

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
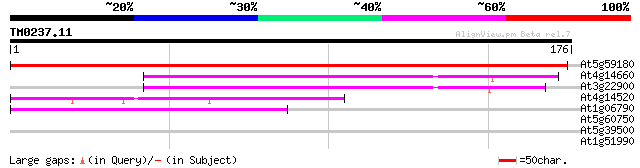
Query= TM0237.11
(176 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59180 RNA polymerase II 334 2e-92
At4g14660 RNA polymerase II fifth largest subunit like protein 76 1e-14
At3g22900 unknown protein 75 1e-14
At4g14520 unknown protein 48 2e-06
At1g06790 RNA polymerase, putative 44 3e-05
At5g60750 unknown protein 28 2.5
At5g39500 Sec7/gnom -like protein 26 9.6
At1g51990 26 9.6
>At5g59180 RNA polymerase II
Length = 176
Score = 334 bits (856), Expect = 2e-92
Identities = 159/175 (90%), Positives = 170/175 (96%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPR+FGRNL++NLV+KLMKDVEGTCSGRHGFVVA+TGI+ +GKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRFFGRNLKENLVSKLMKDVEGTCSGRHGFVVAITGIDTIGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
DGTGFVTFPVKYQCVVFRPFKGEILEAVVT+VNKMGFFAEAGPVQIFVS HLIPDDMEFQ
Sbjct: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTLVNKMGFFAEAGPVQIFVSKHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVA 175
+GDMPNYTTSDGSVKIQK+ EVRLKIIGTRVDAT IFC+GTIKDDFLGVINDP A
Sbjct: 121 AGDMPNYTTSDGSVKIQKECEVRLKIIGTRVDATAIFCVGTIKDDFLGVINDPAA 175
>At4g14660 RNA polymerase II fifth largest subunit like protein
Length = 161
Score = 75.9 bits (185), Expect = 1e-14
Identities = 43/132 (32%), Positives = 75/132 (56%), Gaps = 3/132 (2%)
Query: 43 GFVVAVTGIENVGKGLIRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAG 102
G+ VAVT ++ +G+G IR+ TG V FPV + + F+ FKGEI+ VV V K G F G
Sbjct: 30 GYYVAVTTLDKIGEGKIREHTGEVLFPVMFSGMTFKIFKGEIIHGVVHKVLKHGVFMRCG 89
Query: 103 PVQIFVSNHLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGTR--VDATEIFCIG 160
P++ ++ D ++ G+ P + S +IQ ++ VR+ +IG + E +
Sbjct: 90 PIENVYLSYTKMPDYKYIPGENPIFMNEKTS-RIQVETTVRVVVIGIKWMEVEREFQALA 148
Query: 161 TIKDDFLGVIND 172
+++ D+LG +++
Sbjct: 149 SLEGDYLGPLSE 160
>At3g22900 unknown protein
Length = 174
Score = 75.5 bits (184), Expect = 1e-14
Identities = 43/128 (33%), Positives = 71/128 (54%), Gaps = 3/128 (2%)
Query: 43 GFVVAVTGIENVGKGLIRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAG 102
G+++ T +EN+G+G I++ TG + FPV + + F+ FKGEI+ VV V+K G F ++G
Sbjct: 46 GYLITPTILENIGEGKIKEQTGEIQFPVVFNGICFKMFKGEIVHGVVHKVHKTGVFLKSG 105
Query: 103 PVQIFVSNHLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGT--RVDATEIFCIG 160
P +I +H+ EF G+ P + S +IQ + VR ++ T R + +
Sbjct: 106 PYEIIYLSHMKMPGYEFIPGENPFFMNQYMS-RIQIGARVRFVVLDTEWREAEKDFMALA 164
Query: 161 TIKDDFLG 168
+I D LG
Sbjct: 165 SIDGDNLG 172
>At4g14520 unknown protein
Length = 200
Score = 48.1 bits (113), Expect = 2e-06
Identities = 29/115 (25%), Positives = 59/115 (51%), Gaps = 11/115 (9%)
Query: 1 MFFHIVLERNMQLHPRYF-GRNLRDNLVAKLMKDV--EGTCSGRHGFVVAVTGIENVGKG 57
MF + + R++ + ++ G++ ++ +L++D+ E C HGF + +T ++++G
Sbjct: 1 MFSEVEMARDVAICAKHLNGQSPHQPILCRLLQDLIHEKACR-EHGFYLGITALKSIGNN 59
Query: 58 LIRD-------GTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQ 105
+ +TFPV + C F P +G+IL+ V V G F +GP++
Sbjct: 60 KNNNIDNENNHQAKILTFPVSFTCRTFLPARGDILQGTVKKVLWNGAFIRSGPLR 114
>At1g06790 RNA polymerase, putative
Length = 204
Score = 44.3 bits (103), Expect = 3e-05
Identities = 26/87 (29%), Positives = 42/87 (47%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+ LE ++++ P L D + + L G V++ I++V G +
Sbjct: 1 MFYLSELEHSLRVPPHLLNLPLEDAIKSVLQNVFLDKVLADLGLCVSIYDIKSVEGGFVL 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEA 87
G G T+ V + VVFRPF GE++ A
Sbjct: 61 PGDGAATYKVGLRIVVFRPFVGEVIAA 87
>At5g60750 unknown protein
Length = 347
Score = 28.1 bits (61), Expect = 2.5
Identities = 13/40 (32%), Positives = 20/40 (49%)
Query: 111 HLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGTR 150
H +P+++ D P +D QK ++ LK IGTR
Sbjct: 73 HFVPEELVKPDQDSPCTDKTDWKATFQKAADAVLKAIGTR 112
>At5g39500 Sec7/gnom -like protein
Length = 1443
Score = 26.2 bits (56), Expect = 9.6
Identities = 16/52 (30%), Positives = 24/52 (45%), Gaps = 6/52 (11%)
Query: 86 EAVVTMVNKMGFFAEAGPVQIFV------SNHLIPDDMEFQSGDMPNYTTSD 131
EAV + NK G + AG I H++PD + + D P +TS+
Sbjct: 874 EAVFLIANKYGDYISAGWKNILECVLSLNKLHILPDHIASDAADDPELSTSN 925
>At1g51990
Length = 363
Score = 26.2 bits (56), Expect = 9.6
Identities = 17/41 (41%), Positives = 21/41 (50%), Gaps = 7/41 (17%)
Query: 29 KLMKDVEGTCSGRHGFVVAVTGIENVGKGLIRDGTGFVTFP 69
KL+KD EG S +G + VGK LI+D GF P
Sbjct: 90 KLVKDEEGRESRAYG-------LGKVGKKLIKDEDGFSIAP 123
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,944,452
Number of Sequences: 26719
Number of extensions: 156865
Number of successful extensions: 379
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 373
Number of HSP's gapped (non-prelim): 8
length of query: 176
length of database: 11,318,596
effective HSP length: 93
effective length of query: 83
effective length of database: 8,833,729
effective search space: 733199507
effective search space used: 733199507
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0237.11


