

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0238.4
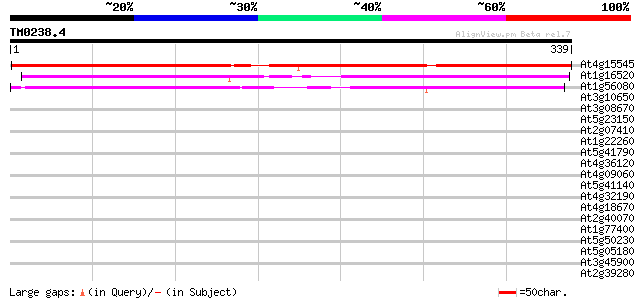
(339 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g15545 unknown protein 375 e-104
At1g16520 unknown protein 219 2e-57
At1g56080 unknown protein 212 3e-55
At3g10650 hypothetical protein 40 0.001
At3g08670 hypothetical protein 40 0.001
At5g23150 transcription factor-like protein (gb|AAD31171.1) 39 0.005
At2g07410 putative retroelement pol polyprotein 38 0.007
At1g22260 hypothetical protein 38 0.007
At5g41790 myosin heavy chain-like protein 38 0.009
At4g36120 myosin-like protein 37 0.012
At4g09060 unknown protein 37 0.020
At5g41140 putative protein 36 0.026
At4g32190 unknown protein 36 0.026
At4g18670 extensin-like protein 36 0.026
At2g40070 En/Spm-like transposon protein 36 0.026
At1g77400 unknown protein 36 0.026
At5g50230 putative protein 36 0.034
At5g05180 unknown protein 36 0.034
At3g45900 unknown protein 36 0.034
At2g39280 unknown protein 36 0.034
>At4g15545 unknown protein
Length = 337
Score = 375 bits (963), Expect = e-104
Identities = 209/340 (61%), Positives = 254/340 (74%), Gaps = 18/340 (5%)
Query: 2 AAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKD 61
+A +G +FDLPD+L+QVLPSDPFEQLD+ARKITSIALS RV+ALESESS+LR +AEK+
Sbjct: 14 SAITGSRSFDLPDELLQVLPSDPFEQLDVARKITSIALSTRVSALESESSDLRELLAEKE 73
Query: 62 HLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRK 121
ELQS ++SL+A+LS L A+ +KE+LI ENASLSNTV++L RDVSKLE FRK
Sbjct: 74 KEFEELQSHVESLEASLSDAFHKLSLADGEKENLIRENASLSNTVKRLQRDVSKLEGFRK 133
Query: 122 TLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPS--ISDTGN 179
TLM SLQ++D N+ G ++AK D++ PS SS+ S S+
Sbjct: 134 TLMMSLQDDDQNA-GTTQIIAKPTP----------NDDDTPFQPSRHSSIQSQQASEAIE 182
Query: 180 SFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSIS 239
A D+E+DA +P + SL L SQ+TTPRLTPPGSPP LSAS +P TS+P+SPRRHS+S
Sbjct: 183 PAATDNENDAPKPSLSASLPLVSQTTTPRLTPPGSPPILSASGTPKTTSRPISPRRHSVS 242
Query: 240 FATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVK 299
FAT+RGM DD S S S S GSQT RTRVDGKEFFRQVR+RLSYEQFGAFL NVK
Sbjct: 243 FATTRGMFDDTRS-----SISISEPGSQTARTRVDGKEFFRQVRSRLSYEQFGAFLGNVK 297
Query: 300 ELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNVH 339
+LN+HKQTRE TL+KA+EIFG +N+DLY IFEGLITRN H
Sbjct: 298 DLNAHKQTREETLRKAEEIFGGDNRDLYVIFEGLITRNAH 337
>At1g16520 unknown protein
Length = 325
Score = 219 bits (558), Expect = 2e-57
Identities = 137/344 (39%), Positives = 198/344 (56%), Gaps = 38/344 (11%)
Query: 8 LNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAEL 67
L+F+LP++++ V+P DPFEQLDLARKITS+A+++RV+ L+SE ELR ++ K+ ++ EL
Sbjct: 6 LDFELPEEVLSVIPMDPFEQLDLARKITSMAIASRVSNLDSEVVELRQKLLGKESVVREL 65
Query: 68 QSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSL 127
+ + L+ L +D +L E SL+ TV KL RD++KLE F++ L++SL
Sbjct: 66 EEKASRLERDCREADSRLKVVLEDNMNLTKEKDSLAMTVTKLTRDLAKLETFKRQLIKSL 125
Query: 128 QEED------------DNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSIS 175
+E D G P ++ + S + S G + + P +S
Sbjct: 126 SDESGPQTEPVDIRTCDQPGSYPGKDGRINAHSIKQAYS--GSTDTNNPVVEASKY---- 179
Query: 176 DTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRR 235
TGN F+ + +PRLTP +P +S SVSP S SP+R
Sbjct: 180 -TGNKFS------------------MTSYISPRLTPTATPKIISTSVSPRGYSAAGSPKR 220
Query: 236 HSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRT-RVDGKEFFRQVRNRLSYEQFGAF 294
S + + ++ +S SS ++S + RT R+DGKEFFRQ R+RLSYEQF +F
Sbjct: 221 TSGAVSPTKATLWYPSSQQSSAANSPPRNRTLPARTPRMDGKEFFRQARSRLSYEQFSSF 280
Query: 295 LANVKELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNV 338
LAN+KELN+ KQTRE TL+KADEIFG ENKDLY F+GL+ RN+
Sbjct: 281 LANIKELNAQKQTREETLRKADEIFGEENKDLYLSFQGLLNRNM 324
>At1g56080 unknown protein
Length = 310
Score = 212 bits (539), Expect = 3e-55
Identities = 132/340 (38%), Positives = 193/340 (55%), Gaps = 38/340 (11%)
Query: 1 MAAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEK 60
M+ GD F+L D+++ V+P+DP++QLDLARKITS+A+++RV+ LES+ S LR ++ EK
Sbjct: 1 MSQSGGD--FNLSDEILAVIPTDPYDQLDLARKITSMAIASRVSNLESQVSGLRQKLLEK 58
Query: 61 DHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFR 120
D L+ EL+ ++ S + +L + L E SL+ T +KL RD +KLE F+
Sbjct: 59 DRLVHELEDRVSSFERLYHEADSSLKNVVDENMKLTQERDSLAITAKKLGRDYAKLEAFK 118
Query: 121 KTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNS 180
+ LM+SL +++ + D V V S + +NE
Sbjct: 119 RQLMQSLNDDNPSQTETAD-VRMVPRGKDENSNGSYSNNE-------------------G 158
Query: 181 FAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISF 240
+E + ++ P+ +P TP G+P LS + SP S SP+ S +
Sbjct: 159 LSEARQRQSMTPQF-----------SPAFTPSGTPKILSTAASPRSYSAASSPKLFSGAA 207
Query: 241 ATSRGMHDDR----TSVFSSMSSSDSGAGSQTGR-TRVDGKEFFRQVRNRLSYEQFGAFL 295
+ + +D R TS SS+++S + S + R R+DGKEFFRQ R+RLSYEQF AFL
Sbjct: 208 SPTSSHYDIRMWSSTSQQSSVANSPPRSHSVSARHPRIDGKEFFRQARSRLSYEQFSAFL 267
Query: 296 ANVKELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLIT 335
AN+KELN+ KQ RE TLQKA+EIFG EN DLY F+GL+T
Sbjct: 268 ANIKELNARKQGREETLQKAEEIFGKENNDLYISFKGLLT 307
>At3g10650 hypothetical protein
Length = 1309
Score = 40.4 bits (93), Expect = 0.001
Identities = 44/171 (25%), Positives = 68/171 (39%), Gaps = 17/171 (9%)
Query: 120 RKTLMRSLQEEDDNSGGAPDM-----------VAKVQSQSSLTSTSQFGDNEASLPPSVS 168
+KT S + ++ AP + V S SLTS+ F + +++P
Sbjct: 724 KKTFSNSASGAESSTSAAPTLNGSIFSAGANAVTPPPSNGSLTSSPSFPPSISNIPSD-- 781
Query: 169 SSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTS 228
+SV + T SFA H S +I ++P S SQST+ SP +P ++
Sbjct: 782 NSVGDMPSTVQSFAATHNSSSIFGKLPTSNDSNSQSTSASPLSSTSPFKFGQPAAPF-SA 840
Query: 229 KPVSPRRHSISFAT---SRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGK 276
VS IS T + + T F M+S+D G G + K
Sbjct: 841 PAVSESSGQISKETEVKNATFGNTSTFKFGGMASADQSTGIVFGAKSAENK 891
>At3g08670 hypothetical protein
Length = 567
Score = 40.4 bits (93), Expect = 0.001
Identities = 40/138 (28%), Positives = 60/138 (42%), Gaps = 13/138 (9%)
Query: 152 STSQFGDNEASLPPS--VSSSVPSISD-TGNSFAEDHESDAIRPRVPYSLLLASQSTTPR 208
STSQ+ + PS +++S S+S S S + RP P AS+S+TP
Sbjct: 163 STSQYSSFTSGRSPSSILNTSSASVSSYIRPSSPSSRSSSSARPSTPTRTSSASRSSTPS 222
Query: 209 LTPPGSPPSLSASVSPTRTSKPVSP-RRHSISFAT---------SRGMHDDRTSVFSSMS 258
PGS S P+ +S+P +P R +S ++ SR R S S+
Sbjct: 223 RIRPGSSSSSMDKARPSLSSRPSTPTSRPQLSASSPNIIASRPNSRPSTPTRRSPSSTSL 282
Query: 259 SSDSGAGSQTGRTRVDGK 276
S+ SG GR +G+
Sbjct: 283 SATSGPTISGGRAASNGR 300
>At5g23150 transcription factor-like protein (gb|AAD31171.1)
Length = 1392
Score = 38.5 bits (88), Expect = 0.005
Identities = 66/297 (22%), Positives = 109/297 (36%), Gaps = 43/297 (14%)
Query: 47 ESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTV 106
+ E +L EK E S LD L+ ++D HR +D + +
Sbjct: 966 DEEDDDLPTSQKEKSTSAGERVSALDDLEIH-DTSSDKCHRVLEDVDHELEME------- 1017
Query: 107 RKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPS 166
DVS RK + S E+ + D++ V +S T + ++ LP
Sbjct: 1018 -----DVSGQ---RKDVAPSSFCENKTKEQSLDVMEPVAEKS--TEFNPLPEDSPPLPQE 1067
Query: 167 VSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLA-----SQSTTPRLTPPGSPPSLSAS 221
+P + + + ++ P P +L SQ P L+PP SPP
Sbjct: 1068 SPPPLPPLPPSPPPPSPPLPPSSLPPPPPAALFPPLPPPPSQPPPPPLSPPPSPPPPPPP 1127
Query: 222 VSPTRTSKPVSPRRHSISFATS--------------RGMHDDRTSVFS-SMSSSDSGAGS 266
S + T++ H I F M DR+S+F+ G S
Sbjct: 1128 PSQSLTTQLSIASHHQIPFQPGFPPPTYPLSHQTYPGSMQQDRSSIFTGDQIVQGPGNSS 1187
Query: 267 QTGRTRVDGK-EFFRQVRNRLSYEQFGAFLANVKELNSHKQTREVTLQKADEIFGPE 322
+ G GK E+F Q + S A + + +E +S +R++ +D +F PE
Sbjct: 1188 RGGLVEGAGKPEYFVQQSSSFS----PAGVCSSREPSSFTSSRQLEFGNSDVLFNPE 1240
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 38.1 bits (87), Expect = 0.007
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query: 204 STTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDS 262
++ P + P S SL+ S SP TSKP P+ H + + +G D R+SV SS ++ S
Sbjct: 292 TSAPSSSSPSSNCSLTLSASPFITSKPTKPQEH--TRPSIQGFRDTRSSVLSSRKTTRS 348
>At1g22260 hypothetical protein
Length = 851
Score = 38.1 bits (87), Expect = 0.007
Identities = 26/94 (27%), Positives = 48/94 (50%), Gaps = 10/94 (10%)
Query: 39 LSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINE 98
++A ALES +EL +I E + L SQL L + S T D L E + + L+++
Sbjct: 350 VAATKEALESAGNELNEKIVELQNDKESLISQLSGLRCSTSQTIDKL---ESEAKGLVSK 406
Query: 99 NASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
+A + + +L ++ +TL+ S++ +D
Sbjct: 407 HADAESAISQLKEEM-------ETLLESVKTSED 433
Score = 30.0 bits (66), Expect = 1.9
Identities = 22/88 (25%), Positives = 41/88 (46%), Gaps = 7/88 (7%)
Query: 38 ALSARVNALESESSELRARIAEKDHLIAELQSQL-------DSLDATLSATADNLHRAEQ 90
AL ++ +E+++LR R E + L L+S+ D L TL A + AE+
Sbjct: 78 ALEEKLQNAFNENAKLRVRKKEDEKLWRGLESKFSSTKTLCDQLTETLQHLASQVQDAEK 137
Query: 91 DKESLINENASLSNTVRKLNRDVSKLEV 118
DK + ++ S + LN+ + + +
Sbjct: 138 DKGFFETKFSTSSEAIDSLNQQMRDMSL 165
Score = 28.1 bits (61), Expect = 7.2
Identities = 25/88 (28%), Positives = 39/88 (43%), Gaps = 7/88 (7%)
Query: 51 SELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASL----SNTV 106
++L A AE+ I L SQL+ + L+ D + +E L E S+ N
Sbjct: 214 TKLEASAAERKLNIENLNSQLEKVHLELTTKEDEVKDLVSIQEKLEKEKTSVQLSADNCF 273
Query: 107 RKL---NRDVSKLEVFRKTLMRSLQEED 131
KL ++V KL+ + L+ L E D
Sbjct: 274 EKLVSSEQEVKKLDELVQYLVAELTELD 301
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 37.7 bits (86), Expect = 0.009
Identities = 33/143 (23%), Positives = 60/143 (41%), Gaps = 19/143 (13%)
Query: 37 IALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLI 96
+ LSA +NA E E L + I E + + QS++ L L+ + D L + E + S +
Sbjct: 502 VDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTELAESKDTLTQKENELSSFV 561
Query: 97 NEN------------------ASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAP 138
+ S V++LN++++ E +K L + + E A
Sbjct: 562 EVHEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQISEMSIKIKRAE 621
Query: 139 DMVAKVQSQSS-LTSTSQFGDNE 160
+ ++ S+S L + DNE
Sbjct: 622 STIQELSSESERLKGSHAEKDNE 644
Score = 35.0 bits (79), Expect = 0.059
Identities = 35/149 (23%), Positives = 66/149 (43%), Gaps = 13/149 (8%)
Query: 47 ESESSEL----RARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASL 102
ESE S L + E + EL++ ++S + ++ +L+ AE++K+ L + A L
Sbjct: 47 ESEHSSLVELHKTHERESSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAEL 106
Query: 103 SNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEAS 162
SN +++ + +L L S ++ D + ++ + S T S+
Sbjct: 107 SNEIQEAQNTMQELMSESGQLKESHSVKERELFSLRD-IHEIHQRDSSTRASE------- 158
Query: 163 LPPSVSSSVPSISDTGNSF-AEDHESDAI 190
L + SS +SD S A + E+ AI
Sbjct: 159 LEAQLESSKQQVSDLSASLKAAEEENKAI 187
Score = 33.9 bits (76), Expect = 0.13
Identities = 22/107 (20%), Positives = 49/107 (45%)
Query: 26 EQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNL 85
E+++ S S ++ L+ E + LR ++A D AEL+ QL+ +S +
Sbjct: 850 EEVEKQMVCKSEEASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEKKSEEISEYLSQI 909
Query: 86 HRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
+++ + + + S+ + L+ + E+ +TL + E D+
Sbjct: 910 TNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDE 956
Score = 33.1 bits (74), Expect = 0.22
Identities = 27/99 (27%), Positives = 43/99 (43%), Gaps = 18/99 (18%)
Query: 32 RKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQD 91
+K ++ + ++N SE L LI L+++LDSL S T L R +Q+
Sbjct: 961 KKEENVQMHDKINVASSEIMALT-------ELINNLKNELDSLQVQKSETEAELEREKQE 1013
Query: 92 KESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEE 130
K L N+ DV K V ++ +L+EE
Sbjct: 1014 KSELSNQIT-----------DVQKALVEQEAAYNTLEEE 1041
Score = 33.1 bits (74), Expect = 0.22
Identities = 26/107 (24%), Positives = 49/107 (45%), Gaps = 3/107 (2%)
Query: 43 VNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASL 102
+ L E EL+ R EK+ +EL S + S D ++ +L AE++K+ L +
Sbjct: 381 IKELMDELGELKDRHKEKE---SELSSLVKSADQQVADMKQSLDNAEEEKKMLSQRILDI 437
Query: 103 SNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSS 149
SN +++ + + + + L S ++ G D+ Q +SS
Sbjct: 438 SNEIQEAQKTIQEHMSESEQLKESHGVKERELTGLRDIHETHQRESS 484
Score = 32.3 bits (72), Expect = 0.38
Identities = 29/113 (25%), Positives = 51/113 (44%), Gaps = 8/113 (7%)
Query: 64 IAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTL 123
+ EL+ Q++S ++ L+ AE++K+ L + A LSN +++ + +L L
Sbjct: 244 VKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQL 303
Query: 124 MRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISD 176
S +D + D+ Q +SS T S+ L + SS ISD
Sbjct: 304 KESHSVKDRDLFSLRDIHETHQRESS-TRVSE-------LEAQLESSEQRISD 348
Score = 31.6 bits (70), Expect = 0.65
Identities = 29/140 (20%), Positives = 63/140 (44%), Gaps = 23/140 (16%)
Query: 15 DLVQVLPSDPFEQLDLARKITSIALSAR-----VNALESESSELRARIAEKDHLIAEL-- 67
+L Q L S E+ L+++I+ +++ + + L SES L+ AEKD+ + L
Sbjct: 591 ELNQNLNSSEEEKKILSQQISEMSIKIKRAESTIQELSSESERLKGSHAEKDNELFSLRD 650
Query: 68 ----------------QSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNR 111
++QL+S + + +++L AE++ ++ + + S+ + +
Sbjct: 651 IHETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISETSDELERTQI 710
Query: 112 DVSKLEVFRKTLMRSLQEED 131
V +L L L E++
Sbjct: 711 MVQELTADSSKLKEQLAEKE 730
Score = 30.8 bits (68), Expect = 1.1
Identities = 28/133 (21%), Positives = 63/133 (47%), Gaps = 30/133 (22%)
Query: 15 DLVQVLPSDPFEQLDLARKITSIALSAR-----VNALESESSELRARIAEKDH------- 62
+L Q L + E+ L++KI ++ + + L SES +L+ + KD
Sbjct: 260 ELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRD 319
Query: 63 -----------LIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASL-------SN 104
++EL++QL+S + +S +L AE++ +++ ++N + N
Sbjct: 320 IHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQN 379
Query: 105 TVRKLNRDVSKLE 117
T+++L ++ +L+
Sbjct: 380 TIKELMDELGELK 392
Score = 30.0 bits (66), Expect = 1.9
Identities = 18/86 (20%), Positives = 42/86 (47%)
Query: 65 AELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLM 124
+EL++QL+S +S + +L AE++ +++ ++N N + + + +L L
Sbjct: 157 SELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMAELGKLK 216
Query: 125 RSLQEEDDNSGGAPDMVAKVQSQSSL 150
S +E++ ++ Q SS+
Sbjct: 217 DSHREKESELSSLVEVHETHQRDSSI 242
Score = 28.9 bits (63), Expect = 4.2
Identities = 31/130 (23%), Positives = 63/130 (47%), Gaps = 13/130 (10%)
Query: 59 EKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVR----KLNRDVS 114
E ++EL++QL L+ + + +L+ AE++K+SL + +++ ++ K+ V+
Sbjct: 482 ESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVT 541
Query: 115 KLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSI 174
+L + TL Q+E++ S +V S+SQ + EA + S V +
Sbjct: 542 ELAESKDTL---TQKENELSS-----FVEVHEAHKRDSSSQVKELEARV-ESAEEQVKEL 592
Query: 175 SDTGNSFAED 184
+ NS E+
Sbjct: 593 NQNLNSSEEE 602
>At4g36120 myosin-like protein
Length = 981
Score = 37.4 bits (85), Expect = 0.012
Identities = 28/93 (30%), Positives = 46/93 (49%), Gaps = 4/93 (4%)
Query: 26 EQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNL 85
EQL L ++ ++ LS + LES +A + EK+ LI++L+SQL S + S L
Sbjct: 748 EQLKLEKENIAVELSRCLQNLEST----KAWLEEKEQLISKLKSQLTSSEDLQSLAETQL 803
Query: 86 HRAEQDKESLINENASLSNTVRKLNRDVSKLEV 118
+ +SL L V+ L + +LE+
Sbjct: 804 KCVTESYKSLDLHAKELEAKVKSLEEETKRLEM 836
>At4g09060 unknown protein
Length = 341
Score = 36.6 bits (83), Expect = 0.020
Identities = 34/143 (23%), Positives = 60/143 (41%), Gaps = 15/143 (10%)
Query: 24 PFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATAD 83
P L +K+ + S R L + L +R+ EKDH+I ++S+ S A
Sbjct: 38 PISMESLQKKLYTAEESQR--RLREQYQGLISRLKEKDHVIDRVRSEA-------SMNAQ 88
Query: 84 NLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAK 143
L + ++ + L +E +L + +KL ++ R LM E D+ + A V +
Sbjct: 89 ALKKFVEENQKLASECGNLLSQCKKLEKECLLYHQDRDALMEFGNESDERAREAEARVRE 148
Query: 144 VQSQSSLTSTS------QFGDNE 160
++ + S Q GD E
Sbjct: 149 LEDEIGRMSEEMQRFKRQIGDGE 171
>At5g41140 putative protein
Length = 983
Score = 36.2 bits (82), Expect = 0.026
Identities = 40/146 (27%), Positives = 71/146 (48%), Gaps = 26/146 (17%)
Query: 7 DLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESS---ELRARIAEKDHL 63
D+N DL ++ + +LDL R +++E+E+S EL+ I EK+ +
Sbjct: 724 DVNADLTHEITRRKDEIEILRLDLEE--------TRKSSMETEASLSEELQRIIDEKEAV 775
Query: 64 IAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTL 123
I L+SQL+ T A DNL K SL N + + N +++ + S+LE ++
Sbjct: 776 ITALKSQLE----TAIAPCDNL------KHSLSNNESEIENLRKQVVQVRSELEK-KEEE 824
Query: 124 MRSLQEEDDNSGGAPDMVAKVQSQSS 149
M +L +N + D + K + +S+
Sbjct: 825 MANL----ENREASADNITKTEQRSN 846
>At4g32190 unknown protein
Length = 783
Score = 36.2 bits (82), Expect = 0.026
Identities = 29/106 (27%), Positives = 47/106 (43%), Gaps = 6/106 (5%)
Query: 27 QLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLH 86
Q +L R +A AR E EL+ ++ E+D A LQS L + L +
Sbjct: 167 QEELKRANVELASQAR------EIEELKHKLRERDEERAALQSSLTLKEEELEKMRQEIA 220
Query: 87 RAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
++ I+E S S + K N V + E L R+L+E+++
Sbjct: 221 NRSKEVSMAISEFESKSQLLSKANEVVKRQEGEIYALQRALEEKEE 266
>At4g18670 extensin-like protein
Length = 839
Score = 36.2 bits (82), Expect = 0.026
Identities = 31/83 (37%), Positives = 38/83 (45%), Gaps = 13/83 (15%)
Query: 156 FGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSP 215
FG ++ PP V S P+ G S S +I P P + + S TTP +P GSP
Sbjct: 398 FGCGRSTRPPVVVPSPPTTPSPGGS----PPSPSISPSPP--ITVPSPPTTP--SPGGSP 449
Query: 216 PSLSASVSPTRT-----SKPVSP 233
PS S SP T S P SP
Sbjct: 450 PSPSIVPSPPSTTPSPGSPPTSP 472
Score = 30.4 bits (67), Expect = 1.4
Identities = 30/86 (34%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 148 SSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTP 207
SS T+ + G +S P S T S S +I P P + + S +TP
Sbjct: 483 SSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPP--ITVPSPPSTP 540
Query: 208 RLTPPGSPPSLSASVSPTRTSKPVSP 233
T PGSPPS S SPT +S SP
Sbjct: 541 --TSPGSPPSPS---SPTPSSPIPSP 561
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 36.2 bits (82), Expect = 0.026
Identities = 41/153 (26%), Positives = 59/153 (37%), Gaps = 8/153 (5%)
Query: 122 TLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPS--ISDTGN 179
TL L S + ++ Q+ S S+S + AS PS S S + TG
Sbjct: 110 TLTSRLANSSTESAARNHLTSRQQTSSPGLSSS----SGASRRPSSSGGPGSRPATPTGR 165
Query: 180 SFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSIS 239
S S + RP P S S +T P LT S ++SA+ PT S+ S ++
Sbjct: 166 SSTLTANSKSSRPSTPTSRATVSSATRPSLT--NSRSTVSATTKPTPMSRSTSLSSSRLT 223
Query: 240 FATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTR 272
S+ S S S+ S G +R
Sbjct: 224 PTASKPTTSTARSAGSVTRSTPSTTTKSAGPSR 256
Score = 28.5 bits (62), Expect = 5.5
Identities = 30/129 (23%), Positives = 47/129 (36%), Gaps = 21/129 (16%)
Query: 133 NSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRP 192
+S G P + S T T+ N S PS +S ++S + S
Sbjct: 150 SSSGGPGSRPATPTGRSSTLTA----NSKSSRPSTPTSRATVSSATRPSLTNSRSTVSAT 205
Query: 193 RVPYSLLLASQSTTPRLTPPGSPPSLSA-----------------SVSPTRTSKPVSPRR 235
P + ++ ++ RLTP S P+ S S P+R++ P+S
Sbjct: 206 TKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTKSAGPSRSTTPLSRST 265
Query: 236 HSISFATSR 244
S TSR
Sbjct: 266 ARSSTPTSR 274
>At1g77400 unknown protein
Length = 232
Score = 36.2 bits (82), Expect = 0.026
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 7/67 (10%)
Query: 172 PSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTP-RLTPPGSPPSLSASVSPTRTSKP 230
P++S +SF + +S + P P+S STTP +L PP +P SLS SP + +
Sbjct: 63 PALSPPSSSFISNSKSRPLSPLTPHSF-----STTPSKLKPPRTPSSLSGFYSPGPSFRS 117
Query: 231 VSPRRHS 237
SPR S
Sbjct: 118 -SPRAFS 123
>At5g50230 putative protein
Length = 515
Score = 35.8 bits (81), Expect = 0.034
Identities = 33/139 (23%), Positives = 60/139 (42%), Gaps = 13/139 (9%)
Query: 48 SESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVR 107
+ES +A + EK+ LI +LQ +L + + L + + LI EN + + +
Sbjct: 83 AESRTSKAILQEKELLINDLQKELTQRREDCTRLQEELEEKTKTVDVLIAENLEIRSQLE 142
Query: 108 KLNRDVSKLEVFRKTL--------MRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGD- 158
++ V K E K L M+ + ++ + +M+AK+++ T Q D
Sbjct: 143 EMTSRVQKAETENKMLIDRWMLQKMQDAERLNEANDLYEEMLAKLKANGLETLARQQVDG 202
Query: 159 ----NEASLPPSVSSSVPS 173
NE V S++PS
Sbjct: 203 IVRRNEDGTDHFVESTIPS 221
>At5g05180 unknown protein
Length = 432
Score = 35.8 bits (81), Expect = 0.034
Identities = 29/124 (23%), Positives = 58/124 (46%), Gaps = 23/124 (18%)
Query: 16 LVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARI------------------ 57
LV + E++++ RK + L A ++AL+++ + I
Sbjct: 233 LVDQIKHSEAEKMEMQRK--EVELQAEISALKTDLATRGEHIEALNKDFDKHKLRYDMLM 290
Query: 58 AEKDHLIAE---LQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVS 114
AEKD + AE L++++ S D + + L++ + L++E+ + NTV +L V
Sbjct: 291 AEKDGVCAEVDNLKAEMRSRDIQIQQMEEQLNQLVYKQTELVSESGNAKNTVEELKAVVK 350
Query: 115 KLEV 118
+LE+
Sbjct: 351 ELEI 354
>At3g45900 unknown protein
Length = 389
Score = 35.8 bits (81), Expect = 0.034
Identities = 32/149 (21%), Positives = 67/149 (44%), Gaps = 14/149 (9%)
Query: 12 LPDDLVQVLPSDPFEQLDLARKITSIALSAR-VNALESESSELRARIAEKDHLIAELQSQ 70
LP+D + ++P + +L R++ + R + + E S+ AR+ E + A ++
Sbjct: 45 LPNDAISLVPRLTSQVTELKRRLATAEEQVRQMKSRRVEDSKANARVVE---IFASHRNA 101
Query: 71 LDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEE 130
+ L + +H E+++E +N + +L R+VS+ + + R EE
Sbjct: 102 WQEEEKRL---LNRIHEIEEEREDFMNR-------ISELEREVSERDEMIGFMSRREIEE 151
Query: 131 DDNSGGAPDMVAKVQSQSSLTSTSQFGDN 159
+D+ G D ++ + LT +S N
Sbjct: 152 EDDDDGQGDESSERYAVDHLTLSSSPSPN 180
>At2g39280 unknown protein
Length = 579
Score = 35.8 bits (81), Expect = 0.034
Identities = 28/92 (30%), Positives = 47/92 (50%), Gaps = 5/92 (5%)
Query: 15 DLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSL 74
D V +L S D ++ + + + + N ES+ ELR++ + +IA + +L L
Sbjct: 439 DAVTLLQSMTGSTFDSSQLVFTACMGYQ-NVHESKLQELRSK--HRPAVIAAFEERLKGL 495
Query: 75 DATLSA--TADNLHRAEQDKESLINENASLSN 104
A + TA LH ++QD S++ ASLSN
Sbjct: 496 QAWRDSKDTATKLHNSKQDPNSVLASKASLSN 527
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.125 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,964,804
Number of Sequences: 26719
Number of extensions: 280777
Number of successful extensions: 1838
Number of sequences better than 10.0: 225
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 1566
Number of HSP's gapped (non-prelim): 378
length of query: 339
length of database: 11,318,596
effective HSP length: 100
effective length of query: 239
effective length of database: 8,646,696
effective search space: 2066560344
effective search space used: 2066560344
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0238.4


