR+GIG+ + + A
Sbjct: 365 GKNFEIPSASLSLFDTVSVLFWTPVYDQFIIPLARKFTRNERGFTQLQRMGIGLVVSIFA 424
Query: 419 MAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQS 478
M +E R+ +K H+ ++ + MSIFW +PQ +++G + F G LEFFYDQ+
Sbjct: 425 MITAGVLEVVRLDYVKTHNAY-DQKQIHMSIFWQIPQYLLIGCAEVFTFIGQLEFFYDQA 483
Query: 479 PEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLF 531
P+ M+ L + L + VA G+Y++T LVT++ K+ WI +NLN HLDY++ L
Sbjct: 484 PDAMRSLCSALSLTTVALGNYLSTVLVTVVMKITKKNGKPGWIPDNLNRGHLDYFFYLLA 543
Query: 532 VISALNFGVFLWVSSGYIYKK 552
+S LNF V+LW+S Y YKK
Sbjct: 544 TLSFLNFLVYLWISKRYKYKK 564
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 402 bits (1033), Expect = e-112
Identities = 217/568 (38%), Positives = 335/568 (58%), Gaps = 16/568 (2%)
Query: 2 EGKGYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNK 61
E K Y DG+VD G P L TG KAC +IL ER AYYG+ NL+ ++TT+L++
Sbjct: 20 EVKLYAEDGSVDFNGNPPLKEKTGNWKACPFILGNECCERLAYYGIAGNLITYLTTKLHQ 79
Query: 62 DVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSL 121
VS+ T+ W G LTP++GA +AD Y GR+WTI IY IG+ L L+ ++ +L
Sbjct: 80 GNVSAATNVTTWQGTCYLTPLIGAVLADAYWGRYWTIACFSGIYFIGMSALTLSASVPAL 139
Query: 122 RPA-CENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVS 180
+PA C C A+ Q A+F+ LY IA+G+G +KP +S+FGADQFDD E+ +K S
Sbjct: 140 KPAECIGDFCPSATPAQYAMFFGGLYLIALGTGGIKPCVSSFGADQFDDTDSRERVRKAS 199
Query: 181 FFNWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQ 240
FFNW+ F+ G+L+++ +V+IQE GWGL + + + L+ FF+G+P+YR + +
Sbjct: 200 FFNWFYFSINIGALVSSSLLVWIQENRGWGLGFGIPTVFMGLAIASFFFGTPLYRFQ-KP 258
Query: 241 ARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAI 300
SP +V + +FR +++P + + L+E Q +G+RKI HT +LD+AA+
Sbjct: 259 GGSPITRISQVVVASFRKSSVKVPEDATLLYETQDKNSAIAGSRKIEHTDDCQYLDKAAV 318
Query: 301 ----RESNTDLSNP--PCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTM 354
+ D SN CTVTQVE K+++ MF IW +I + +A ST+FV+QG M
Sbjct: 319 ISEEESKSGDYSNSWRLCTVTQVEELKILIRMFPIWASGIIFSAVYAQMSTMFVQQGRAM 378
Query: 355 DRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAI 414
+ +G F+LP A+L F + +I +P+YD + +P R+ TG +G +QR+GIG+ +
Sbjct: 379 NCKIG-SFQLPPAALGTFDTASVIIWVPLYDRFIVPLARKFTGVDKGFTEIQRMGIGLFV 437
Query: 415 QVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFF 474
V+ MA VE R+ + +V VP+S+ W +PQ ILG + F G LEFF
Sbjct: 438 SVLCMAAAAIVEIIRLHMANDLGLVESGAPVPISVLWQIPQYFILGAAEVFYFIGQLEFF 497
Query: 475 YDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMI-------DKLNWIGNNLNDSHLDYYY 527
YDQSP+ M+ L + L A G+Y+++ ++T++ + WI +NLN HLDY++
Sbjct: 498 YDQSPDAMRSLCSALALLTNALGNYLSSLILTLVTYFTTRNGQEGWISDNLNSGHLDYFF 557
Query: 528 AFLFVISALNFGVFLWVSSGYIYKKENT 555
L +S +N V+ + ++ Y KK ++
Sbjct: 558 WLLAGLSLVNMAVYFFSAARYKQKKASS 585
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 395 bits (1016), Expect = e-110
Identities = 214/561 (38%), Positives = 334/561 (59%), Gaps = 27/561 (4%)
Query: 9 DGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSIT 68
DG++D+ G P TG KAC +IL ER AYYG+ NL+ + T++L++ VS+ +
Sbjct: 38 DGSIDIYGNPPSKKKTGNWKACPFILGNECCERLAYYGIAKNLITYYTSELHESNVSAAS 97
Query: 69 SFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENG 128
W G +TP++GA IAD+Y GR+WTI IY IG+ LL L+ +L L+PA G
Sbjct: 98 DVMIWQGTCYITPLIGAVIADSYWGRYWTIASFSAIYFIGMALLTLSASLPVLKPAACAG 157
Query: 129 I----CREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNW 184
+ C A+ +Q A+F+T LY IA+G+G +KP +S+FGADQFDD E+ +K SFFNW
Sbjct: 158 VAAALCSPATTVQYAVFFTGLYLIALGTGGIKPCVSSFGADQFDDTDPRERVRKASFFNW 217
Query: 185 WAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSP 244
+ F+ GS +++ +V++QE GWGL + + + +S FF G+P+YR + + SP
Sbjct: 218 FYFSINIGSFISSTLLVWVQENVGWGLGFLIPTVFMGVSIASFFIGTPLYRFQ-KPGGSP 276
Query: 245 SMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAI---- 300
+V + A+R KL LP + S L+E + + +G+RKI HT + FLD+AA+
Sbjct: 277 ITRVCQVLVAAYRKLKLNLPEDISFLYETREKNSMIAGSRKIQHTDGYKFLDKAAVISEY 336
Query: 301 RESNTDLSNP--PCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL 358
+ SNP CTVTQVE K ++ MF IW ++ + ++ ST+FV+QG +M+R +
Sbjct: 337 ESKSGAFSNPWKLCTVTQVEEVKTLIRMFPIWASGIVYSVLYSQISTLFVQQGRSMNRII 396
Query: 359 GPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIA 418
F +P AS F L LI +PIYD + +PF+RR TG +G+ LQR+GIG+ + V++
Sbjct: 397 -RSFEIPPASFGVFDTLIVLISIPIYDRFLVPFVRRFTGIPKGLTDLQRMGIGLFLSVLS 455
Query: 419 MAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQS 478
+A VET R+ + ++ V MSIFW +PQ I++G++ F G +EFFYD+S
Sbjct: 456 IAAAAIVETVRLQL--------AQDFVAMSIFWQIPQYILMGIAEVFFFIGRVEFFYDES 507
Query: 479 PEEMKGLGTTLCTSCVAAGSYINTFLVTMI-------DKLNWIGNNLNDSHLDYYYAFLF 531
P+ M+ + + L A GSY+++ ++T++ K W+ ++LN HLDY++ L
Sbjct: 508 PDAMRSVCSALALLNTAVGSYLSSLILTLVAYFTALGGKDGWVPDDLNKGHLDYFFWLLV 567
Query: 532 VISALNFGVFLWVSSGYIYKK 552
+ +N V+ + + KK
Sbjct: 568 SLGLVNIPVYALICVKHTKKK 588
>At2g37900 putative peptide/amino acid transporter
Length = 575
Score = 387 bits (993), Expect = e-107
Identities = 212/564 (37%), Positives = 315/564 (55%), Gaps = 26/564 (4%)
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
+ LD ++D GR L + TG +A +I+ ER +Y+G+ NLV ++TT LN+D+
Sbjct: 21 WVLDSSLDSRGRVPLRARTGAWRAALFIIAIEFSERLSYFGLATNLVVYLTTILNQDLKM 80
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPAC 125
+I + N WSG+ TL P+LG +IAD Y GR+ T+ + IY +GLVLL ++ + L+P C
Sbjct: 81 AIRNVNYWSGVTTLMPLLGGFIADAYLGRYATVLVATTIYLMGLVLLTMSWFIPGLKP-C 139
Query: 126 ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWW 185
+C E F+ ++Y I++G+G KP++ +FGADQFDD EE++ K+SFFNWW
Sbjct: 140 HQEVCVEPRKAHEVAFFIAIYLISIGTGGHKPSLESFGADQFDDDHVEERKMKMSFFNWW 199
Query: 186 AFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPS 245
+ G L A V YI+++ GWG+A + + +S IIFF G P YR+++ + SP
Sbjct: 200 NVSLCAGILTAVTAVAYIEDRVGWGVAGIILTVVMAISLIIFFIGKPFYRYRT-PSGSPL 258
Query: 246 MNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNT 305
++V + A R L P +PS LHE + S R + HT H FLD+AAI E
Sbjct: 259 TPILQVFVAAIAKRNLPYPSDPSLLHEVSKTEFTS--GRLLCHTEHLKFLDKAAIIEDKN 316
Query: 306 DLSNPP------CTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLG 359
L+ T+T+VE TKL++ + IW L C ST F++Q TMDR +G
Sbjct: 317 PLALEKQSPWRLLTLTKVEETKLIINVIPIWFSTLAFGICATQASTFFIKQAITMDRHIG 376
Query: 360 PKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAM 419
F +P AS++ LT +I L +Y+ +P +R T N RGI +LQR+G GM +I M
Sbjct: 377 -GFTVPPASMFTLTALTLIISLTVYEKLLVPLLRSITRNQRGINILQRIGTGMIFSLITM 435
Query: 420 AVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSP 479
+ VE QR+ + PMS+ WL PQ +++G ++AF G+ E+FY Q P
Sbjct: 436 IIAALVEKQRLDRTNNNK--------PMSVIWLAPQFMVIGFADAFTLVGLQEYFYHQVP 487
Query: 480 EEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFV 532
+ M+ LG S + A S++N L+T +D L +W G +LN S LD +Y FL
Sbjct: 488 DSMRSLGIAFYLSVIGAASFLNNLLITAVDTLAENFSGKSWFGKDLNSSRLDRFYWFLAG 547
Query: 533 ISALNFGVFLWVSSGYIYKKENTS 556
+ A N VF+ V+ YK S
Sbjct: 548 VIAANICVFVIVAKRCPYKSVQPS 571
>At3g53960 transporter like protein
Length = 620
Score = 383 bits (984), Expect = e-106
Identities = 208/572 (36%), Positives = 319/572 (55%), Gaps = 22/572 (3%)
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
+ LD + D G L + TG +A +I+ ER +Y+G+ NLV ++TT L++D+
Sbjct: 20 WVLDSSTDSRGEIPLRAQTGAWRAALFIIGIEFSERLSYFGISTNLVVYLTTILHQDLKM 79
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPAC 125
++ + N WSG+ TL P+LG ++AD Y GR+ T+ + IY +GL+LL L+ + L+ AC
Sbjct: 80 AVKNTNYWSGVTTLMPLLGGFVADAYLGRYGTVLLATTIYLMGLILLTLSWFIPGLK-AC 138
Query: 126 ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWW 185
+C E F+ ++Y I++G+G KP++ +FGADQF+D EE++ K+S+FNWW
Sbjct: 139 HEDMCVEPRKAHEIAFFIAIYLISIGTGGHKPSLESFGADQFEDGHPEERKMKMSYFNWW 198
Query: 186 AFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPS 245
G L A +VYI+++ GWG+A + I S IF G P YR+++ + SP
Sbjct: 199 NAGLCAGILTAVTVIVYIEDRIGWGVASIILTIVMATSFFIFRIGKPFYRYRA-PSGSPL 257
Query: 246 MNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAI---RE 302
++V + A R L P + S LHE Y + R + + + FLD+AA+ R
Sbjct: 258 TPMLQVFVAAIAKRNLPCPSDSSLLHELTNEEY--TKGRLLSSSKNLKFLDKAAVIEDRN 315
Query: 303 SNTDLSNPP----CTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL 358
NT TVT+VE KL++ M IW L C ST+F++Q MDR +
Sbjct: 316 ENTKAEKQSPWRLATVTKVEEVKLLINMIPIWFFTLAFGVCATQSSTLFIKQAIIMDRHI 375
Query: 359 -GPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVI 417
G F +P ASL+ I L+ +I + IY+ +P +RR TGN RGI +LQR+G+GM +
Sbjct: 376 TGTSFIVPPASLFSLIALSIIITVTIYEKLLVPLLRRATGNERGISILQRIGVGMVFSLF 435
Query: 418 AMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQ 477
AM + +E +R+ K+HH+ +T+ +S WL PQ ++LGV++AF G+ E+FYDQ
Sbjct: 436 AMIIAALIEKKRLDYAKEHHM---NKTMTLSAIWLAPQFLVLGVADAFTLVGLQEYFYDQ 492
Query: 478 SPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFL 530
P+ M+ LG S + A S++N L+T+ D L W G +LN S LD +Y L
Sbjct: 493 VPDSMRSLGIAFYLSVLGAASFVNNLLITVSDHLAEEISGKGWFGKDLNSSRLDRFYWML 552
Query: 531 FVISALNFGVFLWVSSGYIYKKENTSTTEVHD 562
++A N F+ V+ Y YK S V D
Sbjct: 553 AALTAANICCFVIVAMRYTYKTVQPSLAVVAD 584
>At1g72125 oligopeptide like transporter
Length = 557
Score = 357 bits (917), Expect = 8e-99
Identities = 193/552 (34%), Positives = 307/552 (54%), Gaps = 27/552 (4%)
Query: 12 VDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFN 71
VD G S TG+ +A +I+ V ERFAYYG+G+NL++++T L + + + N
Sbjct: 18 VDHRGLAARRSNTGRWRAALFIIGVEVAERFAYYGIGSNLISYLTGPLGESTAVAAANVN 77
Query: 72 NWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENGICR 131
WSG+ATL P+LGA++AD + GR+ TI S LIY +GL L TL + +
Sbjct: 78 AWSGIATLLPVLGAFVADAFLGRYRTIIISSLIYVLGLAFL----TLSAFLIPNTTEVTS 133
Query: 132 EASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGAC 191
S+ LF+ SLY +A+G KP + FGADQFD+ +EK + SFFNWW + +
Sbjct: 134 STSSFLNVLFFFSLYLVAIGQSGHKPCVQAFGADQFDEKDSQEKSDRSSFFNWWYLSLSA 193
Query: 192 GSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQAR---SPSMNF 248
G A L VVYIQE+ W + + + ++S ++F G +YR+ R+ +P
Sbjct: 194 GICFAILVVVYIQEEFSWAFGFGIPCVFMVISLVLFVSGRRIYRYSKRRHEEEINPFTRI 253
Query: 249 IRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLS 308
RV VA +N++L + S+L + +L S + SF ++A + +++
Sbjct: 254 GRVFFVALKNQRL----SSSDLCKVELEANTSPEKQ--------SFFNKALLVPNDSSQG 301
Query: 309 NPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAAS 368
+ VE ++ + +W L +A T F +QG TMDRT+ P ++P AS
Sbjct: 302 ENASKSSDVEDATALIRLIPVWFTTLAYAIPYAQYMTFFTKQGVTMDRTILPGVKIPPAS 361
Query: 369 LWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQ 428
L FI ++ ++ +PIYD F+P R T GI L+R+G G+ + I M + VE +
Sbjct: 362 LQVFIGISIVLFVPIYDRVFVPIARLITKEPCGITTLKRIGTGIVLSTITMVIAALVEFK 421
Query: 429 RMSVIKKHHIVG-PEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGT 487
R+ K+H ++ PE T+PMSI+WL+PQ ++LG+++ + GM EFFY Q P E++ +G
Sbjct: 422 RLETAKEHGLIDQPEATLPMSIWWLIPQYLLLGLADVYTLVGMQEFFYSQVPTELRSIGL 481
Query: 488 TLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFVISALNFGV 540
L S + GS +++ L+++ID +W +NLN +HLDY+Y L ++SA+ F
Sbjct: 482 ALYLSALGVGSLLSSLLISLIDLATGGDAGNSWFNSNLNRAHLDYFYWLLAIVSAVGFFT 541
Query: 541 FLWVSSGYIYKK 552
FL++S YIY++
Sbjct: 542 FLFISKSYIYRR 553
>At1g72130 peptide transporter PTR2-B -like protein
Length = 538
Score = 354 bits (908), Expect = 8e-98
Identities = 192/561 (34%), Positives = 305/561 (54%), Gaps = 45/561 (8%)
Query: 5 GYTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVV 64
G T +G +++ + +G K+ I+V ++ ERFAY+G+ +NL+ ++T L +
Sbjct: 12 GNTTEGFLEIR-----ENTSGGWKSARLIIVVQMAERFAYFGIASNLIMYLTGPLGESTA 66
Query: 65 SSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPA 124
++ + N W+G P+LG ++AD+Y GRF TI S +Y +GL LL +T + S +
Sbjct: 67 AAAANVNAWTGTVAFLPLLGGFLADSYLGRFRTIIISSSLYILGLGLLSFSTMIPSHQS- 125
Query: 125 CENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNW 184
++++ LQ +F+ SLY +A+G G P + FGADQFD H+E K SFFNW
Sbjct: 126 ------KDSNQLQETIFFFSLYLVAIGQGGYNPCIKVFGADQFDGNDHKEARDKSSFFNW 179
Query: 185 WAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQA--R 242
F L L YIQE W L + + ++ LLS +F G+ YR + + +
Sbjct: 180 LMFGNCISILTTRLVSTYIQENLSWSLGFGIPSVSMLLSLFLFLLGTTSYRFSTERVGKK 239
Query: 243 SPSMNFIRVPLVAFRNRKL-QLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIR 301
+P RV + A +NR+ L + +E L S + F FLDRAAI
Sbjct: 240 NPFARISRVFMEALKNRRQPDLDIANANANETLLLLAHQS-------SKQFRFLDRAAIS 292
Query: 302 ESNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPK 361
C + ++E K VL + IW+ ++ T A T F +QG TMDR++ P
Sbjct: 293 ----------CELAEIEEAKAVLRLIPIWITSVVYTIVHAQSPTFFTKQGATMDRSISPG 342
Query: 362 FRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAV 421
+PAA+L FI L+ ++ +PIYD +PF R T N GI LQR+G G+ + ++AM +
Sbjct: 343 LLVPAATLQSFINLSVVVFIPIYDRLLVPFARSFTQNSSGITTLQRIGTGIFLSILAMVL 402
Query: 422 TYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEE 481
VET+R+ + E ++PMS++WL+PQ +I GVS+ F G+ EFFY Q P E
Sbjct: 403 AALVETKRLQAAR------DELSIPMSVWWLIPQYVIFGVSDMFTMVGLQEFFYGQVPSE 456
Query: 482 MKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFVIS 534
++ +G L S AG+Y+++F++++IDK+ +W N+L+ +HLDY+Y L +
Sbjct: 457 LRSVGMALNLSIYGAGNYLSSFMISVIDKITNQYGQRSWFDNDLDQAHLDYFYWLLACLG 516
Query: 535 ALNFGVFLWVSSGYIYKKENT 555
+ F +LW + Y+Y + NT
Sbjct: 517 FIGFAFYLWFAKSYVYSRSNT 537
>At1g68570 peptide transporter like
Length = 596
Score = 353 bits (906), Expect = 1e-97
Identities = 202/539 (37%), Positives = 307/539 (56%), Gaps = 21/539 (3%)
Query: 32 YILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYIADTY 91
+I + E+ A G AN+++++TTQL+ + + + N++G ++LTP+LGA+IAD++
Sbjct: 32 FIFANEICEKLAVVGFHANMISYLTTQLHLPLTKAANTLTNFAGTSSLTPLLGAFIADSF 91
Query: 92 TGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENG--ICREASNLQVALFYTSLYTIA 149
GRFWTITF+ +IY IG+ LL ++ + +LRP G +C A Q+++ Y +L A
Sbjct: 92 AGRFWTITFASIIYQIGMTLLTISAIIPTLRPPPCKGEEVCVVADTAQLSILYVALLLGA 151
Query: 150 VGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGW 209
+GSG ++P + FGADQFD+ + + ++FNW+ F L+A +V+IQ+ GW
Sbjct: 152 LGSGGIRPCVVAFGADQFDESDPNQTTKTWNYFNWYYFCMGAAVLLAVTVLVWIQDNVGW 211
Query: 210 GLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSE 269
GL + + LS I F G +YRH A SP I+V + AFR RKL++ +PS
Sbjct: 212 GLGLGIPTVAMFLSVIAFVGGFQLYRHLV-PAGSPFTRLIQVGVAAFRKRKLRMVSDPSL 270
Query: 270 LH-EFQLNYYISSGARKIHHTSHFSFLDRAAI--RESNTDLSNPP-----CTVTQVEGTK 321
L+ +++ IS G K+ HT H SFLD+AAI E N P TV +VE K
Sbjct: 271 LYFNDEIDAPISLGG-KLTHTKHMSFLDKAAIVTEEDNLKPGQIPNHWRLSTVHRVEELK 329
Query: 322 LVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICL 381
V+ M I ++ +A + T ++Q TM+R L F++PA S+ F + L +
Sbjct: 330 SVIRMGPIGASGILLITAYAQQGTFSLQQAKTMNRHLTNSFQIPAGSMSVFTTVAMLTTI 389
Query: 382 PIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVG- 440
YD F+ R+ TG RGI L R+GIG I +IA V VE +R SV +H ++
Sbjct: 390 IFYDRVFVKVARKFTGLERGITFLHRMGIGFVISIIATLVAGFVEVKRKSVAIEHGLLDK 449
Query: 441 PEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYI 500
P VP+S WL+PQ + GV+ AF++ G LEFFYDQ+PE M+ T L ++ G+Y+
Sbjct: 450 PHTIVPISFLWLIPQYGLHGVAEAFMSIGHLEFFYDQAPESMRSTATALFWMAISIGNYV 509
Query: 501 NTFLVTMIDKL-------NWI-GNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYK 551
+T LVT++ K NW+ NNLN L+Y+Y + V+ A+N +LW + Y YK
Sbjct: 510 STLLVTLVHKFSAKPDGSNWLPDNNLNRGRLEYFYWLITVLQAVNLVYYLWCAKIYTYK 568
>At1g22570 Similar to LeOPT1 [Lycopersicon esculentum
Length = 565
Score = 352 bits (904), Expect = 2e-97
Identities = 189/553 (34%), Positives = 310/553 (55%), Gaps = 20/553 (3%)
Query: 11 TVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSF 70
+VD G P S TG ++ +I+ V ERFAY+G+ NL+ ++T L + + +
Sbjct: 18 SVDHRGFPAGKSSTGGWRSAWFIIGVEVAERFAYFGIACNLITYLTGPLGQSTAKAAVNV 77
Query: 71 NNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENGIC 130
N WSG A++ PILGA++AD Y GR+ TI + LIY +GL LL L+ +L + + +
Sbjct: 78 NTWSGTASILPILGAFVADAYLGRYRTIVVASLIYILGLGLLTLSASLIIMGLSKQRNDA 137
Query: 131 REASNLQV-ALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNG 189
++ V LF+ SLY +A+G G KP + FGADQFD +E + SFFNWW +
Sbjct: 138 SAKPSIWVNTLFFCSLYLVAIGQGGHKPCVQAFGADQFDAEDPKEVIARGSFFNWWFLSL 197
Query: 190 ACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYR-----HKSRQARSP 244
+ G ++ + V Y+QE W + + + +++ IF G +YR H+ + +
Sbjct: 198 SAGISISIIVVAYVQENVNWAFGFGIPCLFMVMALAIFLLGRKIYRYPKGHHEEVNSSNT 257
Query: 245 SMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESN 304
RV ++AF+NRKL+L E +L+ + + +FL +A I
Sbjct: 258 FARIGRVFVIAFKNRKLRL-----EHSSLELDQGLLEDGQSEKRKDRLNFLAKAMISREG 312
Query: 305 TDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRL 364
+ PC+ V+ K ++ + IW+ ++ T +A T F +QG T+DR + P +
Sbjct: 313 VE----PCSGRDVDDAKALVRLIPIWITYVVSTIPYAQYITFFTKQGVTVDRRILPGVEI 368
Query: 365 PAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYA 424
PAASL F+ ++ LI +P+Y+ F+P R+ T GI +LQR+G GM + V M +
Sbjct: 369 PAASLLSFVGVSILISVPLYERVFLPIARKITKKPFGITMLQRIGAGMVLSVFNMMLAAL 428
Query: 425 VETQRMSVIKKHHIVG-PEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMK 483
VE++R+ + ++H +V P+ TVPMSI+W +PQ ++LG+ + F G EFFYDQ P E++
Sbjct: 429 VESKRLKIAREHGLVDKPDVTVPMSIWWFVPQYLLLGMIDLFSMVGTQEFFYDQVPTELR 488
Query: 484 GLGTTLCTSCVAAGSYINTFLVTMID----KLNWIGNNLNDSHLDYYYAFLFVISALNFG 539
+G +L S + S+++ FL+++ID K W +NLN +H+DY+Y L +A+ F
Sbjct: 489 SIGLSLSLSAMGLSSFLSGFLISLIDWATGKDGWFNSNLNRAHVDYFYWLLAAFTAIAFF 548
Query: 540 VFLWVSSGYIYKK 552
FL++S Y+Y++
Sbjct: 549 AFLFISKMYVYRR 561
>At1g72115 oligopeptide like transporter
Length = 561
Score = 351 bits (901), Expect = 5e-97
Identities = 196/558 (35%), Positives = 308/558 (55%), Gaps = 24/558 (4%)
Query: 6 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 65
Y +D VD G S+TG+ +A +I+ V ERFA YG+G+NL++++T L +
Sbjct: 13 YVIDA-VDHRGFSARRSITGRWRAAWFIIGVEVAERFANYGIGSNLISYLTGPLGQSTAV 71
Query: 66 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPAC 125
+ + N WSG++T+ P+LGA++AD + GR+ TI + IY +GL L TL +
Sbjct: 72 AAANVNAWSGISTILPLLGAFVADAFLGRYITIIIASFIYVLGLAFL----TLSAFLIPN 127
Query: 126 ENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWW 185
+ S+ ALF+ SLY +A+G KP + FGADQFD+ +E + SFFNWW
Sbjct: 128 NTEVTSSPSSFLNALFFFSLYLVAIGQSGHKPCVQAFGADQFDEKNPQENSDRSSFFNWW 187
Query: 186 AFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYR-HKSRQAR-- 242
+ G +A L VVYIQE W L + + + ++S ++F G YR K+RQ
Sbjct: 188 YLSMCAGIGLAILVVVYIQENVSWALGFGIPCVFMVISLVLFVLGRKSYRFSKTRQEEET 247
Query: 243 SPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRE 302
+P RV VAF+N++L N S+L + +L R SFL++A +
Sbjct: 248 NPFTRIGRVFFVAFKNQRL----NSSDLCKVEL----IEANRSQESPEELSFLNKALLVP 299
Query: 303 SNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKF 362
+++D C VE ++ + +WL L +A T F +QG TM+RT+ P
Sbjct: 300 NDSDEGEVACKSRDVEDATALVRLIPVWLTTLAYAIPFAQYMTFFTKQGVTMERTIFPGV 359
Query: 363 RLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVT 422
+P ASL I ++ ++ +PIYD +P R T + GI L+R+G GM + + M V
Sbjct: 360 EIPPASLQVLISISIVLFVPIYDRVLVPIGRSITKDPCGITTLKRIGTGMVLATLTMVVA 419
Query: 423 YAVETQRMSVIKKHHIVG-PEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEE 481
VE++R+ K++ ++ P+ T+PMSI+WL PQ ++LG+++ GM EFFY Q P E
Sbjct: 420 ALVESKRLETAKEYGLIDQPKTTLPMSIWWLFPQYMLLGLADVHTLVGMQEFFYSQVPTE 479
Query: 482 MKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFVIS 534
++ LG + S + GS +++ L+ +ID +W +NLN +HLDY+Y L V+S
Sbjct: 480 LRSLGLAIYLSAMGVGSLLSSLLIYLIDLATGGDAGNSWFNSNLNRAHLDYFYWLLAVVS 539
Query: 535 ALNFGVFLWVSSGYIYKK 552
A+ F FL++S YIY++
Sbjct: 540 AVGFFTFLFISKSYIYRR 557
>At1g22540 Similar to peptide transporter
Length = 557
Score = 348 bits (892), Expect = 6e-96
Identities = 191/558 (34%), Positives = 317/558 (56%), Gaps = 30/558 (5%)
Query: 11 TVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSF 70
TVD +P + S +G ++ +I+ V ERFAYYG+ +NL+ ++T L + ++ +
Sbjct: 18 TVDYRNKPAVKSSSGGWRSAGFIIGVEVAERFAYYGISSNLITYLTGPLGQSTAAAAANV 77
Query: 71 NNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACE--NG 128
N WSG A+L P+LGA++AD++ GRF TI + +Y +GL +L L+ + S C+ N
Sbjct: 78 NAWSGTASLLPLLGAFVADSFLGRFRTILAASALYIVGLGVLTLSAMIPS---DCKVSNL 134
Query: 129 ICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFN 188
+ + QV F+++LY +A+ G KP + FGADQFD+ EE + K SFFNWW F
Sbjct: 135 LSSCSPRFQVITFFSALYLVALAQGGHKPCVQAFGADQFDEKEPEECKAKSSFFNWWYF- 193
Query: 189 GACGSLMATLFVV-YIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHK-SRQARSPSM 246
G C + TL+V+ YIQ+ W L + + I +++ ++ G+ YR R+ +SP +
Sbjct: 194 GMCFGTLTTLWVLNYIQDNLSWALGFGIPCIAMVVALVVLLLGTCTYRFSIRREDQSPFV 253
Query: 247 NFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTD 306
V + A +N + E +L S +++ FSFL++A + +
Sbjct: 254 RIGNVYVAAVKNWSVS--ALDVAAAEERLGLVSCSSSQQ------FSFLNKALVAK---- 301
Query: 307 LSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPA 366
N C++ ++E K VL + IWL L+ +A T F +QG TM+R++ P +++
Sbjct: 302 --NGSCSIDELEEAKSVLRLAPIWLTCLVYAVVFAQSPTFFTKQGATMERSITPGYKISP 359
Query: 367 ASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVE 426
A+L FI L+ +I +PIYD IP R T GI +LQR+G G+ + +AM V VE
Sbjct: 360 ATLQSFISLSIVIFIPIYDRVLIPIARSFTHKPGGITMLQRIGTGIFLSFLAMVVAALVE 419
Query: 427 TQRMSVIKKHHIV-GPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGL 485
+R+ + +V P+ TVPMS++WL+PQ ++ G+++ F G+ EFFYDQ P E++ +
Sbjct: 420 MKRLKTAADYGLVDSPDATVPMSVWWLVPQYVLFGITDVFAMVGLQEFFYDQVPNELRSV 479
Query: 486 GTTLCTSCVAAGSYINTFLVTMIDK-------LNWIGNNLNDSHLDYYYAFLFVISALNF 538
G L S G+++++F++++I+K +W NNLN +HLDY+Y L +S +
Sbjct: 480 GLALYLSIFGIGNFLSSFMISIIEKATSQSGQASWFANNLNQAHLDYFYWLLACLSFIGL 539
Query: 539 GVFLWVSSGYIYKKENTS 556
+L+V+ Y+ K+ +TS
Sbjct: 540 ASYLYVAKSYVSKRLDTS 557
>At1g22550 Similar to LeOPT1 [Lycopersicon esculentum
Length = 564
Score = 341 bits (875), Expect = 6e-94
Identities = 194/561 (34%), Positives = 305/561 (53%), Gaps = 29/561 (5%)
Query: 7 TLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSS 66
++ +VD G P S TG ++ YI+ V ERFAY+G+G+NL+ ++T L + ++
Sbjct: 14 SVSDSVDHRGLPAGKSSTGGWRSAWYIIGVEVGERFAYFGIGSNLITYLTGPLGQSTATA 73
Query: 67 ITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACE 126
+ N WSG A++ P+LGA+IAD Y GR+ TI + LIY +GL LL L++ L + + +
Sbjct: 74 AVNVNTWSGTASILPVLGAFIADAYLGRYRTIVVASLIYILGLGLLTLSSILILMGLSEQ 133
Query: 127 NGICREASNLQV----ALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFF 182
R AS LF+ SLY +A+G G KP + FGADQFD +E+ + SFF
Sbjct: 134 RQHNRNASAKPFFWVNILFFCSLYLVAIGQGGHKPCVQAFGADQFDVGDPKERISRGSFF 193
Query: 183 NWWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQAR 242
NWW + + G ++ + VVY+Q+ W L + + + +++ +F +G YR+
Sbjct: 194 NWWFLSLSAGITLSIIVVVYVQDNVNWALGFGIPCLFMVMALALFLFGRKTYRYPRGDRE 253
Query: 243 SPSMNFIRVP---LVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAA 299
+ F R+ LVAF+NRKL+L H QL G+ K FL +A
Sbjct: 254 GKNNAFARIGRVFLVAFKNRKLKL------THSGQLEV----GSYK-KCKGQLEFLAKAL 302
Query: 300 IRESNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLG 359
+ PC+ VE ++ + IW+ ++ T +A +T F +QG T+DR +
Sbjct: 303 LPGEG---GVEPCSSRDVEDAMALVRLIPIWITSVVSTIPYAQYATFFTKQGVTVDRKIL 359
Query: 360 PKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAM 419
P F +P AS I L+ I +P Y+ F+P R T GI +LQR+G GM + + M
Sbjct: 360 PGFEIPPASFQALIGLSIFISVPTYERVFLPLARLITKKPSGITMLQRIGAGMVLSSLNM 419
Query: 420 AVTYAVETQRMSVIKKHHIVG-PEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQS 478
V VE +R+ K+H +V P+ T+PMSI+W +PQ ++LG+ + F G EFFYDQ
Sbjct: 420 VVAALVEMKRLETAKEHGLVDRPDATIPMSIWWFVPQYLLLGMIDVFSLVGTQEFFYDQV 479
Query: 479 PEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLF 531
P E++ +G L S + S+++ FL+T+I+ +W NLN +H+DY+Y L
Sbjct: 480 PTELRSIGLALSLSAMGLASFLSGFLITVINWATGKNGGDSWFNTNLNRAHVDYFYWLLA 539
Query: 532 VISALNFGVFLWVSSGYIYKK 552
+A+ F FL +S Y+Y++
Sbjct: 540 AFTAIGFLAFLLLSRLYVYRR 560
>At1g72140 peptide transporter like protein (PTR2-B)
Length = 555
Score = 336 bits (862), Expect = 2e-92
Identities = 192/557 (34%), Positives = 298/557 (53%), Gaps = 44/557 (7%)
Query: 11 TVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSF 70
+VD G P + S +G K+ + + V E+FAY+G+ +NL+ + T L + + ++
Sbjct: 22 SVDFRGNPSIRSSSGAWKSSGFTMCAEVAEKFAYFGIASNLITYFTEALGESTAVAASNV 81
Query: 71 NNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENGIC 130
N W G A P++ IAD++ GRF TI + Y +GL LL + T+ SL C +
Sbjct: 82 NLWLGTAAFLPLIWGSIADSFLGRFRTILLTSSFYIMGLGLLTFSATIPSL---CNDQET 138
Query: 131 REA--SNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFN 188
RE+ S ++V +F+ +LY IA+G G K + FGADQFD+ E + K S+FNW F
Sbjct: 139 RESCVSQVKVIIFFCALYLIALGEGGFKVCLRAFGADQFDEQDPNESKAKSSYFNWLYFA 198
Query: 189 GACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKS----RQARSP 244
+ G L L Y+QE W L Y++ + +L+ +F G YR + RQ +
Sbjct: 199 ISIGILTTRLVTNYVQENLSWALGYAIPCLSMMLALFLFLLGIKTYRFSTGGEGRQGKKH 258
Query: 245 SMNFIRVPLV---AFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIR 301
F+R+ V A RNR+ PS+ N T F FLDRA I
Sbjct: 259 DNPFVRIGRVFVAAARNRRQ----TPSDTCLLLPN----------ESTKKFRFLDRAVIS 304
Query: 302 ESNTDLSNPPCTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPK 361
C +VE K VL + IWL L+ +A T F +QG+TMDR++
Sbjct: 305 ----------CDSYEVEEAKAVLSLIPIWLCSLVFGIVFAQSPTFFTKQGSTMDRSISST 354
Query: 362 FRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAV 421
++PAA+L CFI L L+ +PIYD F+P R T GI LQR+ G+ + +I+M +
Sbjct: 355 LQVPAATLQCFISLAILVFIPIYDRLFVPIARSITRKPAGITTLQRISTGIFLSIISMVI 414
Query: 422 TYAVETQRMSVIKKHHIV-GPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPE 480
VE +R+ + H +V P+ TVPMS+ WL+PQ I+ GVS+ F G+ EFFY + P
Sbjct: 415 AALVEMKRLKTARDHGLVDSPKATVPMSVCWLIPQYILFGVSDVFTMVGLQEFFYGEVPP 474
Query: 481 EMKGLGTTLCTSCVAAGSYINTFLVTMID-------KLNWIGNNLNDSHLDYYYAFLFVI 533
+++ +G L S + G+++++F+V++I+ +++W NNLN +HLDY+Y L +
Sbjct: 475 QLRSMGLALYLSIIGIGNFLSSFMVSVIEEATSQSGQVSWFSNNLNQAHLDYFYWLLACL 534
Query: 534 SALNFGVFLWVSSGYIY 550
S+L F ++ + Y+Y
Sbjct: 535 SSLAFIFTVYFAKSYLY 551
>At1g69870 unknown protein
Length = 620
Score = 323 bits (827), Expect = 2e-88
Identities = 196/571 (34%), Positives = 294/571 (51%), Gaps = 26/571 (4%)
Query: 25 GKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILG 84
G +A ++IL LER G+ AN + ++T + + V + N WSG LTP++G
Sbjct: 53 GGWRAVSFILGNETLERLGSIGLLANFMVYLTKVFHLEQVDAANVINIWSGFTNLTPLVG 112
Query: 85 AYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPACENG----ICREASNLQVAL 140
AYI+DTY GRF TI F+ +GL+ + LT + L PA N C + LQ+ +
Sbjct: 113 AYISDTYVGRFKTIAFASFATLLGLITITLTASFPQLHPASCNSQDPLSCGGPNKLQIGV 172
Query: 141 FYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFV 200
L ++VGSG ++P FG DQFD E + SFFNW+ ++ V
Sbjct: 173 LLLGLCFLSVGSGGIRPCSIPFGVDQFDQRTEEGVKGVASFFNWYYMTFTVVLIITQTVV 232
Query: 201 VYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRK 260
VYIQ++ W + +S+ L+ ++FF G Y + + S +V + A + RK
Sbjct: 233 VYIQDQVSWIIGFSIPTGLMALAVVMFFAGMKRYVYVKPEG-SIFSGIAQVIVAARKKRK 291
Query: 261 LQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIR-ESNTDLSNPP------CT 313
L+LP + SS K+H ++ F LD+AA+ E + PP C+
Sbjct: 292 LKLPAEDDGTVTYYDPAIKSSVLSKLHRSNQFRCLDKAAVVIEGDLTPEGPPADKWRLCS 351
Query: 314 VTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFI 373
V +VE K ++ + IW +I + T V Q MDR LGPKF +PA SL
Sbjct: 352 VQEVEEVKCLIRIVPIWSAGIISLAAMTTQGTFTVSQALKMDRNLGPKFEIPAGSLSVIS 411
Query: 374 VLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVI 433
+LT I LP YD F+PFMRR TG+ GI LLQR+G G+ + +M V VE RM I
Sbjct: 412 LLTIGIFLPFYDRVFVPFMRRITGHKSGITLLQRIGTGIVFAIFSMIVAGIVE--RMRRI 469
Query: 434 KKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSC 493
+ + P PMS+FWL PQ I++G+ AF G +EFF Q PE M+ + +L +
Sbjct: 470 RSINAGDPTGMTPMSVFWLSPQLILMGLCEAFNIIGQIEFFNSQFPEHMRSIANSLFSLS 529
Query: 494 VAAGSYINTFLVTMI-------DKLNWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSS 546
A SY+++FLVT++ D+ +W+ NLN LDY+Y + V+ +N F + +
Sbjct: 530 FAGSSYLSSFLVTVVHKFSGGHDRPDWLNKNLNAGKLDYFYYLIAVLGVVNLVYFWYCAR 589
Query: 547 GYIYK-----KENTSTTEVHDIEMSAEKTVK 572
GY YK ++ D+EM+++K++K
Sbjct: 590 GYRYKVGLPIEDFEEDKSSDDVEMTSKKSMK 620
>At3g21670 nitrate transporter
Length = 590
Score = 317 bits (811), Expect = 1e-86
Identities = 194/578 (33%), Positives = 307/578 (52%), Gaps = 35/578 (6%)
Query: 13 DLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNN 72
D G P S TG IL + ER G+ NLV ++ L+ S T N
Sbjct: 19 DYRGNPPDKSKTGGWLGAGLILGSELSERICVMGISMNLVTYLVGDLHISSAKSATIVTN 78
Query: 73 WSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPA-CEN---- 127
+ G L +LG ++AD GR+ + S + A+G++LL + TT+ S+RP C++
Sbjct: 79 FMGTLNLLGLLGGFLADAKLGRYKMVAISASVTALGVLLLTVATTISSMRPPICDDFRRL 138
Query: 128 -GICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWA 186
C EA+ Q+AL Y +LYTIA+G G +K N+S FG+DQFD +E++Q + FFN +
Sbjct: 139 HHQCIEANGHQLALLYVALYTIALGGGGIKSNVSGFGSDQFDTSDPKEEKQMIFFFNRFY 198
Query: 187 FNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSM 246
F+ + GSL A + +VY+Q+ G G Y +SA ++++I+ G+ YR K + SP
Sbjct: 199 FSISVGSLFAVIALVYVQDNVGRGWGYGISAATMVVAAIVLLCGTKRYRFKKPKG-SPFT 257
Query: 247 NFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTD 306
RV +A++ RK P +PS L+ + + HT LD+AAI ++ +
Sbjct: 258 TIWRVGFLAWKKRKESYPAHPSLLNGYD--------NTTVPHTEMLKCLDKAAISKNESS 309
Query: 307 LSNPP---------CTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRT 357
S+ TVTQVE KLV+ + IW ++ ++ +T V Q T MDR
Sbjct: 310 PSSKDFEEKDPWIVSTVTQVEEVKLVMKLVPIWATNILFWTIYSQMTTFTVEQATFMDRK 369
Query: 358 LGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVI 417
LG F +PA S F++LT L+ + + F+P RR T +GI LQR+G+G+ +
Sbjct: 370 LG-SFTVPAGSYSAFLILTILLFTSLNERVFVPLTRRLTKKPQGITSLQRIGVGLVFSMA 428
Query: 418 AMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQ 477
AMAV +E R + V ++ + S FWL+PQ ++G AF G LEFF +
Sbjct: 429 AMAVAAVIENAR-----REAAVNNDKKI--SAFWLVPQYFLVGAGEAFAYVGQLEFFIRE 481
Query: 478 SPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKL---NWIGNNLNDSHLDYYYAFLFVIS 534
+PE MK + T L S ++ G ++++ LV+++D++ +W+ +NLN + L+Y+Y L V+
Sbjct: 482 APERMKSMSTGLFLSTISMGFFVSSLLVSLVDRVTDKSWLRSNLNKARLNYFYWLLVVLG 541
Query: 535 ALNFGVFLWVSSGYIYKKENTSTTEVHDIEMSAEKTVK 572
ALNF +F+ + + YK + + D + E T K
Sbjct: 542 ALNFLIFIVFAMKHQYKADVITVVVTDDDSVEKEVTKK 579
>At4g21680 peptide transporter - like protein
Length = 589
Score = 312 bits (799), Expect = 4e-85
Identities = 189/579 (32%), Positives = 305/579 (52%), Gaps = 35/579 (6%)
Query: 7 TLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSS 66
T DG+VD G P + + TGK ILV + L A++GVG NLV F+T + +D +
Sbjct: 12 TQDGSVDRHGNPAIRANTGKWLTAILILVNQGLATLAFFGVGVNLVLFLTRVMGQDNAEA 71
Query: 67 ITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRPA-- 124
+ + W+G + +LGA+++D+Y GR+ T + GL++L L+T L P+
Sbjct: 72 ANNVSKWTGTVYIFSLLGAFLSDSYWGRYKTCAIFQASFVAGLMMLSLSTGALLLEPSGC 131
Query: 125 -CENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFN 183
E+ C+ S + LFY S+Y IA+G G +PN++TFGADQFD E K++FF+
Sbjct: 132 GVEDSPCKPHSTFKTVLFYLSVYLIALGYGGYQPNIATFGADQFDAEDSVEGHSKIAFFS 191
Query: 184 WWAFNGACGSLMATLFVVYIQEKNGWGLAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARS 243
++ GSL + + Y +++ W L + SA ++F G+P YRH + + S
Sbjct: 192 YFYLALNLGSLFSNTVLGYFEDQGEWPLGFWASAGSAFAGLVLFLIGTPKYRHFTPR-ES 250
Query: 244 PSMNFIRVPLVAFRNRKLQLPCNPSELHEFQLNYYIS----SGARKIHHTSHFSFLDRAA 299
P F +V + A R K+ ++H +LN Y S +G +KI HT F FLDRAA
Sbjct: 251 PWSRFCQVLVAATRKAKI-------DVHHEELNLYDSETQYTGDKKILHTKGFRFLDRAA 303
Query: 300 IRESNTDLSNPP----------CTVTQVEGTKLVLGMFQIWLLMLIPTNCWALESTIFVR 349
I + + C+VTQVE K VL + IWL ++ + + +++FV
Sbjct: 304 IVTPDDEAEKVESGSKYDPWRLCSVTQVEEVKCVLRLLPIWLCTILYSVVFTQMASLFVV 363
Query: 350 QGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRR--RTGNHRGIKLLQR 407
QG M + FR+PA+S+ F +L+ + Y + P R +T ++G+ LQR
Sbjct: 364 QGAAMKTNI-KNFRIPASSMSSFDILSVAFFIFAYRRFLDPLFARLNKTERNKGLTELQR 422
Query: 408 VGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLPQNIILGVSNAFLA 467
+GIG+ I ++AM VE R+ + + +SIFW +PQ +++G S F+
Sbjct: 423 MGIGLVIAIMAMISAGIVEIHRLKNKEPESATSISSSSTLSIFWQVPQYMLIGASEVFMY 482
Query: 468 TGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDKLN-------WIGNNLND 520
G LEFF Q+P +K + LC + ++ G+Y+++ LV+++ K++ WI NLN
Sbjct: 483 VGQLEFFNSQAPTGLKSFASALCMASISLGNYVSSLLVSIVMKISTTDDVHGWIPENLNK 542
Query: 521 SHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTE 559
HL+ +Y L ++A +F V+L + Y Y K S +E
Sbjct: 543 GHLERFYFLLAGLTAADFVVYLICAKWYKYIKSEASFSE 581
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,296,705
Number of Sequences: 26719
Number of extensions: 510621
Number of successful extensions: 1716
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1370
Number of HSP's gapped (non-prelim): 70
length of query: 573
length of database: 11,318,596
effective HSP length: 105
effective length of query: 468
effective length of database: 8,513,101
effective search space: 3984131268
effective search space used: 3984131268
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0248.4


