

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
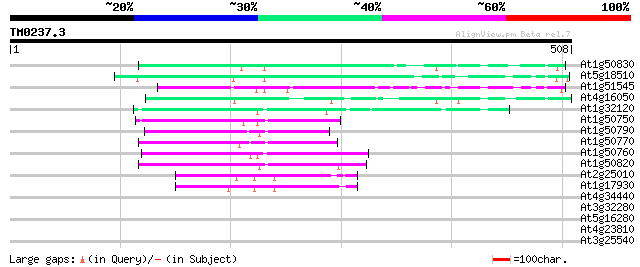
Query= TM0237.3
(508 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g50830 hypothetical protein 103 3e-22
At5g18510 putative protein 87 2e-17
At1g51545 unknown protein 82 7e-16
At4g16050 hypothetical protein 72 6e-13
At1g32120 hypothetical protein 70 2e-12
At1g50750 hypothetical protein 66 5e-11
At1g50790 hypothetical protein 65 1e-10
At1g50770 hypothetical protein 65 1e-10
At1g50760 hypothetical protein 62 1e-09
At1g50820 hypothetical protein 61 2e-09
At2g25010 unknown protein 47 2e-05
At1g17930 unknown protein 42 8e-04
At4g34440 serine/threonine protein kinase - like 32 0.83
At3g32280 hypothetical protein 31 1.8
At5g16280 putative protein 29 5.4
At4g23810 unknown protein 28 9.1
At3g25540 unknown protein 28 9.1
>At1g50830 hypothetical protein
Length = 768
Score = 103 bits (256), Expect = 3e-22
Identities = 108/408 (26%), Positives = 161/408 (38%), Gaps = 57/408 (13%)
Query: 117 DKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPF 176
D ++ WL ++E Q W+ GI + I++S ++ NP+++LS W T P+
Sbjct: 79 DAPFVSWLAKMEALHAQTWRKAGIFEAIKVSTYSITKNPSLILSVSEKWCPETKSFVFPW 138
Query: 177 GMITPTLLDVAAITGLWPIGDDYHAAPATAKP---------ISIPTGNISFSRFVKDHYV 227
G T TL DV + G +G A T++ + I N S R V
Sbjct: 139 GEATITLEDVMVLLGFSVLGSPVFAPLETSETRDSVKKLENVRIQHMNSSTDRRVSQKSW 198
Query: 228 ES---GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPAKLLPLANLLHEGRQLAMARLVLGN 283
S G D EHVAFL+ WLS +VF KS R I + P+A L G ++A+A +L
Sbjct: 199 VSTFLGRGGDMEHVAFLVLWLSLFVFPVKSRRNISNHVFPIAVRLARGERIALAPAILAI 258
Query: 284 LYQMLNEAVEDIRNVKTVSLNAAGPLWLFQLWLNAVFESLLPARDNPPAVSNTRIDAHRL 343
LY+ L+ E R + L Q+W F ++ P + P H L
Sbjct: 259 LYRDLDRIHEVSREDCVDKFHLESLFKLVQVWTWERFRNIRPKASDIPKGEPRIAQWHGL 318
Query: 344 ETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPYSKATRG----SFWLRNSYPNASDS 399
+ DA DF + PY+KA F+L + D
Sbjct: 319 HRRSK--DAWFCFDDF---------------EWRPYTKALNNWNPFRFYLEEAIWVTVDE 361
Query: 400 ALPKEHHITLWRTILSPRVLTVGFASSDYTLCGYNPQLVSRQFGLSQALPNTLFDKSLVL 459
++ E + R + +++ GF Y P V+RQFGL Q LP
Sbjct: 362 SIDDE-FASFARCVTVSQIVGDGFVED------YFPNRVARQFGLDQDLPGL-------- 406
Query: 460 YPGAVKRAYAFDTTVHFYNKRLLNLSPFTYAPSYY----ATNAFKAWW 503
+R + YNK L+ L+ Y PS T ++ WW
Sbjct: 407 --ATCQRNSTEKEAWNDYNKSLIGLN--LYMPSRLDQGSVTARYRVWW 450
>At5g18510 putative protein
Length = 702
Score = 87.0 bits (214), Expect = 2e-17
Identities = 106/436 (24%), Positives = 172/436 (39%), Gaps = 56/436 (12%)
Query: 96 MPFLNDPKRAFRSAPPNPS-----ANDKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQ 150
+P L+D +R S+ N S A+ +L WL +++ WK GI + I+ S +
Sbjct: 31 LPRLDDQQRLSDSSLHNTSSGFWAADHHFFLSWLGKMQALYEPIWKKAGIFEAIKASTYK 90
Query: 151 VSYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWPIGDDYHA-------AP 203
+ + + +LS W T P+G T TL DV + G +G +
Sbjct: 91 IIKDTSSILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPVFSPLECSEMRD 150
Query: 204 ATAKPISIPTGNISFSRFVKDHYVES---GEVSDAEHVAFLLYWLSAYVFCTKSLR-IPA 259
+ K + ++ ++ V S G EH AFL+ WLS +VF K R I
Sbjct: 151 SAEKLEKVRRDSLGKAKRVSQRSWTSSFMGRGGQMEHEAFLVLWLSLFVFPGKFCRSIST 210
Query: 260 KLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNVKTVSLNAAGPLWLFQLWLNAV 319
++P+A L G ++A+A VL LY+ L+ + R +N L Q+W
Sbjct: 211 NVIPIAVRLARGERIALAPAVLAFLYKDLDRICDFSRGKCAGKVNLKSLFKLVQVWTWER 270
Query: 320 FESLLPARDNPPAVSNTRIDAHRLETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPY 379
F ++ P P L+ ++ S ++R Y +++P
Sbjct: 271 FSNIRPKAKEIPKGEPRIAQWDGLQQISKNVKLSFDVFEWRPYTKPL-------KNWNPL 323
Query: 380 SKATRGSFWLRNSYPNASDSALPKEHHITLWRTILSPRVLTVGFASSDYTLCGYNPQLVS 439
+ WL + ++ D A R + V + + + + Y P V+
Sbjct: 324 RFYVDEAMWL--TVDDSVDDAFAS-----------FARCVKVSYLAGNGFVEDYFPYRVA 370
Query: 440 RQFGLSQALPNTLFDKSLVLYPGAVKRAYAFDTTVHFYNKRLLNLSPFTYAPSY----YA 495
RQFGLSQ LP LV + A+D Y+ L L+ Y PS Y
Sbjct: 371 RQFGLSQDLP------GLVTRRRKITEKDAWDD----YSNSLEGLN--LYLPSQLDRGYV 418
Query: 496 TNAFKAWW----SEYW 507
T ++ WW SEY+
Sbjct: 419 TARYQDWWFKSASEYF 434
>At1g51545 unknown protein
Length = 629
Score = 82.0 bits (201), Expect = 7e-16
Identities = 104/388 (26%), Positives = 159/388 (40%), Gaps = 57/388 (14%)
Query: 135 WKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWP 194
W+ GI + I+ S ++ N ++LL+ + W T P+G T TL DV + G
Sbjct: 9 WRKAGIFEAIKASMYKIRKNQSLLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSV 68
Query: 195 IGDDYHAAPATAKPISIPTGNISFSRFV---------KDHYVES--GEVSDAEHVAFLLY 243
G A P + + + +R ++ +V S G EH AFL +
Sbjct: 69 QGSPVFA-PLESSEMRDSVEKLEKARLENRGQDGLVRQNLWVSSFLGRGDQMEHEAFLAF 127
Query: 244 WLSAYVF---CTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNVKT 300
WLS +VF C +S I K+LP+A L G ++A A VL LY+ L + R T
Sbjct: 128 WLSQFVFPDMCRRS--ISTKVLPMAVRLARGERIAFAPAVLARLYRDLGQIQASAREKST 185
Query: 301 VSLNAAGPLWLFQLWLNAVFESLLP-ARDNPPAVSNTRIDAHRLETLTPAYDASSFEADF 359
++ L QLW F+S P AR P RI +T + + + D+
Sbjct: 186 PNVTLKSLFKLVQLWAWERFKSTSPKARVIPK--GEPRISRWHSQT-SKNVRLNLVDFDW 242
Query: 360 RKYFTMFLELKHYRSSFSPYSKATRGSFWLRNSYPNASDSALPKEHHITLWRTILSPRVL 419
R Y T L++ ++P + W+ N D ++ R + +++
Sbjct: 243 RPY-TKPLQI------WNPPRFYPEEAMWMTVD-DNLDDG------FVSFARCMRVSQLV 288
Query: 420 TVGFASSDYTLCGYNPQLVSRQFGLSQALPNTLFDKSLVLYPGAVKRAYAFDTTVHFYNK 479
VG Y P V+ QFGL+Q LP + D S + A+D YNK
Sbjct: 289 GVGIVED------YYPNRVAMQFGLAQDLPGLVTDHS------SFTEKEAWDG----YNK 332
Query: 480 RLLNLSPFTYAPSYYATNA----FKAWW 503
L L Y PS AT + ++ WW
Sbjct: 333 SLDGL--MLYIPSRVATTSVTERYRDWW 358
>At4g16050 hypothetical protein
Length = 900
Score = 72.4 bits (176), Expect = 6e-13
Identities = 91/398 (22%), Positives = 155/398 (38%), Gaps = 63/398 (15%)
Query: 124 LDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPFGMITPTL 183
+D + + W GI + I+ S ++ NP+++LS W TN P+G T TL
Sbjct: 330 IDGFQALHKPTWLKSGIFEAIKASTYRIHKNPSLILSLAQNWCPETNTFVFPWGEATITL 389
Query: 184 LDVAAITGLWPIGDDYHAA--PATAKPISIPTGNISFSRFVKDHYVESGEVSDAEHVAFL 241
DV + G G A+ + K ++ ++ S + EH AFL
Sbjct: 390 EDVNVLLGFSISGSSVFASLQSSEMKEAVEKLQKRCQGSMKQESWISSFVDDEMEHEAFL 449
Query: 242 LYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNE--AVEDIRNVK 299
+ WLS +VF K G ++A A VL NLY L + I+NV
Sbjct: 450 VLWLSKFVFPDK-------------FCTRGERIAFAPAVLANLYNDLGHICVLASIQNVL 496
Query: 300 TVSLNAAGPLWLFQLWLNAVFESLLPARDNPPAVSNTRIDAHRLETLTPAYDASSFEADF 359
SL L Q+W+ F+S+ P P RI + +
Sbjct: 497 ASSL-----FKLVQVWIWERFKSIRPEAKVIPR-GQPRI--------------AQWSGLK 536
Query: 360 RKYFTMFLELKHYRSSFSPYSKATRG----SFWLRNSYPNASDSALPKEH-----HITLW 410
+++ + L + H + PYS+ F++ + D +L ++ ++
Sbjct: 537 QRFKNVGLIIFHGNFDWRPYSEPLENWNPPRFYVEEAKWVRIDESLDGDYDDDDEFVSFA 596
Query: 411 RTILSPRVLTVGFASSDYTLCGYNPQLVSRQFGLSQALPNTLFDKSLVLYPGAVKRAYAF 470
R + +++ +G + Y P V+ QFGL+Q +P + +R +
Sbjct: 597 RCVRVSKLVGIGVVEN------YYPNRVAMQFGLAQDVP---------VLGTNHRRNFTE 641
Query: 471 DTTVHFYNKRLLNLSPFTYAPSYYATNAFKAWWSEYWA 508
+ YNK L+ L Y PS AT + + ++WA
Sbjct: 642 EEAWDDYNKPLVGLK--LYFPSRVATASVTTRYRDWWA 677
>At1g32120 hypothetical protein
Length = 1206
Score = 70.5 bits (171), Expect = 2e-12
Identities = 79/356 (22%), Positives = 139/356 (38%), Gaps = 33/356 (9%)
Query: 113 PSANDKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCL 172
PS N +++W++ + ++ WK G+ D I SR Q+ + ++++ + W TN
Sbjct: 80 PSLN---WIEWVNVMAKSHATVWKKSGVYDAILASRYQIKRHDDLIVALVEKWCIETNTF 136
Query: 173 HVPFGMITPTLLDVAAITGLWPIGDDYHAAPATAKPISIPTGNISFSRFVK--------- 223
P+G T TL D+ + GL G++ A + R+++
Sbjct: 137 VFPWGEATLTLEDMIVLGGLSVTGNNALAPVKRDGMKEVEEKMKEAKRYIEVSLEKKCCV 196
Query: 224 ----DHYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARL 279
+ SG ++ EH AF++ WLS +VF + KL P A L +G +LA+A
Sbjct: 197 SMWMKEMMNSG--NEIEHEAFMVSWLSRFVFTNSGDVLREKLFPAAVQLAKGVRLALAPA 254
Query: 280 VLGNLY---QMLNEAVEDIRNVKTVSLNAAGPLWLFQLWLNAVFESLLPARDNPPAVSNT 336
VL +Y +L E + +TV + + P Q+W F +L P P +
Sbjct: 255 VLARIYGDLGVLKEFLTGYSEKETVVVKS--PFQFVQVWALERFMALQPP-GQPSQLKTG 311
Query: 337 RIDAHRLETLTPAYDASSFEADFRKYFTMFLELKHYRSSFSPYSKATRGSFWLRNSYPNA 396
R D + + R E YR P + F+ + +
Sbjct: 312 EPRIARWHHYGGGQDVYGYPENIRAVLDSAKESFDYRPYTKPVNNFRFPKFYFED---DC 368
Query: 397 SDSALPKEHHITLWRTILSPRVLTVGFASSDYTLCGYNPQLVSRQFGLSQALPNTL 452
P E+ + R + +++ + Y P V+ QFG Q +P +
Sbjct: 369 WVRVRPDENIVAFGRCLRFAKLVGLDCIEP------YYPHRVALQFGYDQDVPGVV 418
>At1g50750 hypothetical protein
Length = 816
Score = 65.9 bits (159), Expect = 5e-11
Identities = 50/197 (25%), Positives = 90/197 (45%), Gaps = 15/197 (7%)
Query: 115 ANDKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHV 174
AN K + W + W++ GI + + S ++ NP ++L W T
Sbjct: 35 ANTK-FESWAIEMAALHEPTWREAGIFEAVMASIYRIPKNPDLILGIAEKWCPYTKTFVF 93
Query: 175 PFGMITPTLLDVAAITGLWPIGDDYHAA-PATAKPIS---------IPTGNISFSRFVK- 223
P+G TL DV ++G +G A ++ K + I ++F V
Sbjct: 94 PWGETAVTLEDVMVLSGFSVLGSPVFATLDSSGKEVKAKLDKEWKKIKKAKVNFVTQVAW 153
Query: 224 -DHYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLG 282
+ +++SG+ + EHVAFL+ WL+ +VF ++ + + P+ L G ++A+A VL
Sbjct: 154 MERFMDSGD--ELEHVAFLVLWLNYFVFPSRLYHLYKAVFPIVVHLSTGTRIALALAVLA 211
Query: 283 NLYQMLNEAVEDIRNVK 299
+LY L + D+ V+
Sbjct: 212 HLYAELIASSLDMGEVQ 228
>At1g50790 hypothetical protein
Length = 812
Score = 64.7 bits (156), Expect = 1e-10
Identities = 49/180 (27%), Positives = 81/180 (44%), Gaps = 16/180 (8%)
Query: 123 WLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPFGMITPT 182
W ++ W+ GI + I S ++ N +++ W TN +G T T
Sbjct: 67 WARKMSALHEPIWRKAGIFEAILASTYKIFKNTDLVMGIAEKWCPDTNTFVFSWGEATIT 126
Query: 183 LLDVAAITGLWPIGDDYHAA-PATAKPISIPTGNISFSRFVKD------------HYVES 229
L DV + G +G A ++ K I G + + KD +++S
Sbjct: 127 LEDVMVLLGFSVLGSPVFATLDSSGKEIMAKLGK-EWLKIKKDKGTFVTQIAWIERFMDS 185
Query: 230 GEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLN 289
G+ + EH+AFL+ WLS +VF T+ I + P+A L G ++A+A VL +LY L+
Sbjct: 186 GD--ELEHLAFLVLWLSYFVFPTRYYHIYEAIWPIAIHLSNGTKMALAPAVLAHLYADLS 243
>At1g50770 hypothetical protein
Length = 632
Score = 64.7 bits (156), Expect = 1e-10
Identities = 46/193 (23%), Positives = 82/193 (41%), Gaps = 15/193 (7%)
Query: 117 DKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPF 176
+K + W ++ W+ GI + + S +++ N ++L W T P+
Sbjct: 60 NKNFKSWARKMAALHEPIWRKAGIFEAVTASTYKINPNTELVLGIAEKWCPDTKTFVFPW 119
Query: 177 GMITPTLLDVAAITGLWPIGDDYHAAPATA------------KPISIPTGNISFSRFVKD 224
G T TL DV + G +G ++ K + GN + R K+
Sbjct: 120 GETTITLEDVMLLLGFSVLGSPVFVTLDSSGEIIREKLEKEWKKVKKDKGNAT-QRTWKE 178
Query: 225 HYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNL 284
+++SG+ + EHVAFL+ WLS +VF ++ I +LP+A L RL + +
Sbjct: 179 RFMDSGD--ELEHVAFLVLWLSYFVFPSRYYHIYGAILPIAVHLSSDNDEVDGRLTIAQM 236
Query: 285 YQMLNEAVEDIRN 297
+ D++N
Sbjct: 237 VGPSKKKYSDVKN 249
>At1g50760 hypothetical protein
Length = 649
Score = 61.6 bits (148), Expect = 1e-09
Identities = 53/220 (24%), Positives = 91/220 (41%), Gaps = 16/220 (7%)
Query: 120 YLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPFGMI 179
+ +W ++ W+ GI + + S ++ N ++L W T +G
Sbjct: 53 FKRWARKMSALHEPIWRKAGIFEAVNASTYKIHKNTDLVLGVAEKWSPDTKTFVFSWGEA 112
Query: 180 TPTLLDVAAITGLWPIGDDYHAA-PATAKPISIPTGNI--SFSRFVK----------DHY 226
T TL DV ++G +G A ++ K I N + R K D +
Sbjct: 113 TITLEDVMVLSGFSVLGSPAFATLDSSGKKIMERLKNAWQTIKREGKVNLLTQVAWMDRF 172
Query: 227 VESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQ 286
+ G + EHVAFL+ WL +V ++ + + P+A L G ++A+A VL +LY
Sbjct: 173 MNCG--GELEHVAFLVLWLGYFVLPSRFHHLCETIFPIAVNLSSGTRIALAPAVLAHLYA 230
Query: 287 MLNEAVEDI-RNVKTVSLNAAGPLWLFQLWLNAVFESLLP 325
L+ I ++ V + + L Q+W F L P
Sbjct: 231 DLSLLKTHISKSPPRVEIRLSALFMLVQVWTWERFRELQP 270
>At1g50820 hypothetical protein
Length = 528
Score = 60.8 bits (146), Expect = 2e-09
Identities = 53/223 (23%), Positives = 91/223 (40%), Gaps = 18/223 (8%)
Query: 117 DKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFWERSTNCLHVPF 176
+K + W ++ W+ GI + + S ++ + ++L W T P+
Sbjct: 57 NKKFKSWARKMASLHEPIWRKAGIFEAVIASTYKIPKDTDLVLGLAEKWCPDTKTFIFPW 116
Query: 177 GMITPTLLDVAAITGLWPIGDDYHAAPATAKPISIPTGNISFSRFVKD------------ 224
G T TL DV + G +G A ++ + + + D
Sbjct: 117 GEATITLEDVMVLLGFSVLGLPVFATVDSSGKEIMAKLEKEWKKIKNDKVCLVTKLAWME 176
Query: 225 HYVESGEVSDAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARLVLGNL 284
++ SG+ + EHV FL+ WLS + F + I +LP+A L G ++A+A VL +L
Sbjct: 177 RFMNSGD--ELEHVGFLVLWLSYFAFPSHLFHISEAILPVAVHLSSGTKMALAPAVLAHL 234
Query: 285 YQMLNEAVEDIR----NVKTVSLNAAGPLWLFQLWLNAVFESL 323
Y L+ IR ++ V L+ L Q+W F L
Sbjct: 235 YADLSLLQGHIRVFSESLIKVQLDLNALFKLVQVWAWERFREL 277
>At2g25010 unknown protein
Length = 509
Score = 47.4 bits (111), Expect = 2e-05
Identities = 50/187 (26%), Positives = 80/187 (42%), Gaps = 28/187 (14%)
Query: 151 VSYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWPIGDDYHAAPA------ 204
+S N +++ + + W R TN H+P G +T TL +VA + GL GD +
Sbjct: 69 MSLNNSLISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAM 128
Query: 205 ------TAKPISIPTGNISFSR----FVKDHYVESGEVSDAEHV-----AFLLYWLSAYV 249
K S ++ SR ++K + E E + + V A+LLY + + +
Sbjct: 129 DMCGRLLGKLPSAANKEVNCSRVKLNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTI 188
Query: 250 FC-TKSLRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNVKTVSLNAAGP 308
F T ++ K LPL + + A L LY+ L A ++K+ S N G
Sbjct: 189 FATTDGDKVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNA-----SLKSQS-NICGC 242
Query: 309 LWLFQLW 315
L L Q W
Sbjct: 243 LTLLQCW 249
>At1g17930 unknown protein
Length = 478
Score = 42.0 bits (97), Expect = 8e-04
Identities = 42/184 (22%), Positives = 78/184 (41%), Gaps = 25/184 (13%)
Query: 151 VSYNPAMLLSALYFWERSTNCLHVPFGMITPTLLDVAAITGLWPIG---------DDYHA 201
+S N +++ + + W R TN H P G +T TL +V+ I GL G D+ +
Sbjct: 60 ISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEKDEDPS 119
Query: 202 APATAKPISIPTGNISFSR----FVKDHYVESGEVSDAEHV-----AFLLYWLSAYVFCT 252
+P G +S +R ++K+ + E + + + + A+L+Y + + +F T
Sbjct: 120 QVCLRLLGKLPKGELSGNRVTAKWLKESFAECPKGATMKEIEYHTRAYLIYIVGSTIFAT 179
Query: 253 KS-LRIPAKLLPLANLLHEGRQLAMARLVLGNLYQMLNEAVEDIRNVKTVSLNAAGPLWL 311
+I L L + + A L LY+ + A + +++ G L L
Sbjct: 180 TDPSKISVDYLILFEDFEKAGEYAWGAAALAFLYRQIGNASQRSQSI------IGGCLTL 233
Query: 312 FQLW 315
Q W
Sbjct: 234 LQCW 237
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 32.0 bits (71), Expect = 0.83
Identities = 14/31 (45%), Positives = 22/31 (70%)
Query: 345 TLTPAYDASSFEADFRKYFTMFLELKHYRSS 375
+++ YDASS+ AD +K+ + LE K Y+SS
Sbjct: 609 SVSSEYDASSYTADMKKFKKLALENKEYQSS 639
>At3g32280 hypothetical protein
Length = 474
Score = 30.8 bits (68), Expect = 1.8
Identities = 16/46 (34%), Positives = 25/46 (53%)
Query: 234 DAEHVAFLLYWLSAYVFCTKSLRIPAKLLPLANLLHEGRQLAMARL 279
+ EHVAFL+ WL +VF + + + P+A L G + A+ L
Sbjct: 10 ELEHVAFLVLWLRYFVFPSGFHYLYVTMFPIAIHLSSGTKTALVVL 55
>At5g16280 putative protein
Length = 1265
Score = 29.3 bits (64), Expect = 5.4
Identities = 21/70 (30%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query: 434 NPQLVSRQFGLSQALPNTLFDKSLVLYPGAVKRA-YAFDTTVHFYNKRLLNLSPFTYAPS 492
N Q+ + + GL N L+ K P A K + YAF + FY+ + +S +A +
Sbjct: 292 NQQISATRKGLKNQFKNFLWRKGKDDNPDATKGSMYAFLNSCSFYDYLDIFISSTDFAST 351
Query: 493 YYATNAFKAW 502
+ N KAW
Sbjct: 352 FEDYNIDKAW 361
>At4g23810 unknown protein
Length = 324
Score = 28.5 bits (62), Expect = 9.1
Identities = 16/47 (34%), Positives = 27/47 (57%)
Query: 39 KSQVLIPFSVNGTLRAILGPLKVVKHDRPNSLPEEHESFHPSVRGEE 85
+S +L+ +S + +++ I P+ VV P S+PE S + S R EE
Sbjct: 66 RSLLLLNWSSSPSVQLIPTPVTVVPVANPGSVPESPASINGSPRSEE 112
>At3g25540 unknown protein
Length = 310
Score = 28.5 bits (62), Expect = 9.1
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 1/54 (1%)
Query: 112 NPSANDKAYLKWLDRVEEAKRQHWKDVGILDLIQLSRSQVSYNPAMLLSALYFW 165
N S N K K +V + K WK + L L+ S V+YN + LYFW
Sbjct: 60 NKSDNIKDRKKNSPKVRKFKESAWKCIYYLSAELLALS-VTYNEPWFSNTLYFW 112
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,868,910
Number of Sequences: 26719
Number of extensions: 496981
Number of successful extensions: 1112
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 1079
Number of HSP's gapped (non-prelim): 23
length of query: 508
length of database: 11,318,596
effective HSP length: 104
effective length of query: 404
effective length of database: 8,539,820
effective search space: 3450087280
effective search space used: 3450087280
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0237.3


