

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0237.12
(2349 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
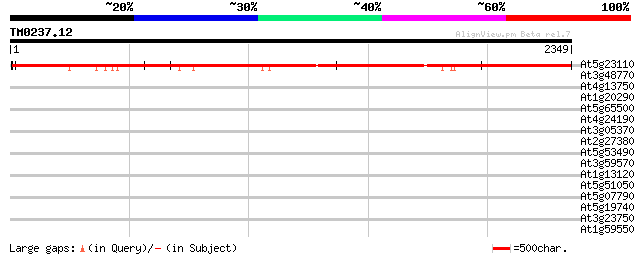
Score E
Sequences producing significant alignments: (bits) Value
At5g23110 putative protein 3091 0.0
At3g48770 hypothetical protein 43 0.002
At4g13750 hypothetical protein 42 0.003
At1g20290 hypothetical protein 36 0.23
At5g65500 putative protein 35 0.40
At4g24190 HSP90-like protein 35 0.52
At3g05370 putative disease resistance protein 33 2.6
At2g27380 hypothetical protein 33 2.6
At5g53490 thylakoid lumenal 17.4 kD protein, chloroplast precurs... 32 4.4
At3g59570 putative protein 32 4.4
At1g13120 unknown protein 32 4.4
At5g51050 calcium-binding transporter-like protein 32 5.8
At5g07790 putative protein 32 5.8
At5g19740 peptidase-like protein 31 7.5
At3g23750 receptor-like kinase, putative 31 9.8
At1g59550 hypothetical protein 31 9.8
>At5g23110 putative protein
Length = 4706
Score = 3091 bits (8014), Expect = 0.0
Identities = 1532/2349 (65%), Positives = 1860/2349 (78%), Gaps = 17/2349 (0%)
Query: 7 ESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDS 66
+S+ LEDFGQ VDLTRRIREVLLNYPEGTTVLKELIQNADDAGAT V LCLDRR H S
Sbjct: 2 DSLLLEDFGQKVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATKVRLCLDRRVHGSGS 61
Query: 67 LLSSSLSQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTD 126
LLS SL+QWQGP+LLAYNDAVFTE+DF+SIS+IGGS KHGQA KTGRFGVGFNSVYHLTD
Sbjct: 62 LLSDSLAQWQGPSLLAYNDAVFTEEDFVSISRIGGSGKHGQAWKTGRFGVGFNSVYHLTD 121
Query: 127 LPSFVSGKYVVLFDPQGVYLPRVSAANPGKRIDFTGSSALSLYKDQFSPYCAFGCDMQSP 186
+PSFVSGKYVVLFDPQG YLP +SAANPGKRID+ GSSALS YKDQF PYCAFGCDM+SP
Sbjct: 122 IPSFVSGKYVVLFDPQGAYLPNISAANPGKRIDYVGSSALSQYKDQFLPYCAFGCDMRSP 181
Query: 187 FAGTLFRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTLLFLKSVLCIEMYVW 246
F GTLFRFPLRN +QA+ S+LSRQAY +DIS MF QLFEEGV +LLFLK VL IEMY W
Sbjct: 182 FNGTLFRFPLRNTEQAASSRLSRQAYFEDDISLMFDQLFEEGVFSLLFLKCVLSIEMYTW 241
Query: 247 DAGEPKPKKIHSCSVSSVSDDTIWHRQALLRLSK-SLNTTTEVDAFPLEFVTEAVRGVET 305
D G+ +PKK++SCSVSS ++DT+WHRQA+LRLSK S++ E+DAF LEF++E+ +G +T
Sbjct: 242 DDGDSEPKKLYSCSVSSPNNDTVWHRQAVLRLSKTSISGDREMDAFTLEFLSESEKGNQT 301
Query: 306 VRQVDRFYIVQTMASASSRIGSFAITASKEYDIQLLPWASIAACISDNSLNNDVLRTGQA 365
R+ DRFYIVQTMASASS+IG FA TASKEYDI LLPWAS+AACISD+S N++L+ G A
Sbjct: 302 KRRTDRFYIVQTMASASSKIGLFAATASKEYDIHLLPWASVAACISDDSSENNILKLGHA 361
Query: 366 FCFLPLPVRTGLSVQVNGFFEVSSNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPAFVH 425
FCFLPLPVRTGL+VQVNG+FEVSSNRRGIWYG+DMDRSGKVRS WNRLLLED+VAP+F
Sbjct: 362 FCFLPLPVRTGLTVQVNGYFEVSSNRRGIWYGEDMDRSGKVRSAWNRLLLEDVVAPSFAR 421
Query: 426 MLHGVKELLGPTDIYYSLWPTGSFEEPWSILVQQIYINICNAPVIYSNLGGGRWVSPSEA 485
+L ++E+L D Y+SLWP+GSFE PWSILV+QIY NICNAPV++S+L GG+WVSP++A
Sbjct: 422 LLLCLREVLDSRDSYFSLWPSGSFEAPWSILVEQIYKNICNAPVLFSDLDGGKWVSPADA 481
Query: 486 FLHDEKFTKSKDLSLALMQLGMPVVHLPNSLFDMLLKYNS---SKVITPGTVRQFLRECE 542
+LHDE+F+ SKDL AL+QL MP+V LP +FDMLLK+ S KV+TP VR FL+EC+
Sbjct: 482 YLHDEEFSGSKDLGDALLQLEMPIVCLPRLVFDMLLKHPSFLLPKVVTPDRVRNFLKECK 541
Query: 543 SCNHLSRAHKLLLLEYCLEDLVDDDVGKAAYNLPLLPLANGNFASFLEASKGIPYFICDE 602
+ + L ++ KL+LLEYCL+DL DD V A NL LLPLANG+F F + + YFICDE
Sbjct: 542 TLSALKKSLKLVLLEYCLDDLTDDSVCTQASNLKLLPLANGDFGFFSGRTGSVSYFICDE 601
Query: 603 LEYKLLEPVSDRVIDQSIPPNILTRLSGIAMSSNTNIALFSIHHFAHLFPVFMPDDWKYK 662
LE+ LL+ V DRVID++IPP + TRL IA S N+A+FSIH+ LFP +P +WK++
Sbjct: 602 LEHMLLQKVYDRVIDKNIPPPLYTRLFAIAESRTANVAIFSIHNLLQLFPRLVPAEWKHR 661
Query: 663 CKVFWDPDSCQK-PTSSWFVLFWQYLGKQSEILPLFKDWPILPSTSGHLLRPSRQLKMIN 721
K+ W P+S + P+SSWFVLFWQYL K+ + L LF DWPILPSTSG+L S Q K+IN
Sbjct: 662 SKISWHPESNRDHPSSSWFVLFWQYLDKRCQSLSLFCDWPILPSTSGYLYIASPQSKLIN 721
Query: 722 GSTLSDTVQDILVKIGCHILKPGYVVEHPDLFSYLCGGNAAGVLESIFNAFSS-AENMQV 780
L V+++L KIG IL VEH DL S++ + GVLESIF+A SS + +Q
Sbjct: 722 AEKLPAAVRNVLEKIGGKILNNNIKVEHSDLSSFVSDASYTGVLESIFDAASSDLDGVQN 781
Query: 781 SFSSLIAEERNELRRFLLDPQWYVGHSMDEFNIRFCRRLPIYQVYHREPTQDSQFSDLEN 840
L A+E++ELR FLLDP+W++GH + + +R C+ LPI+++Y Q+S++SDL N
Sbjct: 782 LIYDLNAQEKDELRSFLLDPKWHIGHQIGDLYLRICKILPIHRIYGETSAQESKYSDLVN 841
Query: 841 PRKYLPPLDVPEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQA 900
P K+LPPLDVP +L G EFI+ +EED+LSRYYG+ERM K+ FY+++VF+R+ LQ
Sbjct: 842 PPKHLPPLDVPACLL-GCEFILCCQGSEEDVLSRYYGIERMRKSNFYRQNVFNRIEVLQP 900
Query: 901 EDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIGTLKCPSVLYDPRNEEIYALLE 960
E RD +M+S+LQ+LP L LED +R+ L+NL+F+PTV G LK PSVL+DPRNEE+YALLE
Sbjct: 901 EIRDQVMISILQDLPQLCLEDRLLREELQNLEFVPTVNGPLKRPSVLHDPRNEELYALLE 960
Query: 961 DSDSFPSGVFRESETLDIMRGLGLKTSVSPDTVLESARCIEHLMHEDQQKAYLKGKVLFS 1020
DSD FP+ F+ S LD+++GLGLKT+VSP+T+LESAR +E LMH+D +KA+ +GKVLFS
Sbjct: 961 DSDCFPASGFQGSAILDMLQGLGLKTTVSPETILESARLVERLMHKDLEKAHSRGKVLFS 1020
Query: 1021 YLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSDIEKFWNDLQLISWCPVLVSP 1080
+LEVNA+KWLPD+ + GA+NRI S+AATAFR RN ++ KFW++L++I WCPVLVS
Sbjct: 1021 FLEVNAVKWLPDQSSEDDGAINRIFSRAATAFRPRNLTCNLVKFWSELKMICWCPVLVSA 1080
Query: 1081 PFHSLPWPVVSSMVAPPKVVRPPNDLWLVSAGMRILDGECSSTALLYCLGWMCPPGGGVI 1140
PF +LPWPVV+S VAPPK+VRP D+WLVSA MRILDGECSSTAL Y LGW+ PGG I
Sbjct: 1081 PFQTLPWPVVTSTVAPPKLVRPKTDMWLVSASMRILDGECSSTALAYNLGWLSHPGGSAI 1140
Query: 1141 AAQLLELGKNNEIVTDQVLRQELALAMPRIYSILTGMIGSDEIEIVKAVLEGCRWIWVGD 1200
AAQLLELGKNNEI+ DQVLRQELALAMP+IYSIL ++GSDE++IVKAVLEG RWIWVGD
Sbjct: 1141 AAQLLELGKNNEILIDQVLRQELALAMPKIYSILARLLGSDEMDIVKAVLEGSRWIWVGD 1200
Query: 1201 GFATSDEVVLDGPLHLAPYIRVIPVDLAVFKNLFLELGIREFLQPSDYVNILFRMANKKG 1260
GFAT EVVLDGPL L PYIRVIP DLAVF+ LF+ELG+REFL PSDY ++L R+A +KG
Sbjct: 1201 GFATLSEVVLDGPLQLVPYIRVIPTDLAVFRGLFVELGVREFLTPSDYADVLCRIAVRKG 1260
Query: 1261 SSPLDTQEIRAVMLIVHHLAEVYLHGQKVQLYLPDVSGRLFLAGDLVYNDAPWLLGSEDP 1320
+SPLD QEIRA +LI LAE KV +YLPDVSGRLF + DLVYNDAPWL S++
Sbjct: 1261 TSPLDPQEIRAAVLIAQQLAEAQFL-DKVTIYLPDVSGRLFPSSDLVYNDAPWLTASDNL 1319
Query: 1321 DGSFGNAPSVTWNAKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADSMNFGLSGAAE 1380
+ SF ++ NAKRT+QKFVHGNISN+VAEKLGVRSLRR+LLAESADSMNF LSGAAE
Sbjct: 1320 NSSFSAESTMLLNAKRTMQKFVHGNISNEVAEKLGVRSLRRVLLAESADSMNFSLSGAAE 1379
Query: 1381 AFGQHEALTTRLKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYGTSSVLSPEM 1440
AFGQHEALTTRLKHILEMYADGPG LFELVQNAEDAGASEV FLLDK+ YGTSS+LSPEM
Sbjct: 1380 AFGQHEALTTRLKHILEMYADGPGILFELVQNAEDAGASEVTFLLDKTHYGTSSLLSPEM 1439
Query: 1441 ADWQGPALYCFNDSVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPMFVS 1500
ADWQGPALYCFN+SVFT QD+YAISRIGQ SKLEK FAIGRFGLGFNCVYHFTDIP FVS
Sbjct: 1440 ADWQGPALYCFNNSVFTQQDMYAISRIGQASKLEKPFAIGRFGLGFNCVYHFTDIPGFVS 1499
Query: 1501 GENIVMFDPHASNLPGISPSHPGLRIKFAGRKILEQFPDQFSSLLHFGCDLQHPFPGTLF 1560
GENIVMFDPHA++LPGISP+HPGLRIKFAGR IL+QFPDQF+ LHFGCDL+H FPGTLF
Sbjct: 1500 GENIVMFDPHANHLPGISPTHPGLRIKFAGRYILDQFPDQFAPFLHFGCDLEHTFPGTLF 1559
Query: 1561 RFPLRTAGVASRSQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKSISIFLKEGTGHE 1620
RFPLR A VA RS IKKE Y PEDV SLF +FS VVSE L+FLRNVK++SIF KEG GHE
Sbjct: 1560 RFPLRNASVAPRSHIKKETYAPEDVLSLFTSFSGVVSEALIFLRNVKTVSIFTKEGAGHE 1619
Query: 1621 MRLLHRVSRASLGESEIGSAEVQDVFNFFKEDRLVGMNRAQFLKKLSLSIDRDLPYKCQK 1680
M+LLHRV + + VF+ E+ GMN+ Q LKKLS ++ +DLPYKCQK
Sbjct: 1620 MQLLHRVCKDHNVGQDTEPKPSSQVFSLLDENIFAGMNKDQLLKKLSNTVVKDLPYKCQK 1679
Query: 1681 ILITEQGTHGRNSHYWIMTECLGGGNVLKGTSEASTSNSHNFVPWACVAAYLNSVKHGED 1740
I++TEQ + G H WI ECL G K + S H +PWA VA ++NSVK+
Sbjct: 1680 IVVTEQDSSGCILHGWITGECLNAGVSKKNLNLPEMS--HKLIPWASVAVHINSVKN--- 1734
Query: 1741 LVDSAEVEDDCLVSSDLFQFASLPMHPRENFEGRAFCFLPLPISTGLPAHVNAYFELSSN 1800
VED S++F +++ + R NF GRAFCFLPLPI+TGLPAH+NAYFELSSN
Sbjct: 1735 ----ENVEDLAASISNIFGPSTISIQNRRNFGGRAFCFLPLPITTGLPAHINAYFELSSN 1790
Query: 1801 RRDIWFGSDMTGGGRKRSDWNIYLLENVVAPAYGRLLEKVALEIGPCYLFFSLWPKTLGL 1860
RRD+WFG+DM G G+ RSDWN+YL+E VV PAYG LLEK+A E+GPC LFFS+WP TLG
Sbjct: 1791 RRDLWFGNDMAGDGKVRSDWNLYLIEEVVVPAYGHLLEKIASELGPCDLFFSVWPVTLGT 1850
Query: 1861 EPWASVIRKLYQFVAEFNLRVLYTEARGGQWISTKHAIFPDFSFPKADELIKALSGASLP 1920
EPWAS++RKLY F+A LRVLYT+ARGGQWISTK AI+PDFSFPKADEL+ L+ A LP
Sbjct: 1851 EPWASLVRKLYSFIANNGLRVLYTKARGGQWISTKQAIYPDFSFPKADELVDVLADAGLP 1910
Query: 1921 VITLPQSLLERFMEICPSLHFLKPKLLRTLLIRRKREFKDRDAMILTLEYCLHDFEESMQ 1980
VI + +++ ERF E C SLH + P+LLRTLL RRKREF+DR+ + L LEYCL D +
Sbjct: 1911 VINISKTVAERFGEACSSLHLMTPQLLRTLLTRRKREFRDRNGLALALEYCLLDLKVPFL 1970
Query: 1981 FDTLGGLPLLPLADGSFTSIEMKGIGERVYIARGDEYGLLRDSIPHQLVDCVIPKEVHRK 2040
D L GLPLLPLADGSFT+ G ER++ A Y LL+DS+PHQLVD +P+ V+ K
Sbjct: 1971 ADLLYGLPLLPLADGSFTTFNKNGTAERIFFAEEIGYELLKDSLPHQLVDREVPEGVYSK 2030
Query: 2041 LCYIAQTDSTNISFLSCQLLEKLLVKLLPVEWQHASLVSWTPGIHGQPSLEWLQLLWNYL 2100
L +AQ+ + I LSC LLEKL KLLP +W + + WTPG G P++EW+++LW+YL
Sbjct: 2031 LLAVAQSGESCICLLSCNLLEKLFFKLLPADWHLSEKILWTPGQRGHPTVEWIRVLWSYL 2090
Query: 2101 KANCDDLLMFSKWPILPVGDDCLIQLKPNLNVIKNDGWSEKMSSLLVKVGCLFLRPDLQL 2160
K +CDDL +FSKWPILPV D CL+QL N NVI++DGWSE MSSLL+K GC FL +L +
Sbjct: 2091 KLSCDDLSVFSKWPILPVEDGCLMQLILNSNVIRDDGWSENMSSLLLKCGCRFLNRELPV 2150
Query: 2161 DHPKLECFVQSPTARGVLNVFLAVAGEPQKIEGIFTDASDGELHELRSFILQSKWFSEEQ 2220
+HP+LE FVQ PTA G+LN LA++G + I+GIF + S+GELHELR+FILQSKWFS
Sbjct: 2151 EHPQLETFVQPPTATGILNALLAISGGHENIKGIFLNVSEGELHELRNFILQSKWFSGGH 2210
Query: 2221 IDRSHVETIKHLPMFESYKSRKLVSLIKPIKWLGPTGVGEDLLNDSFIRTESDMERVIMR 2280
++ H ETIKHLP+FESY+SRKLVSL P+KWL P G+ EDLL+D F+R +S+ ER I +
Sbjct: 2211 MNEVHFETIKHLPIFESYRSRKLVSLNCPVKWLKPDGIREDLLDDDFVRLDSERERTIFK 2270
Query: 2281 RYLEIKEPTRVEFYKDHIFNRMSEFLLKPEVVSSILNDVQLLIKEDISLKSSLSAVSFVL 2340
RYL+IKEP+++EFYK + NRMSEFL + E + +IL+D+ L+ D+SL+ ++S FVL
Sbjct: 2271 RYLQIKEPSKMEFYKACVLNRMSEFLSQQEALLAILHDLNDLVVADVSLQCAISTTPFVL 2330
Query: 2341 AADGSWQQP 2349
AA+G WQQP
Sbjct: 2331 AANGLWQQP 2339
Score = 832 bits (2148), Expect = 0.0
Identities = 622/2110 (29%), Positives = 981/2110 (46%), Gaps = 269/2110 (12%)
Query: 12 EDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDSLLSSS 71
E FGQ LT R++ +L Y +G +L EL+QNA+DAGA+ V+ LD+ + SLLS
Sbjct: 1379 EAFGQHEALTTRLKHILEMYADGPGILFELVQNAEDAGASEVTFLLDKTHYGTSSLLSPE 1438
Query: 72 LSQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFV 131
++ WQGPAL +N++VFT+ D +IS+IG +SK + GRFG+GFN VYH TD+P FV
Sbjct: 1439 MADWQGPALYCFNNSVFTQQDMYAISRIGQASKLEKPFAIGRFGLGFNCVYHFTDIPGFV 1498
Query: 132 SGKYVVLFDPQGVYLPRVSAANPGKRIDFTGSSALSLYKDQFSPYCAFGCDMQSPFAGTL 191
SG+ +V+FDP +LP +S +PG RI F G L + DQF+P+ FGCD++ F GTL
Sbjct: 1499 SGENIVMFDPHANHLPGISPTHPGLRIKFAGRYILDQFPDQFAPFLHFGCDLEHTFPGTL 1558
Query: 192 FRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTLLFLKSVLCIEMYVWDAG-- 249
FRFPLRNA A RS + ++ Y PED+ S+F L+FL++V + ++ +
Sbjct: 1559 FRFPLRNASVAPRSHIKKETYAPEDVLSLFTSFSGVVSEALIFLRNVKTVSIFTKEGAGH 1618
Query: 250 ------------------EPKPKKIHSCSVSSVSDDTIWHRQALLRLSKSLNTTTEVDAF 291
EPKP S V S+ D+ I+ +L K L+ T D
Sbjct: 1619 EMQLLHRVCKDHNVGQDTEPKP----SSQVFSLLDENIFAGMNKDQLLKKLSNTVVKD-L 1673
Query: 292 PLEFVTEAVRGVETVRQVDRFYIVQTMASASSRIGSFAITASKEYDIQLLPWASIAACIS 351
P + V ++ + +I +A + + E +L+PWAS+A I
Sbjct: 1674 PYKCQKIVVTEQDSSGCILHGWITGECLNAGVSKKNLNLP---EMSHKLIPWASVAVHI- 1729
Query: 352 DNSLNNDVLRT-----------------------GQAFCFLPLPVRTGLSVQVNGFFEVS 388
NS+ N+ + G+AFCFLPLP+ TGL +N +FE+S
Sbjct: 1730 -NSVKNENVEDLAASISNIFGPSTISIQNRRNFGGRAFCFLPLPITTGLPAHINAYFELS 1788
Query: 389 SNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPAFVHMLHGVKELLGPTDIYYSLWPTGS 448
SNRR +W+G+DM GKVRS WN L+E++V PA+ H+L + LGP D+++S+WP
Sbjct: 1789 SNRRDLWFGNDMAGDGKVRSDWNLYLIEEVVVPAYGHLLEKIASELGPCDLFFSVWPVTL 1848
Query: 449 FEEPWSILVQQIYINICN--APVIYSNLGGGRWVSPSEAFLHDEKFTKSKDLSLALMQLG 506
EPW+ LV+++Y I N V+Y+ GG+W+S +A D F K+ +L L G
Sbjct: 1849 GTEPWASLVRKLYSFIANNGLRVLYTKARGGQWISTKQAIYPDFSFPKADELVDVLADAG 1908
Query: 507 MPVVHLPNSLFDMLLKYNSS-KVITPGTVRQFL----RECESCNHLSRAHKLLLLEYCLE 561
+PV+++ ++ + + SS ++TP +R L RE N L+ L LEYCL
Sbjct: 1909 LPVINISKTVAERFGEACSSLHLMTPQLLRTLLTRRKREFRDRNGLA-----LALEYCLL 1963
Query: 562 DLVDDDVGKAAYNLPLLPLANGNFASFLEASKGIPYFICDELEYKLL-EPVSDRVIDQSI 620
DL + Y LPLLPLA+G+F +F + F +E+ Y+LL + + +++D+ +
Sbjct: 1964 DLKVPFLADLLYGLPLLPLADGSFTTFNKNGTAERIFFAEEIGYELLKDSLPHQLVDREV 2023
Query: 621 PPNILTRLSGIAMSSNTNIALFSIHHFAHLFPVFMPDDWKYKCKVFWDPDSCQKPTSSWF 680
P + ++L +A S + I L S + LF +P DW K+ W P PT W
Sbjct: 2024 PEGVYSKLLAVAQSGESCICLLSCNLLEKLFFKLLPADWHLSEKILWTPGQRGHPTVEWI 2083
Query: 681 VLFWQYLGKQSEILPLFKDWPILPSTSGHLLRPSRQLKMINGSTLSDTVQDILVKIGCHI 740
+ W YL + L +F WPILP G L++ +I S+ + +L+K GC
Sbjct: 2084 RVLWSYLKLSCDDLSVFSKWPILPVEDGCLMQLILNSNVIRDDGWSENMSSLLLKCGCRF 2143
Query: 741 LKPGYVVEHPDLFSYLCGGNAAGVLESIFNAFSSAENMQVSFSSLIAEERNELRRFLLDP 800
L VEHP L +++ A G+L ++ EN++ F ++ E +ELR F+L
Sbjct: 2144 LNRELPVEHPQLETFVQPPTATGILNALLAISGGHENIKGIFLNVSEGELHELRNFILQS 2203
Query: 801 QWYVGHSMDEFNIRFCRRLPIYQVYHREPTQDSQFSDLENPRKYLPPLDVPEFILVGIEF 860
+W+ G M+E + + LPI++ Y + + L P K+L P + E +L +F
Sbjct: 2204 KWFSGGHMNEVHFETIKHLPIFESY-----RSRKLVSLNCPVKWLKPDGIREDLLDD-DF 2257
Query: 861 IVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAEDRDSIMLSVLQNLPLLSLE 920
+ S E I RY ++ K +FYK V +R+ E ++ +L++L +L L +
Sbjct: 2258 VRLDSERERTIFKRYLQIKEPSKMEFYKACVLNRMSEFLSQQE--ALLAILHDLNDLVVA 2315
Query: 921 DASIRDLLRNLKFIPTVIGTLKCPSVLYDPRNEEIYALLEDSDSFPSGVFRESETLDIMR 980
D S++ + F+ G + PS LYDPR + LL FPS F +S+ LD +
Sbjct: 2316 DVSLQCAISTTPFVLAANGLWQQPSRLYDPRVPALQELLHKEVYFPSEKFSDSKILDALV 2375
Query: 981 GLGLKTSVSPDTVLESARCIEHLMHEDQQKAYLKGKVLFSYLEVNALKWLPDKFDDKKGA 1040
GLGL+T++ T L++AR + L +A G+ L ++ K L K K G
Sbjct: 2376 GLGLRTTLDCSTYLDAARSVSMLHDLGDLEASRYGRRLLFHI-----KTLSIKLSSKTGE 2430
Query: 1041 VNRILSQAATAFRSRN---------------------TKSDIEKFWNDLQLISWCPVLVS 1079
N SQ + S + T+ ++FW L+ I WCP+ +
Sbjct: 2431 ANHDESQNIMSITSEDSFDGETYPEYETETSYLGSLLTQQSEDEFWCQLRSIPWCPICLD 2490
Query: 1080 PPFHSLPWPVVSSMVAPPKVVRPPNDLWLVSAGMRILDGECSSTALLYCLGWMCPPGGGV 1139
PP +PW S++VA P VRP + ++LVSA M +LDGEC S+ L LGWM +
Sbjct: 2491 PPIEGIPWLESSNLVASPDRVRPKSQMFLVSATMHLLDGECQSSYLHQKLGWMDCLTIDI 2550
Query: 1140 IAAQLLELGKN-----NEIVTDQVLRQELALAMPRIYSILTGMIGSDEIEIVKAVLEGCR 1194
+ QL+E+ K+ + + L +P +Y+ L + ++ +K+ L G
Sbjct: 2551 LCRQLIEISKSYKEQKSRSSVNPEFESMLQSQIPLLYTRLQELSRENDFLALKSALSGVP 2610
Query: 1195 WIWVGDGFATSDEVVLDGPLHLAPYIRVIPVDLAVFKNLFLELGIREFLQPSDYVNILFR 1254
W+W+GD F ++D + D P+ PY+ V+P +L+ FK L LELG+R +DY+N L
Sbjct: 2611 WVWLGDDFVSADVLSFDSPVKFTPYLYVVPSELSDFKELLLELGVRLSFDAADYMNTLQH 2670
Query: 1255 MANKKGSSPLDTQEIRAVMLIVHHLA----EVYLHGQKVQLYLPDVSGRLFLAGDLVYND 1310
+ N S L ++I V+ ++ +A EV + +PD +G L DLVYND
Sbjct: 2671 LQNDIKGSQLTDEQINFVLCVLEAVADCFSEVSSDSDNNSVLVPDSAGFLVPLEDLVYND 2730
Query: 1311 APWLLGSEDPDGSFGNAPSVTWNAKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADS 1370
APW+ S + + KR FVH +I++D+A +LG++SLR + L ++ +
Sbjct: 2731 APWV-------------DSSSLSGKR----FVHPSINSDMANRLGIQSLRCISLVDNDIT 2773
Query: 1371 MNFGLSGAAEAFGQHEALTTRLKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQY 1430
+ T+LK +L +YA LF+L++ A+ ++ + DK ++
Sbjct: 2774 QDLPCMDF-----------TKLKELLSLYASKDFLLFDLLELADCCKVKKLHIIFDKREH 2822
Query: 1431 GTSSVLSPEMADWQGPALYCFNDSV-FTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCV 1489
++L + ++QGPA+ + V T +++ ++ + Q + +GLG
Sbjct: 2823 PRKTLLQHNLGEFQGPAIVAILEGVTLTREEICSLQLLSQWRIKGETL---NYGLGLLSC 2879
Query: 1490 YHFTDIPMFVSGENIVMFDPHASNLPGISPSHP-GLRIKFAGRKILEQFPDQFSSLLHFG 1548
Y D+ VSG MFDP + L + P G G ++E+F DQF+ +L
Sbjct: 2880 YFMCDLLSIVSGGYFYMFDPQGATLSASTTQAPAGKMFSLIGTNLVERFSDQFNPMLIGQ 2939
Query: 1549 CDLQHPFPGTLFRFPLRTAGVASRSQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKS 1608
T+ R PL T +I K+ + A V + FL N
Sbjct: 2940 DKAWSLTDSTIIRMPLST-------EILKDGFE--------AGLDRVKQISDQFLENASR 2984
Query: 1609 ISIFLKEGTGHEMRLLHRVSRASLGESEIGSAEVQDVFNFFKEDRLVGMNRA---QFLKK 1665
I IFLK VS+ S E G+A+ + + M + LK
Sbjct: 2985 ILIFLKS-----------VSQVSFSTWEQGNAQPHQDYTLHIDSASAIMRNPFAEKNLKT 3033
Query: 1666 LSLSIDRDLPYKCQKILITEQGTH-GRNS--HYWIMTECLGGG---NVLKGTSEASTSNS 1719
LS K I E H G N W++ G G N+ +G +
Sbjct: 3034 SKLSRIFGSSNSGVKSRIIEVNLHIGENKLLDRWLVVLSKGSGQSQNMARGRKYLA---- 3089
Query: 1720 HNFVPWACVAAYLNSVKHGEDLVDSAEVEDDCLVSSDLFQFASLPMHPRENFEGRAFCFL 1779
+N P A VAA+++ D+ AS M P L
Sbjct: 3090 YNLTPVAGVAAHVSRNGRPVDV-----------------HAASPIMSP-----------L 3121
Query: 1780 PLPISTGLPAHVNAYFELSSNRRDIWFGS---------DMTGGGRKRSDWNIYLLENVVA 1830
PL S LP + F + +N F + + G WN L+ + V
Sbjct: 3122 PLSGSVNLPVTILGCFLIRNNCGRFLFKNKNERAMSEPQLDAGDILIDAWNKELM-SCVR 3180
Query: 1831 PAYGRL---LEKVALEIGPC------------------YLFFSLWPKTLG---------L 1860
+Y + +E+++ E + +S WP++ L
Sbjct: 3181 DSYIEIVVEMERLSREHSSSSTESSTARQLALSLKAYGHQLYSFWPRSNQHDDAIEAEVL 3240
Query: 1861 EP-WA----SVIRKLYQFVAEFNLRVLYTEARGGQWISTKHAIFPDFSFPKADELIKALS 1915
+P W VIR Y VA+ L LY+ G + + +F + P ++ + L
Sbjct: 3241 KPEWECLVEQVIRPFYARVADLPLWQLYS----GSLVKAEEGMF--LTQPGSEVAVNLLP 3294
Query: 1916 -------GASLPVITLPQSLLERFMEICPSLHFLKPKLLRTLLIRRKR--EFKDRDAMIL 1966
PV ++P LL + + +KPK++R LL + + + D I
Sbjct: 3295 LTVCSFVKEHYPVFSVPWELLAEVQAVGIPVREVKPKMVRVLLRKSSASIDLRSVDTFID 3354
Query: 1967 TLEYCLHDFE 1976
LEYCL D +
Sbjct: 3355 VLEYCLSDIQ 3364
Score = 107 bits (266), Expect = 1e-22
Identities = 134/597 (22%), Positives = 236/597 (39%), Gaps = 72/597 (12%)
Query: 23 RIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDSLLSSSLSQWQGPALLA 82
+++E+L Y +L +L++ AD + + D+R H +LL +L ++QGPA++A
Sbjct: 2783 KLKELLSLYASKDFLLFDLLELADCCKVKKLHIIFDKREHPRKTLLQHNLGEFQGPAIVA 2842
Query: 83 YNDAV-FTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFVSGKYVVLFDP 141
+ V T ++ S+ + G+ T +G+G S Y + DL S VSG Y +FDP
Sbjct: 2843 ILEGVTLTREEICSLQLLSQWRIKGE---TLNYGLGLLSCYFMCDLLSIVSGGYFYMFDP 2899
Query: 142 QGVYLPRVSAANP-GKRIDFTGSSALSLYKDQFSPYCAFGCDMQSPFAGTLFRFPLRNAD 200
QG L + P GK G++ + + DQF+P S T+ R PL +
Sbjct: 2900 QGATLSASTTQAPAGKMFSLIGTNLVERFSDQFNPMLIGQDKAWSLTDSTIIRMPL--ST 2957
Query: 201 QASRSKLSRQAYTPEDISSMFVQLFEEGVLTLLFLKSVLCIEMYVWDAGEPKPKK---IH 257
+ + + IS F+ E L+FLKSV + W+ G +P + +H
Sbjct: 2958 EILKDGFEAGLDRVKQISDQFL---ENASRILIFLKSVSQVSFSTWEQGNAQPHQDYTLH 3014
Query: 258 SCSVSSVSDDTIWHRQALLRLSKSLNTTTEVDAFPLEFVTEAVRGVETVRQVDRFYIVQT 317
S S++ + + L+ SK ++ + E + + +DR+ +V +
Sbjct: 3015 IDSASAIMRNPFAEKN--LKTSKLSRIFGSSNSGVKSRIIEVNLHIGENKLLDRWLVVLS 3072
Query: 318 MASASSRIGSFAITASKEYDIQLLPWASIAACISDNSLNNDVLRTGQAFCFLPLPVRTGL 377
S S+ + K L P A +AA +S N DV LPL L
Sbjct: 3073 KGSGQSQNMA---RGRKYLAYNLTPVAGVAAHVSRNGRPVDVHAASPIMSPLPLSGSVNL 3129
Query: 378 SVQVNGFFEVSSNRRGIWYGD---------DMDRSGKVRSIWNRLLLEDLVAPAFVHML- 427
V + G F + +N + + +D + WN+ L+ V +++ ++
Sbjct: 3130 PVTILGCFLIRNNCGRFLFKNKNERAMSEPQLDAGDILIDAWNKELM-SCVRDSYIEIVV 3188
Query: 428 ----------------HGVKELLGPTDIY----YSLWPTGS----------FEEPWSILV 457
++L Y YS WP + + W LV
Sbjct: 3189 EMERLSREHSSSSTESSTARQLALSLKAYGHQLYSFWPRSNQHDDAIEAEVLKPEWECLV 3248
Query: 458 QQI----YINICNAPVIYSNLGGGRWVSPSEAFLHDEKFTKSKDLSLALMQLGMPVVHLP 513
+Q+ Y + + P+ L G V E + ++ L L H P
Sbjct: 3249 EQVIRPFYARVADLPL--WQLYSGSLVKAEEGMFLTQPGSEVAVNLLPLTVCSFVKEHYP 3306
Query: 514 --NSLFDMLLKYNSSKV----ITPGTVRQFLRECESCNHLSRAHKLL-LLEYCLEDL 563
+ +++L + + + + P VR LR+ + L + +LEYCL D+
Sbjct: 3307 VFSVPWELLAEVQAVGIPVREVKPKMVRVLLRKSSASIDLRSVDTFIDVLEYCLSDI 3363
Score = 64.7 bits (156), Expect = 6e-10
Identities = 175/779 (22%), Positives = 285/779 (36%), Gaps = 141/779 (18%)
Query: 675 PTSSWFVLFWQYLGKQSEILPLFKDWPILPSTSG----------HLL---RPSRQLKMIN 721
P+ W LFW+ ++ L LF DWP++P+ G HL+ P+ Q +
Sbjct: 3562 PSPEWIQLFWKNFNGSADELSLFSDWPLIPAFLGRPILCRVRERHLIFFPPPALQPVSRS 3621
Query: 722 GSTLSDTVQDILVKI----GCHILKPGYVV-------EHPDLFSYL-------CGGNAAG 763
G+ + T DI L YV +HP L L C
Sbjct: 3622 GTDMHQTDSDISTTSVSGGPLSELTQRYVSGFDLAQSKHPWLILLLNQCNIPVCDTAYID 3681
Query: 764 VLE----------SIFNAFSS--AENMQVSFSSLIAE----ERNELRRFLLDPQWYVGHS 807
E S+ A +S AE + + + IA R+EL L + G S
Sbjct: 3682 CAERCKCLPSPSVSLGQAIASKLAEGKRAGYIADIASFPTFGRDELFTLLANDFSSSGSS 3741
Query: 808 MDEFNIRFCRRLPIYQVYHREPTQDSQFS-DLENPRKYLPPLDVPEFIL----VGIEFIV 862
+ + LPI++ T + + + +L P D F V F+
Sbjct: 3742 YQAYELEVLSSLPIFKTVTGSYTHLQRHGLCIISGDSFLKPYDECCFCYLPDSVECHFLQ 3801
Query: 863 RSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAEDRDSIMLSVLQNLPLLSLE-D 921
T +L + + R G A+F + +R+ I++ V N L LE D
Sbjct: 3802 ALGVT---VLHNHQTLVRFGLAEFESR---------SQSEREDILIYVYGNW--LDLEVD 3847
Query: 922 ASIRDLLRNLKFIPT---VIGTLKCPSVLYDPRNEEIYALL-EDSDSFPSGVFRESETLD 977
+ + + LR KF+ L L+DP + + ++ + FP F L
Sbjct: 3848 SDVIEALREAKFVRNSDEFSSELSKSKDLFDPSDTLLVSVFFGERKRFPGERFSSEGWLR 3907
Query: 978 IMRGLGLKTSVSPDTVLESARCIEHLMHEDQQKAYLKGKVLFSYLEVNALKWLPDKFDDK 1037
I+R GL+T+ D +LE A+ +E L +E + + F V++ K + +
Sbjct: 3908 ILRKAGLRTAAEADVILECAKRVEFLGNERNRSSEEDD---FETDLVHSEKDISVELSTL 3964
Query: 1038 KGAVNRILSQAATAFRSRNTKSDIEKFWNDLQLISWCPVLVSPPFHSL------------ 1085
G+V + F S F N L I+ P F SL
Sbjct: 3965 AGSVIEAILLNFAGFYST-------AFCNTLGQIACVPA--ESGFPSLGGRKGGKRVLTR 4015
Query: 1086 --------PWPVVSSMVAPPKVVR--PPNDLWLVSAGMRILDGECSSTALLYCLGWMCPP 1135
WP+ S V R PP W +R+ ST L + +
Sbjct: 4016 YSEAVLLRDWPLAWSSVPILSTQRFIPPGFSW---TALRLKSPPIFSTVLKHLQ--VIGR 4070
Query: 1136 GGGVIAAQLLELGKNNEIVTDQVLRQELALAMPRIYSILTGMIGSDEIEIVK-AVLEGCR 1194
GG L + ++T V E+ + ++ LT SD +E+ K A L
Sbjct: 4071 NGG--EDTLAHWPNDPNVMTIDVTSCEVLKYLEIVWDSLTT---SDILELQKVAFLPAAN 4125
Query: 1195 WIWVGDGFATSDEVVLDGPLHLAPYIRVIPVDLAVFKNLFLELGIREFLQPSDYVNILFR 1254
G + + + P++L+P+ +P F N+ +LG+ + L + +IL +
Sbjct: 4126 ----GTRLVGASSLFVRLPINLSPFAFELPSLYLPFLNILKDLGLNDVLSVAAAKDILSK 4181
Query: 1255 MANKKGSSPLDTQEIRAVMLIVHHLAEVY-------LHGQKVQLYLPDVSGRLFLAGDLV 1307
+ G L+ E+RAVM I+H L + ++ K + +PD RL A V
Sbjct: 4182 LQKLCGYRRLNPNELRAVMEILHFLCDEINTTKPPEINTIKSDVIVPDDGCRLVHALSCV 4241
Query: 1308 YNDAPWLLGSEDPDGSFGNAPSVTWNAKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAE 1366
Y D SFG+ + R + VH + + LGVR L +++ E
Sbjct: 4242 YVD------------SFGSRYVRYIDTARL--RLVHPLLPERICLDLGVRKLSDVVIEE 4286
Score = 41.6 bits (96), Expect = 0.006
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Query: 2072 WQHASLVSWTPGIHGQPSLEWLQLLWNYLKANCDDLLMFSKWPILP 2117
W+ S S G PS EW+QL W + D+L +FS WP++P
Sbjct: 3550 WESTSSSSDDSG----PSPEWIQLFWKNFNGSADELSLFSDWPLIP 3591
>At3g48770 hypothetical protein
Length = 1899
Score = 43.1 bits (100), Expect = 0.002
Identities = 59/219 (26%), Positives = 93/219 (41%), Gaps = 36/219 (16%)
Query: 1397 EMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYGTSSVLSPEMADWQGPA--LYCFNDS 1454
E+YA L EL+QNAED E + D S + S ++ + PA L N+
Sbjct: 231 ELYAKDVHFLMELIQNAEDNEYPEGV---DPSL--EFVITSEDITNTGAPATLLIFNNEK 285
Query: 1455 VFTPQDLYAISRIGQESKL--EKAFAIGRFGLGFNCVYHFTDIPMFVSGENIVMFDPHAS 1512
F+ +++ +I +G+ +K K IG G+GF V+ T P S + F+
Sbjct: 286 GFSEKNIESICSVGRSTKKGNRKCGYIGEKGIGFKSVFLITSQPYIFSNGYQIRFNEAPC 345
Query: 1513 NLPGISPSHPGLRIKFAGRKILEQFPDQFSSLLHFGCDLQHPFPGTLFRFPLRTAGVASR 1572
SH L I+ ++ DQ SL+ D+Q + G+ P T + +
Sbjct: 346 -------SHCSLGY------IVPEWVDQHPSLV----DIQRMY-GSGSALPTTTIILPLK 387
Query: 1573 SQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKSISI 1611
S + V+ + S V E LLFL +K +SI
Sbjct: 388 S---------DKVKPVKEQLSNVHPEVLLFLSKIKRLSI 417
Score = 41.2 bits (95), Expect = 0.007
Identities = 38/130 (29%), Positives = 60/130 (45%), Gaps = 12/130 (9%)
Query: 17 TVDLTRRIREVLLN-YPEGTTVLKELIQNADDAGATT-VSLCLDRRSHAGDSLLSSSLSQ 74
T DL + ++ + Y + L ELIQNA+D V L+ + S ++
Sbjct: 218 TEDLHQAVKNLSAELYAKDVHFLMELIQNAEDNEYPEGVDPSLEF------VITSEDITN 271
Query: 75 WQGPA--LLAYNDAVFTEDDFISISKIGGSSKHG--QASKTGRFGVGFNSVYHLTDLPSF 130
PA L+ N+ F+E + SI +G S+K G + G G+GF SV+ +T P
Sbjct: 272 TGAPATLLIFNNEKGFSEKNIESICSVGRSTKKGNRKCGYIGEKGIGFKSVFLITSQPYI 331
Query: 131 VSGKYVVLFD 140
S Y + F+
Sbjct: 332 FSNGYQIRFN 341
>At4g13750 hypothetical protein
Length = 2137
Score = 42.4 bits (98), Expect = 0.003
Identities = 39/148 (26%), Positives = 61/148 (40%), Gaps = 27/148 (18%)
Query: 1384 QHEALTTRLKHIL-EMYADGPGTLFELVQNAEDAGASEVI-----FLLDKSQYGTSSVLS 1437
QH L L+ + E+Y+ + ELVQNA+D E + F+L K+ +
Sbjct: 595 QHARLGRALQCLSQELYSQDSHFILELVQNADDNKYPEHVEPTLTFILQKTGIVVLN--- 651
Query: 1438 PEMADWQGPALYCFNDSVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPM 1497
N+ F P+++ A+ +GQ +K IG+ G+GF V+ +D P
Sbjct: 652 --------------NECGFMPENIRALCDVGQSTKKGSGGYIGKKGIGFKSVFRVSDAPE 697
Query: 1498 FVSGENIVMFDPHASN----LPGISPSH 1521
S FD LP + P H
Sbjct: 698 IHSNGFHFKFDISEGQIGYILPTVVPPH 725
Score = 41.6 bits (96), Expect = 0.006
Identities = 30/110 (27%), Positives = 51/110 (46%), Gaps = 12/110 (10%)
Query: 31 YPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDSLLSSSLSQWQGPALLAYNDAVFTE 90
Y + + + EL+QNADD H +L + + Q G +L N+ F
Sbjct: 611 YSQDSHFILELVQNADDNKYP---------EHVEPTL--TFILQKTGIVVLN-NECGFMP 658
Query: 91 DDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFVSGKYVVLFD 140
++ ++ +G S+K G G+ G+GF SV+ ++D P S + FD
Sbjct: 659 ENIRALCDVGQSTKKGSGGYIGKKGIGFKSVFRVSDAPEIHSNGFHFKFD 708
>At1g20290 hypothetical protein
Length = 497
Score = 36.2 bits (82), Expect = 0.23
Identities = 33/134 (24%), Positives = 55/134 (40%), Gaps = 13/134 (9%)
Query: 885 QFYKKHVFDRVGELQAEDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIGTLKCP 944
QF KHV+ +G AE++D + L +L + D + RN + IG
Sbjct: 98 QFKWKHVYSHIGS--AEEKDGAVAIWLHSLNQRNYPDLRSNECKRNEVIVYKKIGEASEE 155
Query: 945 SVLYDP--------RNEEIYALLEDSDSFPSGVFRESETLDIMRGLGLKTSV---SPDTV 993
++ + N E++ + + +D G E T + G S+ SP+TV
Sbjct: 156 NIHEEKITVKENTNGNAELFVVDKVNDETSEGTIMEERTTLEEKATGNDGSIIAFSPETV 215
Query: 994 LESARCIEHLMHED 1007
E AR I H++
Sbjct: 216 TEQARLIPSSFHQE 229
>At5g65500 putative protein
Length = 765
Score = 35.4 bits (80), Expect = 0.40
Identities = 23/83 (27%), Positives = 43/83 (51%), Gaps = 6/83 (7%)
Query: 2269 RTESDMERVIMRRYLEIKEPTRVEFYKDHIFNRMSEFLLKPEVVSSILND-----VQLLI 2323
+ E ++ERV+++R I E ++ +D +FNR EF + EV+ S+ + + +
Sbjct: 352 QAEVELERVVLQRGEMITEIEKLRSQRD-VFNRRIEFCKEREVIGSVSKEEVKCGYREYV 410
Query: 2324 KEDISLKSSLSAVSFVLAADGSW 2346
EDI L + + L + G+W
Sbjct: 411 AEDIRLATETYSDRLRLKSGGNW 433
>At4g24190 HSP90-like protein
Length = 823
Score = 35.0 bits (79), Expect = 0.52
Identities = 35/127 (27%), Positives = 57/127 (44%), Gaps = 17/127 (13%)
Query: 23 RIREVLLN--YPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGD---SLLSSSLSQWQG 77
R+ ++++N Y L+ELI NA DA L L + G+ + L + +
Sbjct: 87 RLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKDVLGEGDTAKLEIQIKLDKA 146
Query: 78 PALLAYNDAVF--TEDDFI----SISKIGGSS------KHGQASKTGRFGVGFNSVYHLT 125
+L+ D T++D I +I+K G S+ G + G+FGVGF S Y +
Sbjct: 147 KKILSIRDRGIGMTKEDLIKNLGTIAKSGTSAFVEKMQSSGDLNLIGQFGVGFYSAYLVA 206
Query: 126 DLPSFVS 132
D +S
Sbjct: 207 DYIEVIS 213
>At3g05370 putative disease resistance protein
Length = 860
Score = 32.7 bits (73), Expect = 2.6
Identities = 31/110 (28%), Positives = 54/110 (48%), Gaps = 25/110 (22%)
Query: 552 KLLLLEY-CLEDL-VDDDVGKAAYNLPLLPLANGNFASFLEASKGI-------------- 595
KL LEY CLED + +V + L ++ L+N +F SF E+S+G+
Sbjct: 397 KLAKLEYFCLEDNNMVGEVPSWLWRLTMVALSNNSFNSFGESSEGLDETQVQWLDLSSNS 456
Query: 596 -----PYFICDELEYKLLEPVSDRVIDQSIPP---NILTRLSGIAMSSNT 637
P++IC ++L +SD + SIPP + + L+ + + +N+
Sbjct: 457 FQGPFPHWICKLRSLEIL-IMSDNRFNGSIPPCLSSFMVSLTDLILRNNS 505
>At2g27380 hypothetical protein
Length = 761
Score = 32.7 bits (73), Expect = 2.6
Identities = 12/36 (33%), Positives = 20/36 (55%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW 1107
++ P + PP H P P+ S + PP V +PP ++
Sbjct: 229 TYSPPVKPPPVHKPPTPIYSPPIKPPPVHKPPTPIY 264
Score = 32.7 bits (73), Expect = 2.6
Identities = 13/35 (37%), Positives = 19/35 (54%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW 1107
+ P + PP H P P+ S V PP V +PP ++
Sbjct: 348 YSPPVKPPPVHKPPTPIYSPPVKPPPVHKPPTPIY 382
Score = 32.3 bits (72), Expect = 3.4
Identities = 13/35 (37%), Positives = 18/35 (51%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW 1107
+ P + PP H P P+ S V PP V PP ++
Sbjct: 247 YSPPIKPPPVHKPPTPIYSPPVKPPPVQTPPTPIY 281
Score = 32.0 bits (71), Expect = 4.4
Identities = 11/30 (36%), Positives = 17/30 (56%)
Query: 1078 VSPPFHSLPWPVVSSMVAPPKVVRPPNDLW 1107
+ PP H P P+ S + PP V +PP ++
Sbjct: 184 IKPPVHKPPTPIYSPPIKPPPVHKPPTPIY 213
Score = 32.0 bits (71), Expect = 4.4
Identities = 13/35 (37%), Positives = 18/35 (51%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW 1107
+ P + PP H P P S V PP V +PP ++
Sbjct: 213 YSPPIKPPPVHKPPTPTYSPPVKPPPVHKPPTPIY 247
Score = 31.6 bits (70), Expect = 5.8
Identities = 13/31 (41%), Positives = 17/31 (53%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
+ P + PP H P P+ S V PP V +PP
Sbjct: 432 YSPPVKPPPVHKPPTPIYSPPVKPPPVHKPP 462
Score = 31.6 bits (70), Expect = 5.8
Identities = 12/31 (38%), Positives = 17/31 (54%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
+ P + PP H P P+ S + PP V +PP
Sbjct: 196 YSPPIKPPPVHKPPTPIYSPPIKPPPVHKPP 226
Score = 31.6 bits (70), Expect = 5.8
Identities = 12/31 (38%), Positives = 17/31 (54%)
Query: 1073 WCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
+ P + PP H P P+ S V PP + +PP
Sbjct: 365 YSPPVKPPPVHKPPTPIYSPPVKPPPIQKPP 395
Score = 31.2 bits (69), Expect = 7.5
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 633 TYSPPIKPPPVHKPPTPTYSPPIKPPPVQKPP 664
Score = 31.2 bits (69), Expect = 7.5
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 548 TYSPPIKPPPIHKPPTPTYSPPIKPPPVHKPP 579
Score = 30.8 bits (68), Expect = 9.8
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 599 TYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPP 630
Score = 30.8 bits (68), Expect = 9.8
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 565 TYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPP 596
Score = 30.8 bits (68), Expect = 9.8
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 616 TYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPP 647
Score = 30.8 bits (68), Expect = 9.8
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 1072 SWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPP 1103
++ P + PP H P P S + PP V +PP
Sbjct: 582 TYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPP 613
>At5g53490 thylakoid lumenal 17.4 kD protein, chloroplast precursor
(P17.4) (sp|P81760)
Length = 236
Score = 32.0 bits (71), Expect = 4.4
Identities = 30/124 (24%), Positives = 58/124 (46%), Gaps = 11/124 (8%)
Query: 832 DSQFSDLENPRKYLPPLDVPEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQFYKKHV 891
D +F D N + L + ++VG +F ++ E ++S+ Y VE K + V
Sbjct: 113 DLRFCDYTNDQTNLKGKTLSAALMVGAKF--DGADMTEVVMSKAYAVEASFKGVNFTNAV 170
Query: 892 FDRVGELQAEDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIGTLKCPSVLYDPR 951
DRV ++ + ++ + + L + E+A++ D++ F T+IG + + R
Sbjct: 171 IDRVNFGKSNLKGAVFRNTV--LSGSTFEEANLEDVV----FEDTIIGYIDLQKIC---R 221
Query: 952 NEEI 955
NE I
Sbjct: 222 NESI 225
>At3g59570 putative protein
Length = 549
Score = 32.0 bits (71), Expect = 4.4
Identities = 36/133 (27%), Positives = 54/133 (40%), Gaps = 7/133 (5%)
Query: 175 PYCAFGCDMQSPFAGTLFRFP-LRNADQASRSKLSRQAY-TPEDISSMFVQLFEEGVLTL 232
P +G PFA +F FP L D RS L ++ TP++ +S L E
Sbjct: 243 PSSPYGFPSPGPFADDIFDFPSLPVTDLFGRSSLGKKGLSTPDEETSTQSDLRSEDERMH 302
Query: 233 LFLKSVLCIEMYVWDAGEPKPKKIHSCSVSSVSDDTIWHRQALLRLSKSLNTTTEVDAFP 292
F I+ EP + + S S+ + H Q++ +LS S+ T+ VD
Sbjct: 303 SFR-----IDKNADLVIEPHGSSYNDVTASINSEIEVVHLQSVEKLSHSVCTSEIVDGLR 357
Query: 293 LEFVTEAVRGVET 305
+ V E ET
Sbjct: 358 ISDVPETPSARET 370
>At1g13120 unknown protein
Length = 611
Score = 32.0 bits (71), Expect = 4.4
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query: 1626 RVSRASLGESEIGSAEVQDVFNFFKEDRL-VGMNRAQFLKKLSLSIDRDLPYKCQKILI 1683
RV R G + S ++ D+ FK+ R V ++ A F KK+ + ++ P+ C +++
Sbjct: 359 RVIRQISGTKDSVSGKINDIVKIFKDPRCPVSISIAAFAKKMVTTKEKPNPFACSYVIV 417
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 31.6 bits (70), Expect = 5.8
Identities = 15/62 (24%), Positives = 29/62 (46%)
Query: 1828 VVAPAYGRLLEKVALEIGPCYLFFSLWPKTLGLEPWASVIRKLYQFVAEFNLRVLYTEAR 1887
V P G L + + + GP + L+P LG+ P+A + Y+ + + + + +A
Sbjct: 340 VAVPRLGTLTKDILVHEGPRAFYKGLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAE 399
Query: 1888 GG 1889
G
Sbjct: 400 PG 401
>At5g07790 putative protein
Length = 616
Score = 31.6 bits (70), Expect = 5.8
Identities = 27/126 (21%), Positives = 56/126 (44%), Gaps = 12/126 (9%)
Query: 2218 EEQIDRSHVETIKHLPMFESYKSRKLVSLIKPI----KWLGPTGVGEDLLND-SFIRTES 2272
EE I R V+ E ++ +K++ L P+ L ++ L + S R
Sbjct: 133 EENISRLLVDN-----KVEKFEKKKVLDLELPVFEYSDMLEEVHEAQNFLEEQSLQRMSL 187
Query: 2273 DMERVIMRRYLEIKEPTRVEFYKDHIFNRMSEFLLKPEV--VSSILNDVQLLIKEDISLK 2330
D + + L++ EP ++E + D++FN+ ++ E+ S N+ + L+K +
Sbjct: 188 DSGKQSSKLQLDLNEPAKIEEHSDYVFNQFLSSVISNEIGEESDTKNEGESLVKGSNGIN 247
Query: 2331 SSLSAV 2336
+ +V
Sbjct: 248 EAQGSV 253
>At5g19740 peptidase-like protein
Length = 681
Score = 31.2 bits (69), Expect = 7.5
Identities = 17/74 (22%), Positives = 37/74 (49%), Gaps = 9/74 (12%)
Query: 13 DFGQTVDLTRRIREVLLNYPEGTTVLK-------ELIQNADDAGATTVSLCLDRRSHAGD 65
++G+ D R +++ +N + + ++++NA +AGA V + D+R + GD
Sbjct: 149 NYGRVEDFVRLKKDMGVNVSGAVVIARYGQIYRGDIVKNAYEAGAVGVVIYTDKRDYGGD 208
Query: 66 SLLSSSLSQWQGPA 79
+ S+W P+
Sbjct: 209 EWFPA--SKWMPPS 220
>At3g23750 receptor-like kinase, putative
Length = 928
Score = 30.8 bits (68), Expect = 9.8
Identities = 15/44 (34%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query: 743 PGYVVEHPDLFS-YLCGGNAAGVLESIFNAFSSAENMQVSFSSL 785
P +V+ L + YL N AGVL IF++ +S +N+++S++++
Sbjct: 151 PSELVDSTSLTTIYLDNTNIAGVLPDIFDSLASLQNLRLSYNNI 194
>At1g59550 hypothetical protein
Length = 307
Score = 30.8 bits (68), Expect = 9.8
Identities = 14/48 (29%), Positives = 26/48 (54%)
Query: 1739 EDLVDSAEVEDDCLVSSDLFQFASLPMHPRENFEGRAFCFLPLPISTG 1786
+D+V +E E++ +SSDLF+F L P+ + + C + + G
Sbjct: 192 DDVVTLSEDEEETCLSSDLFEFPVLTKEPKGDCDRSVVCSISVRFPNG 239
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,858,481
Number of Sequences: 26719
Number of extensions: 2424426
Number of successful extensions: 5356
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 5252
Number of HSP's gapped (non-prelim): 72
length of query: 2349
length of database: 11,318,596
effective HSP length: 115
effective length of query: 2234
effective length of database: 8,245,911
effective search space: 18421365174
effective search space used: 18421365174
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0237.12


