

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0248.1
(119 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
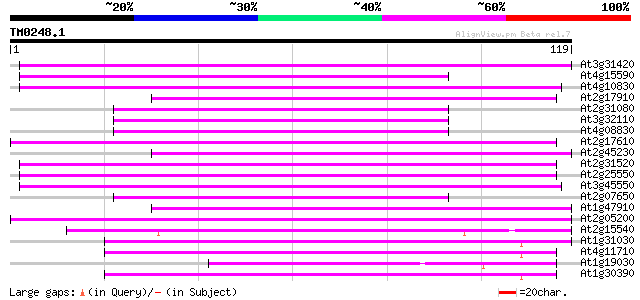
Score E
Sequences producing significant alignments: (bits) Value
At3g31420 hypothetical protein 61 1e-10
At4g15590 reverse transcriptase like protein 59 4e-10
At4g10830 putative protein 55 6e-09
At2g17910 putative non-LTR retroelement reverse transcriptase 54 1e-08
At2g31080 putative non-LTR retroelement reverse transcriptase 54 1e-08
At3g32110 non-LTR reverse transcriptase, putative 53 2e-08
At4g08830 putative protein 52 5e-08
At2g17610 putative non-LTR retroelement reverse transcriptase 52 5e-08
At2g45230 putative non-LTR retroelement reverse transcriptase 51 8e-08
At2g31520 putative non-LTR retroelement reverse transcriptase 50 2e-07
At2g25550 putative non-LTR retroelement reverse transcriptase 50 2e-07
At3g45550 putative protein 49 3e-07
At2g07650 putative non-LTR retrolelement reverse transcriptase 49 3e-07
At1g47910 reverse transcriptase, putative 47 2e-06
At2g05200 putative non-LTR retroelement reverse transcriptase 47 2e-06
At2g15540 putative non-LTR retroelement reverse transcriptase 46 3e-06
At1g31030 F17F8.5 46 3e-06
At4g11710 putative protein 45 4e-06
At1g19030 hypothetical protein 44 1e-05
At1g30390 hypothetical protein 43 2e-05
>At3g31420 hypothetical protein
Length = 1491
Score = 60.8 bits (146), Expect = 1e-10
Identities = 33/117 (28%), Positives = 54/117 (45%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
++P+ N F + E D + W+ G +T + W +S PK+ GGLG +
Sbjct: 957 AMPVYSMNIFKLTKEVCEEIDSLLARFWWSSGNETKGMHWFTWKRMSIPKKEGGLGFKEL 1016
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIVSAK 119
N+A+L K LL+ L RVL +Y +V+ R S++WK I+ +
Sbjct: 1017 ENFNLALLGKQTWHLLQHPNCLMARVLRGRYFPETNVMNAVQGRRASFVWKSILHGR 1073
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 58.9 bits (141), Expect = 4e-10
Identities = 33/91 (36%), Positives = 48/91 (52%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
S+P+ + L + DK R+ +W + +L+ W +V PK GGLGLR++
Sbjct: 597 SIPIHTMSSILLPASLLEQLDKVSRNFLWGSTVEKRKQHLLSWKKVCRPKAAGGLGLRAS 656
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKY 93
N A+L K+ LL DK LW RVL +KY
Sbjct: 657 KDMNRALLAKVGWRLLNDKVSLWARVLRRKY 687
>At4g10830 putative protein
Length = 1294
Score = 55.1 bits (131), Expect = 6e-09
Identities = 29/115 (25%), Positives = 55/115 (47%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
S+P+ + F L++ E + + + W K + + W + K+ GGLG R
Sbjct: 1161 SMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEGGLGFRDL 1220
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIVS 117
+ N A+L K V ++ + L+ R++ +Y S+L K +R SY W +++
Sbjct: 1221 AKFNDALLAKQVWRMINNPNSLFARIMKARYFREDSILDAKRQRYQSYGWTSMLA 1275
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 54.3 bits (129), Expect = 1e-08
Identities = 26/86 (30%), Positives = 41/86 (47%)
Query: 31 WAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKGKLWGRVLS 90
W Q + + W ++ PK+ GG G + N A+L K +L++KG L+ RV
Sbjct: 805 WNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQ 864
Query: 91 QKYLSNKSVLAGKYKRGDSYIWKGIV 116
+Y SN L+ SY W+ I+
Sbjct: 865 SRYFSNSDFLSATRGSRPSYAWRSIL 890
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 53.9 bits (128), Expect = 1e-08
Identities = 26/71 (36%), Positives = 41/71 (57%)
Query: 23 DKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKG 82
D+ R+ +W + +L+ W ++ PK GG+GLRS N A++ K+ LL+DK
Sbjct: 682 DRYSRTFLWGSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKE 741
Query: 83 KLWGRVLSQKY 93
LW RV+ +KY
Sbjct: 742 SLWARVVRKKY 752
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 53.1 bits (126), Expect = 2e-08
Identities = 25/71 (35%), Positives = 38/71 (53%)
Query: 23 DKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKG 82
DK R+ +W + +LV W V P+ GGLG+RS + N A++ K+ +L D
Sbjct: 1358 DKVSRAFLWGSTLEKKKQHLVAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGS 1417
Query: 83 KLWGRVLSQKY 93
LW +V+ KY
Sbjct: 1418 SLWAQVVRSKY 1428
>At4g08830 putative protein
Length = 947
Score = 52.0 bits (123), Expect = 5e-08
Identities = 25/71 (35%), Positives = 37/71 (51%)
Query: 23 DKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKG 82
DK R + + +LV W V PK GGLG+R++ N A++ K+ L+ D+
Sbjct: 496 DKLARVFLLGSSAEKKKLHLVAWDRVCLPKSEGGLGIRTSKCMNKALVSKVGWRLINDRY 555
Query: 83 KLWGRVLSQKY 93
LW R+L KY
Sbjct: 556 SLWARILRSKY 566
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 52.0 bits (123), Expect = 5e-08
Identities = 32/116 (27%), Positives = 51/116 (43%)
Query: 1 IGSLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLR 60
+ ++P + F + L + +R W+ H V W +++ PK+ GGL +R
Sbjct: 252 VTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKLNDPKKMGGLAIR 311
Query: 61 STSLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIV 116
NIA+L K +L+ L RV KY + +L K SY WK I+
Sbjct: 312 DLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQSSYAWKSIL 367
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 51.2 bits (121), Expect = 8e-08
Identities = 26/89 (29%), Positives = 39/89 (43%)
Query: 31 WAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKGKLWGRVLS 90
W + + WC +S PK GGLG + NIA+L K + ++ +K L +V
Sbjct: 827 WKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFK 886
Query: 91 QKYLSNKSVLAGKYKRGDSYIWKGIVSAK 119
+Y S L S+ WK I A+
Sbjct: 887 SRYFSKSDPLNAPLGSRPSFAWKSIYEAQ 915
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/114 (24%), Positives = 51/114 (44%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
++P+ + F + E + + + W K V W + K+ GGLG R
Sbjct: 955 AMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDL 1014
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIV 116
+ N A+L K L++ L+ RV+ +Y + S+L K ++ SY W ++
Sbjct: 1015 AKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL 1068
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 49.7 bits (117), Expect = 2e-07
Identities = 28/114 (24%), Positives = 51/114 (44%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
++P+ + F + E + + + W K V W + K+ GGLG R
Sbjct: 1181 AMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDL 1240
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIV 116
+ N A+L K L++ L+ RV+ +Y + S+L K ++ SY W ++
Sbjct: 1241 AKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL 1294
>At3g45550 putative protein
Length = 851
Score = 49.3 bits (116), Expect = 3e-07
Identities = 27/115 (23%), Positives = 51/115 (43%)
Query: 3 SLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRST 62
++P+ + F + E + + + W K V W + K+ GGLG R
Sbjct: 420 AMPVYAMSCFKLPQGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDL 479
Query: 63 SLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIVS 117
+ N A+L K +++ L+ RV+ +Y + S++ K + SY W ++S
Sbjct: 480 AKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLLS 534
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 49.3 bits (116), Expect = 3e-07
Identities = 24/71 (33%), Positives = 35/71 (48%)
Query: 23 DKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKG 82
DK RS +W +L+ W V P+ GGLG+R N A+L K+ L++D
Sbjct: 344 DKVSRSFLWGSSVTQRKQHLISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYH 403
Query: 83 KLWGRVLSQKY 93
LW R++ Y
Sbjct: 404 SLWARIMRCNY 414
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 47.0 bits (110), Expect = 2e-06
Identities = 23/89 (25%), Positives = 42/89 (46%)
Query: 31 WAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKGKLWGRVLS 90
W+ G + + + W ++ K +GGLG R+ N A+L K + L+ L+ +V
Sbjct: 611 WSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFK 670
Query: 91 QKYLSNKSVLAGKYKRGDSYIWKGIVSAK 119
+Y + L SY W+ ++SA+
Sbjct: 671 GRYFRKSNPLDSIKSYSPSYGWRSMISAR 699
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 46.6 bits (109), Expect = 2e-06
Identities = 31/119 (26%), Positives = 49/119 (41%)
Query: 1 IGSLPLLEKNEFTFKLHLHLEYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLR 60
+ S+PL + F L + + W V W +++ PK GGLG R
Sbjct: 692 LASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFR 751
Query: 61 STSLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSVLAGKYKRGDSYIWKGIVSAK 119
N ++L KL LL L R+L KY + S + K S+ W+ I++ +
Sbjct: 752 DIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMECKLPSQPSHGWRSIIAGR 810
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 46.2 bits (108), Expect = 3e-06
Identities = 33/116 (28%), Positives = 56/116 (47%), Gaps = 10/116 (8%)
Query: 13 TFKLHLHLEYDKNIRSCI--------WAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSL 64
T K+ L + D IRS + W +++ + + W ++ P +GGLG R+
Sbjct: 681 TIKVSLGIHKDGAIRSKLSSVVANFWWKTREESNGIHWIAWDKLCTPFSDGGLGFRTLEE 740
Query: 65 NNIAMLEKLVESLLRDKGKLWGRVLSQKYLS-NKSVLAGKYKRGDSYIWKGIVSAK 119
N+ +L K + L+R L RVL +Y + + GK R S+ W+ I++AK
Sbjct: 741 FNLVLLAKQLWRLIRFPNSLLSRVLRGRYFRYSDPIQIGKANR-PSFGWRSIMAAK 795
>At1g31030 F17F8.5
Length = 872
Score = 45.8 bits (107), Expect = 3e-06
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 1/100 (1%)
Query: 21 EYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRD 80
E DK S +W+ + + W V PK GGLGLR+ N KLV ++ +
Sbjct: 485 EIDKLCSSFLWSGSEMSSHKAKISWDIVCKPKAEGGLGLRNLKEANDVSCLKLVWRIISN 544
Query: 81 KGKLWGRVLSQKYLSNKSVLAGKYKRG-DSYIWKGIVSAK 119
LW + +++ + KS+ + K S+IW+ I+ +
Sbjct: 545 SNSLWTKWVAEYLIRKKSIWSLKQSTSMGSWIWRKILKIR 584
>At4g11710 putative protein
Length = 473
Score = 45.4 bits (106), Expect = 4e-06
Identities = 29/97 (29%), Positives = 43/97 (43%), Gaps = 1/97 (1%)
Query: 21 EYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRD 80
E DK + +W+ G S + W V PKE GGLGLRS N KL+ ++
Sbjct: 87 EIDKMCSAYLWSGGELNTSKAKITWAFVCKPKEEGGLGLRSLKEANDVCCLKLIWRIISH 146
Query: 81 KGKLWGRVLSQKYLSNKSVLAGKYKRG-DSYIWKGIV 116
LW + + L S A + S++W+ I+
Sbjct: 147 ADSLWVKWIQSSLLKKVSFWAVRENTSLGSWMWRKIL 183
>At1g19030 hypothetical protein
Length = 398
Score = 44.3 bits (103), Expect = 1e-05
Identities = 29/76 (38%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Query: 43 VPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRDKGKLWGRVLSQKYLSNKSV--L 100
V W EV PK+ GGLGLRS + N + KL+ +L + LW L +KYL + L
Sbjct: 89 VAWSEVCSPKQEGGLGLRSIAEANKVCVLKLIWRILSARRSLWVE-LVKKYLIRRGSFWL 147
Query: 101 AGKYKRGDSYIWKGIV 116
+ S+IWK I+
Sbjct: 148 VKENTTCGSWIWKKIL 163
>At1g30390 hypothetical protein
Length = 504
Score = 43.1 bits (100), Expect = 2e-05
Identities = 28/97 (28%), Positives = 42/97 (42%), Gaps = 1/97 (1%)
Query: 21 EYDKNIRSCIWAKGGQTHSWYLVPWCEVSCPKENGGLGLRSTSLNNIAMLEKLVESLLRD 80
E DK + +W+ G S + W V PKE GGLGLRS N KL+ ++
Sbjct: 168 EIDKMCSAYLWSGGELNTSKAKIAWAFVCKPKEEGGLGLRSLKEANDVCCLKLIWRIISH 227
Query: 81 KGKLWGRVLSQKYLSNKSVLAGKYKRG-DSYIWKGIV 116
LW + + L A + S++W+ I+
Sbjct: 228 ADSLWVKWIQSSLLKKVFFWAVRENTSLGSWMWRKIL 264
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,865,278
Number of Sequences: 26719
Number of extensions: 112579
Number of successful extensions: 273
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 209
Number of HSP's gapped (non-prelim): 70
length of query: 119
length of database: 11,318,596
effective HSP length: 95
effective length of query: 24
effective length of database: 8,780,291
effective search space: 210726984
effective search space used: 210726984
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0248.1


