

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0234.16
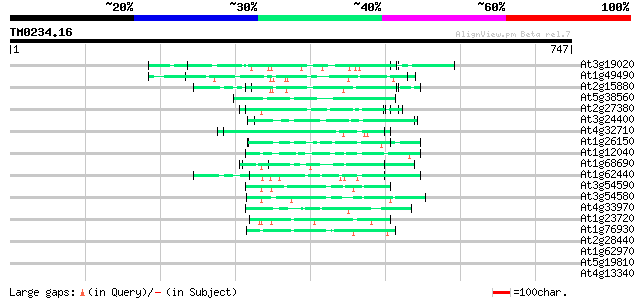
(747 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g19020 hypothetical protein 58 2e-08
At1g49490 hypothetical protein 55 2e-07
At2g15880 unknown protein 54 3e-07
At5g38560 putative protein 52 2e-06
At2g27380 hypothetical protein 50 4e-06
At3g24400 protein kinase, putative 50 6e-06
At4g32710 putative protein kinase 49 8e-06
At1g26150 Pto kinase interactor, putative 49 1e-05
At1g12040 leucine-rich repeat/extensin 1 (LRX1) 49 1e-05
At1g68690 protein kinase, putative 48 2e-05
At1g62440 putative extensin-like protein (gnl|PID|e1310400 47 3e-05
At3g54590 extensin precursor -like protein 45 1e-04
At3g54580 extensin precursor -like protein 44 3e-04
At4g33970 extensin-like protein 44 3e-04
At1g23720 unknown protein 44 3e-04
At1g76930 extensin 43 6e-04
At2g28440 En/Spm-like transposon protein 42 0.001
At1g62970 unknown protein 42 0.001
At5g19810 proline-rich protein 42 0.001
At4g13340 extensin-like protein 41 0.002
>At3g19020 hypothetical protein
Length = 951
Score = 57.8 bits (138), Expect = 2e-08
Identities = 70/297 (23%), Positives = 107/297 (35%), Gaps = 41/297 (13%)
Query: 237 GDDGDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASD 296
G G N P+P + + + + P +E ++ P + S K S
Sbjct: 396 GGGGSNPSPKPTPTPKAPEPKKEINPPNLEEPSK-----PKPEESPKPQQPSPKPETPSH 450
Query: 297 DDLNLFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPN-EAPLWNNF 355
+ N + P S KQ S P T P+ +P + P QP P E+P +
Sbjct: 451 EPSNPKEPKPESP--KQES-PKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESS 507
Query: 356 RTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPL-ENS 414
+ PP P P P P K + P+ Q P+P Q PP P E S
Sbjct: 508 KQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQ-------PPKQETPKPEES 560
Query: 415 WRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDE--------PILANPL----HEAD 462
+ + K T P P P P+P + P + P L +P+ ++A
Sbjct: 561 PKPQPPKQET--------PKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDAS 612
Query: 463 PL----AQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHV 515
P+ Q P+ + + P+ P S V SP P V + P + + P V
Sbjct: 613 PIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPV 669
Score = 57.8 bits (138), Expect = 2e-08
Identities = 101/422 (23%), Positives = 148/422 (34%), Gaps = 81/422 (19%)
Query: 185 PPAPEFDP-PLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDND 243
PP PE P P K + + +E+ P +++ + + ET KP++
Sbjct: 511 PPKPEESPKPEPPKPEESPKPQPPKQETPKP----EESPKPQPPKQETPKPEESPK---- 562
Query: 244 DGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFD 303
P PK++ + KP P E + E P + + +S V +D +D
Sbjct: 563 --PQPPKQETPKPEESPKPQ--PPKQEQPPKTEAP----KMGSPPLESPVPNDP----YD 610
Query: 304 ALPISALLKQSSNPLTP-----IPESQPAAQTTSPP--HSPRSSFFQPSPN----EAPLW 352
A PI Q +P T P+S P PP HSP F P P P++
Sbjct: 611 ASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVY 670
Query: 353 NNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLE 412
+ PP P+ P P P V P + + P P+H P PP+
Sbjct: 671 S-----PPPPVHSPPPPP-----VHSPPPPVHSPPPPVHSPP---------PPV------ 705
Query: 413 NSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAP 472
S V + + P P + P P P P PI + P P
Sbjct: 706 ---HSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPP-----------PPPV 751
Query: 473 HQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSS 532
H P P P S V SP P V + P + + P V SP S +S
Sbjct: 752 HSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVH-----SPPPPSPIYS- 805
Query: 533 NHPASPETNLSIIPYTHLRPTS--LSECINIFHQEASLMLCNVQGQTDLSENADYVDEEW 590
P P + P T L P + ++ E+ + VQ T SE+ + +
Sbjct: 806 --PPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSN 863
Query: 591 HS 592
HS
Sbjct: 864 HS 865
Score = 53.1 bits (126), Expect = 5e-07
Identities = 75/345 (21%), Positives = 118/345 (33%), Gaps = 38/345 (11%)
Query: 186 PAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDG 245
P+P+ P +K I + EE P K +++ + ++ + P +
Sbjct: 402 PSPKPTPTPKAPEPKKEINPPNLEEPSKP--KPEESPKPQQPSPKPETPSHEPSNPKEPK 459
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
P SPK++ + KP P E ++ + +S K
Sbjct: 460 PESPKQESPKTE-QPKPKPESPKQESPKQEAPKPEQPKPKPESPK--------------- 503
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQ---PSPNEAPLWNNFRTNPPDP 362
++SS P PE P + P SP+ + P P E+P + P P
Sbjct: 504 ------QESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKP 557
Query: 363 LIQPHPSPSKHQNVWLPELLIQTLP----QPIHLYPSQLMLLTLLPPITLNPLENS---- 414
P P P K Q PE + P QP ++ L P+ +P + S
Sbjct: 558 EESPKPQPPK-QETPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKK 616
Query: 415 WRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANP--LHEADPLAQQVQPAP 472
R + ST E + SP + P P + P P+ + P +H P P
Sbjct: 617 RRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPV 676
Query: 473 HQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQT 517
H P P S V SP P V + P + + P V +
Sbjct: 677 HSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHS 721
Score = 43.9 bits (102), Expect = 3e-04
Identities = 65/324 (20%), Positives = 101/324 (31%), Gaps = 38/324 (11%)
Query: 185 PPAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDD 244
P +P+ + P T++ + K S + + P + K Q K K + + + +
Sbjct: 460 PESPKQESPKTEQPKPK----PESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPE 515
Query: 245 GPPSPKKQKKQVRIVVKPTRVE-PAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFD 303
P P+ K + +P + E P E + + P + T +S K +
Sbjct: 516 ESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQE------ 569
Query: 304 ALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPL 363
+ ++S P P E P + P S P +A R PP P
Sbjct: 570 ----TPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPS 625
Query: 364 IQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVS 423
+ + S P + P P+H P PP+ P +
Sbjct: 626 TEETKTTSPQS----PPVHSPPPPPPVHSPP---------PPVFSPP---------PPMH 663
Query: 424 TLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQ-QQPVQPEP 482
+ + P P P P P P P+H P P H PV P
Sbjct: 664 SPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPP 723
Query: 483 EPEQSVSNHSSVRSPNPLVATSEP 506
P QS P P S P
Sbjct: 724 PPVQSPPPPPVFSPPPPAPIYSPP 747
>At1g49490 hypothetical protein
Length = 847
Score = 54.7 bits (130), Expect = 2e-07
Identities = 83/368 (22%), Positives = 126/368 (33%), Gaps = 71/368 (19%)
Query: 186 PAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDG 245
P+P+ PP + S + + P NK + Q + + SKP+D +
Sbjct: 431 PSPKPQPP-----------KHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKP 479
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEV---------------IRRVETPARVTRSSAQSSK 290
SPK ++ Q+ KP V P E RR P +V + +
Sbjct: 480 EESPKPEQPQIPEPTKP--VSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQ 537
Query: 291 SAVASDDDLNLFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAP 350
+ S + PI + +P P+ S P SPP S P P+ P
Sbjct: 538 PPMPSPSPPS-----PIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVAS---PPPPSPPP 589
Query: 351 LWNNFRTNPPDPLIQPHP---SPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPIT 407
+ PP P+ P P SP V+ P + P P++ P T PP T
Sbjct: 590 ---PVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPP----TFSPPPT 642
Query: 408 LNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDP------DEPILANPLHEA 461
N + + P+P + P P E P D+ + +P+
Sbjct: 643 HN---------------TNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAP 687
Query: 462 DPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIG 521
P+ Q P+ Q P N S V++P P+ A P + V T
Sbjct: 688 TPV-QSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQA---PTTSSETSQVPTPSSE 743
Query: 522 SPQGASEA 529
S Q S+A
Sbjct: 744 SNQSPSQA 751
Score = 54.3 bits (129), Expect = 2e-07
Identities = 78/345 (22%), Positives = 112/345 (31%), Gaps = 77/345 (22%)
Query: 235 DDGDDGDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSS--AQSSKSA 292
D G N PSP + KP V P + ETP + S Q K
Sbjct: 382 DKCSGGSNGGSSPSPNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHE 441
Query: 293 VASDDDLNLFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPS------- 345
++ LP K+S P PE P + P SP+ QP
Sbjct: 442 SPKPEEPENKHELPKQ---KESPKPQPSKPEDSPKPEQPKPEESPKPE--QPQIPEPTKP 496
Query: 346 ---PNEA------------PLWNNFRTNPPDPLIQ---------PHPSPSKHQNVWLPEL 381
PNEA P+ N R +PP P ++ P PSPS ++ P
Sbjct: 497 VSPPNEAQGPTPDDPYDASPVKN--RRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPP 554
Query: 382 LIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGP 441
+ + P P++ P + + PP+ P + P P P P
Sbjct: 555 PVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSP------------------PPPVHSPPP 596
Query: 442 RPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLV 501
P + P P+ + P P+ P+ PV P P SP P
Sbjct: 597 PP--VFSPPPPVFSPP--PPSPVYSPPPPSHSPPPPVYSPPPP---------TFSPPPTH 643
Query: 502 ATSEPHLG------ASEPHVQTCDIGSPQGASEAHSSNHPASPET 540
T++P +G A P +T + +P S+ P T
Sbjct: 644 NTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPT 688
Score = 42.4 bits (98), Expect = 0.001
Identities = 51/239 (21%), Positives = 82/239 (33%), Gaps = 31/239 (12%)
Query: 185 PPAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDD 244
PP P F PP V S P+ S T++P G
Sbjct: 602 PPPPVFSPPPPSPVYSP---PPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMG--APTPT 656
Query: 245 GPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDA 304
P+P + QV P+ ++++ V+ P V S+ S + V + + A
Sbjct: 657 QAPTPSSETTQVPT---PSSESDQSQILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQA 713
Query: 305 ---------LPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNF 355
P+ A S P P S+ + P P+PN P+ +
Sbjct: 714 PNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQS-- 771
Query: 356 RTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENS 414
P P +P SP + + V PE P P++ PS + + ++ P EN+
Sbjct: 772 ----PTPSSEPVSSPEQSEEVEAPE------PTPVN--PSSVPSSSPSTDTSIPPPENN 818
Score = 39.7 bits (91), Expect = 0.006
Identities = 47/193 (24%), Positives = 73/193 (37%), Gaps = 38/193 (19%)
Query: 319 TPIPESQPAAQTTSPP----HSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
TP P+++TT P S +S P P+ ++ ++ P + P+PS +
Sbjct: 654 TPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQV----PTPSSSE 709
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
+ P L P P+ P T SE +V T PS
Sbjct: 710 SYQAPNLSPVQAPTPVQA------------PTT--------SSETSQVPTPSSESNQSPS 749
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQ-PEPEPEQSVSNHSS 493
P P P+ + +P ++P++ +P Q + V+ PEP P N SS
Sbjct: 750 QAPTPILEPVHAPTPNSKPVQSPTPSSEPVS-----SPEQSEEVEAPEPTP----VNPSS 800
Query: 494 VRSPNPLVATSEP 506
V S +P TS P
Sbjct: 801 VPSSSPSTDTSIP 813
>At2g15880 unknown protein
Length = 727
Score = 53.9 bits (128), Expect = 3e-07
Identities = 54/210 (25%), Positives = 75/210 (35%), Gaps = 20/210 (9%)
Query: 314 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNE----APLWNNFRTNPPDPLIQPHPS 369
S P + P S P T SP H P+ P PN+ +P+ FR +PP P + P
Sbjct: 446 SPPPASSPPTSPPVHSTPSPVHKPQPPKESPQPNDPYDQSPV--KFRRSPPPPPVHSPPP 503
Query: 370 PSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYY 429
PS + P + P P++ P PP+ P S V +
Sbjct: 504 PSPIHSPPPPPVYSPPPPPPVYSPPPP-------PPVYSPPPPPPVHSPPPPVHSPPPPV 556
Query: 430 LTCPSPRRYPGPR----PERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPE 485
+ P P P P P + P P+ + P P+ P PV P P
Sbjct: 557 HSPPPPVHSPPPPVHSPPPPVHSPPPPVYSPP---PPPVHSPPPPVHSPPPPVHSPPPPV 613
Query: 486 QSVSNHSSVRSPNPLVATSEPHLGASEPHV 515
S V SP P V + P + + P V
Sbjct: 614 YSPPPPPPVHSPPPPVFSPPPPVHSPPPPV 643
Score = 49.7 bits (117), Expect = 6e-06
Identities = 72/305 (23%), Positives = 102/305 (32%), Gaps = 32/305 (10%)
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
PP P++ V P P + + +P Q K + +D +D
Sbjct: 433 PPPPQQPHHHVVHSPPPASSPPTSPPVHSTPSPVH----KPQPPKESPQPNDP---YDQS 485
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQ 365
P+ + S P P+ P + SPP P S P P +P PP P+
Sbjct: 486 PVKF---RRSPPPPPVHSPPPPSPIHSPPPPPVYSPPPPPPVYSP-------PPPPPVYS 535
Query: 366 PHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTL 425
P P P H P + + P P+H P + PP ++ S V +
Sbjct: 536 PPPPPPVHS----PPPPVHSPPPPVHSPPPPVHS----PPPPVHSPPPPVHSPPPPVYSP 587
Query: 426 EEYYLTCPSPRRYPGPRPERLVDPD--EPILANPLHEADPLAQQVQPAPHQQQPVQPEPE 483
+ P P + P P P P P+H P P H P P
Sbjct: 588 PPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYSPP 647
Query: 484 PEQSVSNHSSVRSPNPLVATSEPHL--GASEPHVQTCDIGSPQGASEAHSSNHPASPETN 541
P V+SP P S P L S P QT + SP + + + P P +
Sbjct: 648 PPVYSPPPPPVKSPPPPPVYSPPLLPPKMSSPPTQT-PVNSPPPRTPSQTVEAP--PPSE 704
Query: 542 LSIIP 546
IIP
Sbjct: 705 EFIIP 709
Score = 45.1 bits (105), Expect = 1e-04
Identities = 54/206 (26%), Positives = 71/206 (34%), Gaps = 44/206 (21%)
Query: 322 PESQPAAQTTSPPHSPRSSFFQPSP----NEAPLWNNFRTNPPDPLIQPH------PSPS 371
P P+ T P H P+ P P N++P+ FR +PP P QPH P P+
Sbjct: 394 PSKSPSPVPTRPVHKPQPPKESPQPNDPYNQSPV--KFRRSPPPPQ-QPHHHVVHSPPPA 450
Query: 372 KHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLT 431
P + + P P+H PP KE + Y +
Sbjct: 451 SSPPTSPP---VHSTPSPVH---------KPQPP-------------KESPQPNDPYDQS 485
Query: 432 CPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNH 491
RR P P P P PI + P P P P P P P P S
Sbjct: 486 PVKFRRSPPPPPVHSPPPPSPIHSPP----PPPVYSPPPPPPVYSP--PPPPPVYSPPPP 539
Query: 492 SSVRSPNPLVATSEPHLGASEPHVQT 517
V SP P V + P + + P V +
Sbjct: 540 PPVHSPPPPVHSPPPPVHSPPPPVHS 565
>At5g38560 putative protein
Length = 681
Score = 51.6 bits (122), Expect = 2e-06
Identities = 52/219 (23%), Positives = 82/219 (36%), Gaps = 34/219 (15%)
Query: 299 LNLFDALPISALLKQSSNPLTPIP-ESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRT 357
++L LPI + +S+ P P ++QP + PP +P PSP ++P +
Sbjct: 1 MSLVPPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTP-----PPSPPQSPPPVVSSS 55
Query: 358 NPPDPLIQPHPSPSKHQNVWLPELLIQTLPQP-IHLYPSQLMLLTLLPPITLNPLENSWR 416
PP + P PS S P + T P P + P +++ PP T
Sbjct: 56 PPPPVVSSPPPSSSPP-----PSPPVITSPPPTVASSPPPPVVIASPPPSTPAT------ 104
Query: 417 SEKEKVSTLEEYYLTCPSPRRY--PGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQ 474
T P+P + P P P+ P P NP + P P+P
Sbjct: 105 --------------TPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPG 150
Query: 475 QQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEP 513
+ P P+P P ++ P P + S P ++P
Sbjct: 151 ETPSPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPTDP 189
>At2g27380 hypothetical protein
Length = 761
Score = 50.4 bits (119), Expect = 4e-06
Identities = 47/184 (25%), Positives = 64/184 (34%), Gaps = 26/184 (14%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
S P+ P P P T SPP P P+P +P PP P+ P P
Sbjct: 147 SPPVKPPPVQMPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPVHKPPTPIYSPPIKPPPVH 206
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
P P P+H P+ T PP+ P+ P+
Sbjct: 207 KPPTPIYSPPIKPPPVHKPPTP----TYSPPVKPPPVHKP------------------PT 244
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHS-S 493
P P +P + P PI + P+ P Q P P PV+P P + +S
Sbjct: 245 PIYSPPIKPPPVHKPPTPIYSPPV---KPPPVQTPPTPIYSPPVKPPPVHKPPTPTYSPP 301
Query: 494 VRSP 497
V+SP
Sbjct: 302 VKSP 305
Score = 50.1 bits (118), Expect = 5e-06
Identities = 48/193 (24%), Positives = 65/193 (32%), Gaps = 26/193 (13%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTN-PPDPLIQPHPSPSKH 373
S P+ P P +P T SPP P P+P +P + PP P P P
Sbjct: 567 SPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPV 626
Query: 374 QNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCP 433
P P P+H P+ T PPI P++ P
Sbjct: 627 HKPPTPTYSPPIKPPPVHKPPTP----TYSPPIKPPPVQKP------------------P 664
Query: 434 SPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSS 493
+P P +P + P P + P+ P QV P P PV+P P +S
Sbjct: 665 TPTYSPPVKPPPVQLPPTPTYSPPVK---PPPVQVPPTPTYSPPVKPPPVQVPPTPTYSP 721
Query: 494 VRSPNPLVATSEP 506
P P+ P
Sbjct: 722 PIKPPPVQVPPTP 734
Score = 49.7 bits (117), Expect = 6e-06
Identities = 46/195 (23%), Positives = 63/195 (31%), Gaps = 25/195 (12%)
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQ 365
P+ S P+ P P +P T SPP P P+P +P PP P
Sbjct: 474 PVKPPTPTYSPPVQPPPVQKPPTPTYSPPVKPPPIQKPPTPTYSPPIKPPPVKPPTPTYS 533
Query: 366 PHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTL 425
P P P P PIH P+ T PPI P+
Sbjct: 534 PPIKPPPVHKPPTPTYSPPIKPPPIHKPPTP----TYSPPIKPPPVHKP----------- 578
Query: 426 EEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPE 485
P+P P +P + P P + P+ P P P P++P P +
Sbjct: 579 -------PTPTYSPPIKPPPVHKPPTPTYSPPI---KPPPVHKPPTPTYSPPIKPPPVHK 628
Query: 486 QSVSNHSSVRSPNPL 500
+S P P+
Sbjct: 629 PPTPTYSPPIKPPPV 643
Score = 49.3 bits (116), Expect = 8e-06
Identities = 45/186 (24%), Positives = 67/186 (35%), Gaps = 10/186 (5%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
S P+ P P +P T SPP P P+P +P PP P+ P P
Sbjct: 383 SPPVKPPPIQKPPTPTYSPPIKPPPLQKPPTPTYSPPIKLPPVKPPTPIYSPPVKPPPVH 442
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
P P P+H P+ T PPI P++ + V P+
Sbjct: 443 KPPTPIYSPPVKPPPVHKPPTP----TYSPPIKPPPVKPPTPTYSPPVQ--PPPVQKPPT 496
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSV 494
P P +P + P P + P+ + P+ P P P++P P + +S
Sbjct: 497 PTYSPPVKPPPIQKPPTPTYSPPI-KPPPVK---PPTPTYSPPIKPPPVHKPPTPTYSPP 552
Query: 495 RSPNPL 500
P P+
Sbjct: 553 IKPPPI 558
Score = 48.5 bits (114), Expect = 1e-05
Identities = 51/209 (24%), Positives = 72/209 (34%), Gaps = 13/209 (6%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
S P+ P +P T SPP P P+P +P PP P+ P P
Sbjct: 299 SPPVKSPPVQKPPTPTYSPPIKPPPVQKPPTPTYSPPIKPPPVKPPTPIYSPPVKPPPVH 358
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
P P P+H P+ + + PP P ++ S K L++ P
Sbjct: 359 KPPTPIYSPPVKPPPVHKPPTPIYSPPVKPPPIQKPPTPTY-SPPIKPPPLQK-----PP 412
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSV 494
Y P V P PI + P+ P P P PV+P P + +S
Sbjct: 413 TPTYSPPIKLPPVKPPTPIYSPPV---KPPPVHKPPTPIYSPPVKPPPVHKPPTPTYSPP 469
Query: 495 RSPNPLVATSEPHLGASEPHVQTCDIGSP 523
P P+ +P P VQ + P
Sbjct: 470 IKPPPV----KPPTPTYSPPVQPPPVQKP 494
Score = 48.1 bits (113), Expect = 2e-05
Identities = 44/186 (23%), Positives = 64/186 (33%), Gaps = 9/186 (4%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
S P+ P P +P SPP P P+P +P PP P P P Q
Sbjct: 433 SPPVKPPPVHKPPTPIYSPPVKPPPVHKPPTPTYSPPIKPPPVKPPTPTYSPPVQPPPVQ 492
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
P P PI P+ T PPI P++ + + + P+
Sbjct: 493 KPPTPTYSPPVKPPPIQKPPTP----TYSPPIKPPPVKPPTPTYSPPIKPPPVH--KPPT 546
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSV 494
P P +P + P P + P+ P P P P++P P + +S
Sbjct: 547 PTYSPPIKPPPIHKPPTPTYSPPI---KPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPP 603
Query: 495 RSPNPL 500
P P+
Sbjct: 604 IKPPPV 609
Score = 45.8 bits (107), Expect = 9e-05
Identities = 49/206 (23%), Positives = 71/206 (33%), Gaps = 12/206 (5%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPN-EAPLWNNFRTNPPDPLIQPHPSPSKH 373
S P+ P P +P T SPP P P+P P++ PP P P P
Sbjct: 79 SPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPTYSPPIYPPPI 138
Query: 374 QNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCP 433
Q P P P+ + P+ + PP P ++ ++ P
Sbjct: 139 QKPPTPSYSPPVKPPPVQMPPTPTYSPPIKPPPVHKPPTPTYS------PPIKPPVHKPP 192
Query: 434 SPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSS 493
+P P +P + P PI + P+ P P P PV+P P + +S
Sbjct: 193 TPIYSPPIKPPPVHKPPTPIYSPPI---KPPPVHKPPTPTYSPPVKPPPVHKPPTPIYSP 249
Query: 494 VRSPNPLVATSEPHLG--ASEPHVQT 517
P P+ P P VQT
Sbjct: 250 PIKPPPVHKPPTPIYSPPVKPPPVQT 275
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/198 (24%), Positives = 69/198 (34%), Gaps = 22/198 (11%)
Query: 315 SNPLTPIPESQPAAQTTSPP------HSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHP 368
S P+ P P +P SPP H P + + P P+ PP P+ P
Sbjct: 333 SPPIKP-PPVKPPTPIYSPPVKPPPVHKPPTPIYSPPVKPPPVHK-----PPTPIYSPPV 386
Query: 369 SPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEY 428
P Q P P P+ P+ T PPI L P++ V +
Sbjct: 387 KPPPIQKPPTPTYSPPIKPPPLQKPPTP----TYSPPIKLPPVKPPTPIYSPPVKPPPVH 442
Query: 429 YLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSV 488
P+P P +P + P P + P+ + P+ P P PVQP P +
Sbjct: 443 --KPPTPIYSPPVKPPPVHKPPTPTYSPPI-KPPPVKP---PTPTYSPPVQPPPVQKPPT 496
Query: 489 SNHSSVRSPNPLVATSEP 506
+S P P+ P
Sbjct: 497 PTYSPPVKPPPIQKPPTP 514
Score = 42.4 bits (98), Expect = 0.001
Identities = 52/214 (24%), Positives = 79/214 (36%), Gaps = 21/214 (9%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPS---PS 371
S P+ P P +P T SPP P P+P+ +P PP + P P+ P
Sbjct: 113 SPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPSYSP-----PVKPPPVQMPPTPTYSPPI 167
Query: 372 KHQNVWLPELLIQTLP--QPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYY 429
K V P + P P+H P+ + + PP P + S K + +
Sbjct: 168 KPPPVHKPPTPTYSPPIKPPVHKPPTPIYSPPIKPPPVHKPPTPIY-SPPIKPPPVHK-- 224
Query: 430 LTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVS 489
P+P P +P + P PI + P+ P P P PV+P P
Sbjct: 225 --PPTPTYSPPVKPPPVHKPPTPIYSPPI---KPPPVHKPPTPIYSPPVKPPPVQTPPTP 279
Query: 490 NHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSP 523
+S P P+ +P P V++ + P
Sbjct: 280 IYSPPVKPPPV---HKPPTPTYSPPVKSPPVQKP 310
Score = 32.7 bits (73), Expect = 0.76
Identities = 42/180 (23%), Positives = 64/180 (35%), Gaps = 14/180 (7%)
Query: 326 PAAQTTSPP--HSPR---SSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPE 380
P + TT PP +SP +P P++ PP P P P Q P
Sbjct: 52 PPSYTTPPPPIYSPPIYPPPIQKPPTYSPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPT 111
Query: 381 LLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPG 440
P PI P+ + PP P S+ S K ++ P+P P
Sbjct: 112 YSPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPSY-SPPVKPPPVQ----MPPTPTYSPP 166
Query: 441 PRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPL 500
+P + P P + P+ P+ + P P P++P P + +S P P+
Sbjct: 167 IKPPPVHKPPTPTYSPPI--KPPVHK--PPTPIYSPPIKPPPVHKPPTPIYSPPIKPPPV 222
>At3g24400 protein kinase, putative
Length = 694
Score = 49.7 bits (117), Expect = 6e-06
Identities = 56/213 (26%), Positives = 67/213 (31%), Gaps = 43/213 (20%)
Query: 326 PAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQT 385
P T SPP P P P P+ T PP P P P LP
Sbjct: 6 PPGGTPSPPPQPLP--IPPPPQPLPV-----TPPPPPTALPPALPPPPPPTALPP----A 54
Query: 386 LPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPER 445
LP P PP T+ P+ S S P P P P P
Sbjct: 55 LPPPP-------------PPTTVPPIPPSTPS---------------PPPPLTPSPLPPS 86
Query: 446 LVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSE 505
P P+ +P + PL PA P+ P P P + ++ P P +
Sbjct: 87 PTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLP----PSPTTPSPPPPSPSIPS 142
Query: 506 PHLGASEPHVQTCDIGSPQGASEAHSSNHPASP 538
P L S P SP S A S P SP
Sbjct: 143 PPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSP 175
Score = 47.0 bits (110), Expect = 4e-05
Identities = 62/233 (26%), Positives = 79/233 (33%), Gaps = 31/233 (13%)
Query: 317 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNV 376
PL P QP T PP + P P L P + P P P+
Sbjct: 17 PLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTAL---------PPALPPPPPPTT---- 63
Query: 377 WLPELLIQTLPQPIHLYPSQL--MLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
+P + T P L PS L T PP+T +P S T PS
Sbjct: 64 -VPPIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPS------PPLTPSPPPAITPS 116
Query: 435 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSV 494
P P P P P P +P + PL P+P P++P P S + S+
Sbjct: 117 PPLTPSPLPPSPTTPSPP-PPSPSIPSPPLT----PSPPPSSPLRPSSPPPPSPATPSTP 171
Query: 495 --RSPNPLVATSEPHLGASEPHVQTCDIG--SPQGASEAHSSNHPASPETNLS 543
P P T P +G+ P G SP S S+ PA LS
Sbjct: 172 PRSPPPPSTPTPPPRVGSLSPPPPASPSGGRSPSTPSTTPGSSPPAQSSKELS 224
Score = 31.2 bits (69), Expect = 2.2
Identities = 48/196 (24%), Positives = 58/196 (29%), Gaps = 42/196 (21%)
Query: 359 PPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSE 418
PP P P P L I PQP+ + P PP L P
Sbjct: 6 PPGGTPSPPPQP----------LPIPPPPQPLPVTPPP-------PPTALPPA------- 41
Query: 419 KEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPV 478
L P P P P L P P P+ + P P P P+
Sbjct: 42 -----------LPPPPP---PTALPPALPPPPPPTTVPPIPPSTPSP----PPPLTPSPL 83
Query: 479 QPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASP 538
P P S PL + P + S P + SP S S SP
Sbjct: 84 PPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSPLPPSPTTPSPPPPSPSIPSP 143
Query: 539 ETNLSIIPYTHLRPTS 554
S P + LRP+S
Sbjct: 144 PLTPSPPPSSPLRPSS 159
Score = 31.2 bits (69), Expect = 2.2
Identities = 23/66 (34%), Positives = 28/66 (41%), Gaps = 3/66 (4%)
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQ 365
P L + P P+ S P A T SPP +P S PSP P + P P +
Sbjct: 90 PSPPLTPSPTTPSPPLTPSPPPAITPSPPLTP--SPLPPSPT-TPSPPPPSPSIPSPPLT 146
Query: 366 PHPSPS 371
P P PS
Sbjct: 147 PSPPPS 152
Score = 30.0 bits (66), Expect = 4.9
Identities = 20/57 (35%), Positives = 23/57 (40%), Gaps = 6/57 (10%)
Query: 316 NPLTPIPESQPAAQTTSPPHSPR-SSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPS 371
+P TP P P+ T SPP +P PSP P PP P P PS
Sbjct: 86 SPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTP-----SPLPPSPTTPSPPPPS 137
>At4g32710 putative protein kinase
Length = 731
Score = 49.3 bits (116), Expect = 8e-06
Identities = 75/267 (28%), Positives = 96/267 (35%), Gaps = 42/267 (15%)
Query: 277 TPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIPESQP-AAQTTSPPH 335
+PA T A S A S D + A P+S L S+P P+P P +A T SPP
Sbjct: 8 SPAPATSPPAMSLPPA-DSVPDTSSPPAPPLSPLPPPLSSP-PPLPSPPPLSAPTASPPP 65
Query: 336 SPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPS 395
P S P P E+P + PP PL P P PS H + + LP PS
Sbjct: 66 LPVESPPSP-PIESPPPPLLESPPPPPLESPSP-PSPHVSAPSGSPPLPFLPAKPSPPPS 123
Query: 396 QLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRP------------ 443
T+ P T++P S SE P PRR GP+P
Sbjct: 124 SPPSETVPPGNTISPPPRSLPSESTPPVNTASPPPPSP-PRRRSGPKPSFPPPINSSPPN 182
Query: 444 ---------ERLVDPDEPILANPLHEADPLAQQVQPA--------PHQQ-------QPVQ 479
E P P+ P + + PA P Q P+
Sbjct: 183 PSPNTPSLPETSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPAGQLPDGTVAPPIG 242
Query: 480 PEPEPEQSVSNHSSVRSPNPLVATSEP 506
P EP+ S + S +P PLV S P
Sbjct: 243 PVIEPKTSPAESISPGTPQPLVPKSLP 269
Score = 46.2 bits (108), Expect = 7e-05
Identities = 50/219 (22%), Positives = 82/219 (36%), Gaps = 12/219 (5%)
Query: 285 SAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIPE--SQPAAQTTSPPHSPRSSFF 342
S SS + S ++L A + + PL+P+P S P + PP S ++
Sbjct: 4 SPSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASP 63
Query: 343 QPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTL 402
P P E+P + PP L P P P + + P + + P+ P++
Sbjct: 64 PPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPS 123
Query: 403 LPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEAD 462
PP P N+ + + + SP P P P R +P P++ +
Sbjct: 124 SPPSETVPPGNTISPPPRSLPSESTPPVNTASP---PPPSPPRRRSGPKPSFPPPINSSP 180
Query: 463 PLAQQVQPAPHQQQ--PVQPEPEPEQSVSNHSSVRSPNP 499
P P+P+ P P+P S + S +P P
Sbjct: 181 P-----NPSPNTPSLPETSPPPKPPLSTTPFPSSSTPPP 214
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 48.9 bits (115), Expect = 1e-05
Identities = 52/193 (26%), Positives = 72/193 (36%), Gaps = 32/193 (16%)
Query: 317 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNV 376
P TPI P PP SP S P P PL PP L+ P SP +H
Sbjct: 134 PTTPITSPSPPTNPPPPPESP-PSLPAPDPPSNPL-------PPPKLVPPSHSPPRH--- 182
Query: 377 WLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPR 436
LP +P P PS PP + P ++ S+ E S + P R
Sbjct: 183 -LPSPPASEIPPPPRHLPS--------PPASERP--STPPSDSEHPSPPPPGH---PKRR 228
Query: 437 RYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSS--- 493
P P + P P ++ P P+P ++ P+P P+ SN SS
Sbjct: 229 EQPPPPGSKRPTPSPPSPSDSKRPVHP----SPPSPPEETLPPPKPSPDPLPSNSSSPPT 284
Query: 494 VRSPNPLVATSEP 506
+ P+ +V+ P
Sbjct: 285 LLPPSSVVSPPSP 297
Score = 48.5 bits (114), Expect = 1e-05
Identities = 59/231 (25%), Positives = 78/231 (33%), Gaps = 28/231 (12%)
Query: 319 TPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWL 378
+P PE+ P + P P S P P P+ +PP PL P P+ +
Sbjct: 61 SPPPET-PLSSPPPEPSPPSPSLTGPPPTTIPVSPPPEPSPPPPLPTEAPPPANPVSSPP 119
Query: 379 PELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRY 438
PE P P P + + PP P S S L P P
Sbjct: 120 PE--SSPPPPPPTEAPPTTPITSPSPPTNPPPPPESPPS------------LPAPDPPSN 165
Query: 439 PGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQ--QQPVQPEPEPEQSVSNHSSVRS 496
P P P +LV P + P H P A ++ P P P P S S H S
Sbjct: 166 PLP-PPKLVPPSH---SPPRHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEHPSPPP 221
Query: 497 P-NPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLSIIP 546
P +P P G+ P SP S++ HP+ P +P
Sbjct: 222 PGHPKRREQPPPPGSKRP------TPSPPSPSDSKRPVHPSPPSPPEETLP 266
Score = 36.6 bits (83), Expect = 0.053
Identities = 37/162 (22%), Positives = 59/162 (35%), Gaps = 6/162 (3%)
Query: 399 LLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS----PRRYPGPRPERLVDPDEPIL 454
L+ L PP P +N+ +E + P+ P P P P P L
Sbjct: 23 LMALPPPQPSFPGDNATSPTREPTNGNPPETTNTPAQSSPPPETPLSSPPPEPSPPSPSL 82
Query: 455 ANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVAT-SEPHLGASEP 513
P P++ +P+P P + P P VS+ SP P T + P + P
Sbjct: 83 TGPPPTTIPVSPPPEPSPPPPLPTEAPP-PANPVSSPPPESSPPPPPPTEAPPTTPITSP 141
Query: 514 HVQTCDIGSPQGASEAHSSNHPASPETNLSIIPYTHLRPTSL 555
T P+ + + P++P ++P +H P L
Sbjct: 142 SPPTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHL 183
Score = 31.6 bits (70), Expect = 1.7
Identities = 36/148 (24%), Positives = 58/148 (38%), Gaps = 28/148 (18%)
Query: 317 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTN----PPDPLIQPHPSPSK 372
P P P SPP P + P P+ PL +N + PP ++ P PSP +
Sbjct: 241 PSPPSPSDSKRPVHPSPPSPPEETLPPPKPSPDPLPSNSSSPPTLLPPSSVVSP-PSPPR 299
Query: 373 HQNVWLPELLIQTLPQPIHLYPSQ----------------LMLLTLLPPITLNPLENSWR 416
++V P+ P P+ S L+LLTL+ + +
Sbjct: 300 -KSVSGPDNPSPNNPTPVTDNSSSSGISIAAVVGVSIGVALVLLTLIGVVVC-----CLK 353
Query: 417 SEKEKVSTLEEYYLTCPSPRRYPGPRPE 444
K+++ST+ Y+ P+P PR +
Sbjct: 354 KRKKRLSTIGGGYV-MPTPMESSSPRSD 380
>At1g12040 leucine-rich repeat/extensin 1 (LRX1)
Length = 744
Score = 48.5 bits (114), Expect = 1e-05
Identities = 62/245 (25%), Positives = 89/245 (36%), Gaps = 48/245 (19%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPH--PSPSK 372
S+P P P P +SPP P ++ P P PP P+ P SP
Sbjct: 520 SSPPPPPPSPPPPCPESSPP--PPVVYYAPVTQSPP--------PPSPVYYPPVTQSPPP 569
Query: 373 HQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTC 432
V+ P + + P P +Y PP+T +P S + +
Sbjct: 570 PSPVYYPPVT-NSPPPPSPVY---------YPPVTYSPPPPSPVYYPQVTPSPPP----- 614
Query: 433 PSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHS 492
PSP YP P P P+ P+ + P P+P PV P P P V S
Sbjct: 615 PSPLYYPPVTPSP--PPPSPVYYPPVTPSPP-----PPSPVYYPPVTPSPPPPSPVYYPS 667
Query: 493 SVRSPNPLVATSEPHLGASE--PHVQTCDIGSPQGASEAH---------SSNHPASPETN 541
+SP P +E + S+ P + C G P A+ ++ SS P SP +
Sbjct: 668 ETQSPPP---PTEYYYSPSQSPPPTKACKEGHPPQATPSYEPPPEYSYSSSPPPPSPTSY 724
Query: 542 LSIIP 546
+P
Sbjct: 725 FPPMP 729
Score = 42.4 bits (98), Expect = 0.001
Identities = 51/205 (24%), Positives = 67/205 (31%), Gaps = 38/205 (18%)
Query: 314 SSNPLTPIPESQPAAQTTSPPHSPRS-------SFFQPSPNEAPLWNNFRTNPPDPLIQP 366
SS P P + P + SPP P S ++ P P + + + R PP P P
Sbjct: 405 SSPPPPPSSKMSPTVRAYSPPPPPSSKMSPSVRAYSPPPPPYSKMSPSVRAYPPPPPPSP 464
Query: 367 HPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLE 426
P P + P + + P P ++Y S PP
Sbjct: 465 SPPPPYVYSSPPPPYVYSSPPPPPYVYSSP-------PPPPY------------------ 499
Query: 427 EYYLTCPSPRRYPGPRPERLVD---PDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPE 483
Y + P P Y P P + P P P E+ P V AP Q P P P
Sbjct: 500 -VYSSPPPPYVYSSPPPPYVYSSPPPPPPSPPPPCPESSPPPPVVYYAPVTQSPPPPSPV 558
Query: 484 PEQSVSNHSSVRSP--NPLVATSEP 506
V+ SP P V S P
Sbjct: 559 YYPPVTQSPPPPSPVYYPPVTNSPP 583
Score = 33.1 bits (74), Expect = 0.58
Identities = 45/180 (25%), Positives = 64/180 (35%), Gaps = 37/180 (20%)
Query: 358 NPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLY------PSQLMLLTLL----PPIT 407
+PP P + P ++TLP PI++Y PS M T+ PP
Sbjct: 382 SPPPPTFKMSPE-------------VRTLPPPIYVYSSPPPPPSSKMSPTVRAYSPPPPP 428
Query: 408 LNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANP----LHEADP 463
+ + S R+ Y PS R YP P P P + ++P ++ + P
Sbjct: 429 SSKMSPSVRAYSPPP---PPYSKMSPSVRAYPPPPPPSPSPPPPYVYSSPPPPYVYSSPP 485
Query: 464 LAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSP 523
V +P V P P S+ P P V +S P S P C SP
Sbjct: 486 PPPYVYSSPPPPPYVYSSPPPPYVYSS-----PPPPYVYSSPPPPPPSPP--PPCPESSP 538
>At1g68690 protein kinase, putative
Length = 708
Score = 48.1 bits (113), Expect = 2e-05
Identities = 60/229 (26%), Positives = 79/229 (34%), Gaps = 36/229 (15%)
Query: 311 LKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP 370
L +++P TP P + P + PP+ + + P+ N PP PL +P S
Sbjct: 23 LNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANG---APPPPLPKPPESS 79
Query: 371 SKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYL 430
S +P T P P + PS + PP + PL +S
Sbjct: 80 SPPPQPVIPSPPPSTSPPPQPVIPSPPPSAS-PPPALVPPLPSS---------------- 122
Query: 431 TCPSPRRYPGPRPERLVDPDEPILA-NPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVS 489
P P P PRP P PIL +P P+ Q P P +P Q P P
Sbjct: 123 -PPPPASVPPPRP----SPSPPILVRSPPPSVRPI--QSPPPPPSDRPTQSPPPP----- 170
Query: 490 NHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASP 538
S SP T P SE Q+ SP S P P
Sbjct: 171 ---SPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPP 216
Score = 45.8 bits (107), Expect = 9e-05
Identities = 54/208 (25%), Positives = 74/208 (34%), Gaps = 26/208 (12%)
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSP-----------PHSPRSSFFQPSPNEAPLWNN 354
P+++ L S+ P P P T+ P P P SS P P P
Sbjct: 34 PVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSSPPPQP-VIPSPPP 92
Query: 355 FRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLM----LLTLLPPITLNP 410
+ PP P+I P P PS L L + P P + P + +L PP ++ P
Sbjct: 93 STSPPPQPVI-PSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRP 151
Query: 411 LENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQP 470
++ S S PSP P RP + P P P P + P
Sbjct: 152 IQ----SPPPPPSDRPTQSPPPPSPPSPPSERPTQ--SPPSPPSERPTQSPPPPS---PP 202
Query: 471 APHQQQPVQPEPEPEQSVSNHSSVRSPN 498
+P +P Q P P + RSPN
Sbjct: 203 SPPSDRPSQSPPPPPEDTKPQPPRRSPN 230
Score = 44.7 bits (104), Expect = 2e-04
Identities = 48/195 (24%), Positives = 66/195 (33%), Gaps = 12/195 (6%)
Query: 345 SPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLP 404
+P + P+ N+ P P + SP+ V P L + P P P + T P
Sbjct: 4 TPVQPPVSNSPPVTSPPPPLNNATSPATPPPVTSP--LPPSAPPPNRAPPPPPPVTTSPP 61
Query: 405 PITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPL 464
P+ E S + + P P P P +P++ +P A P
Sbjct: 62 PVANGAPPPPLPKPPESSSPPPQPVIPSPPPSTSP---------PPQPVIPSPPPSASPP 112
Query: 465 AQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLV-ATSEPHLGASEPHVQTCDIGSP 523
V P P P P P S S VRSP P V P S+ Q+ SP
Sbjct: 113 PALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSP 172
Query: 524 QGASEAHSSNHPASP 538
+ P SP
Sbjct: 173 PSPPSERPTQSPPSP 187
Score = 43.5 bits (101), Expect = 4e-04
Identities = 50/195 (25%), Positives = 70/195 (35%), Gaps = 32/195 (16%)
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAP-LWNNFRTNPPDPLI 364
P+ + SS P P+ S P + TSPP P PS + P L ++PP P
Sbjct: 71 PLPKPPESSSPPPQPVIPSPPPS--TSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPAS 128
Query: 365 QPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVST 424
P P PS P +L+++ P PS + + PP + P ++
Sbjct: 129 VPPPRPSPS-----PPILVRSPP------PSVRPIQSPPPPPSDRPTQSP---------- 167
Query: 425 LEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEP 484
PSP P RP + P P P P + P+ Q P PE
Sbjct: 168 ------PPPSPPSPPSERPTQ--SPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPED 219
Query: 485 EQSVSNHSSVRSPNP 499
+ S SP P
Sbjct: 220 TKPQPPRRSPNSPPP 234
Score = 40.0 bits (92), Expect = 0.005
Identities = 52/215 (24%), Positives = 78/215 (36%), Gaps = 22/215 (10%)
Query: 319 TPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWL 378
+P+P S P PP P ++ P N AP P PL +P S S +
Sbjct: 37 SPLPPSAPPPNRAPPPPPPVTTSPPPVANGAP---------PPPLPKPPESSSPPPQPVI 87
Query: 379 PELLIQTLPQPIHLYPSQLMLLT----LLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
P T P P + PS + L+PP+ +P + + S + P
Sbjct: 88 PSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPA-SVPPPRPSPSPPILVRSPP 146
Query: 435 PR----RYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQ--PEPEPEQSV 488
P + P P P P + P ++ Q P+P ++P Q P P P
Sbjct: 147 PSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQS-PPSPPSERPTQSPPPPSPPSPP 205
Query: 489 SNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSP 523
S+ S +SP P ++P P+ SP
Sbjct: 206 SDRPS-QSPPPPPEDTKPQPPRRSPNSPPPTFSSP 239
>At1g62440 putative extensin-like protein (gnl|PID|e1310400
Length = 786
Score = 47.4 bits (111), Expect = 3e-05
Identities = 60/244 (24%), Positives = 88/244 (35%), Gaps = 28/244 (11%)
Query: 320 PIPESQPAAQTTSPPH-----SPRSSFFQPSP------NEAPLWNNFRTN----PPDPLI 364
P+ P Q+ PP SPR + PSP + P + T PP P
Sbjct: 527 PVVNCPPTTQSPPPPKYEQTPSPREYYPSPSPPYYQYTSSPPPPTYYATQSPPPPPPPTY 586
Query: 365 QPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVST 424
SP V+ P + P P++ P ++ PP+ +P+ S V
Sbjct: 587 YAVQSPPPPPPVYYPPVTASPPPPPVYYTP--VIQSPPPPPVYYSPVTQS-PPPPPPVYY 643
Query: 425 LEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEP 484
PSP YP P P P+ P+ ++ P P+P PV P P
Sbjct: 644 PPVTQSPPPSPVYYP---PVTQSPPPPPVYYLPVTQSPP-----PPSPVYYPPVAKSPPP 695
Query: 485 EQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGS--PQGASEAHSSNHPASPETNL 542
V +SP P E H AS + S P+G +++ S++H T
Sbjct: 696 PSPVYYPPVTQSPPPPSTPVEYHPPASPNQSPPPEYQSPPPKGCNDSPSNDHHYQTPTPP 755
Query: 543 SIIP 546
S+ P
Sbjct: 756 SLPP 759
Score = 44.3 bits (103), Expect = 3e-04
Identities = 64/286 (22%), Positives = 95/286 (32%), Gaps = 51/286 (17%)
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
PP K VR++ P + R P S++ S S A+
Sbjct: 386 PPPSFKMSPTVRVLPPPPPSSKMSPTFRATPPPP-----SSKMSPSFRATPP-------- 432
Query: 306 PISALLKQSSNPLTPIPESQ--PAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPL 363
P S+ + S P P S+ P+ + PP P P P + + + R PP P
Sbjct: 433 PPSSKMSPSFRATPPPPSSKMSPSVKAYPPPPPPPEYEPSPPPPSSEMSPSVRAYPPPPP 492
Query: 364 IQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVS 423
+ P P ++ P P ++Y S ++ PP T +P + E+
Sbjct: 493 LSPPPPSPPPPYIYSSPPPPSPSPPPPYIYSSPPPVVN-CPPTTQSPPPPKY----EQTP 547
Query: 424 TLEEYYLTCPSPRRY----------------PGPRPE------RLVDPDEPILANPLHEA 461
+ EYY PSP Y P P P + P P+ P+ +
Sbjct: 548 SPREYY-PSPSPPYYQYTSSPPPPTYYATQSPPPPPPPTYYAVQSPPPPPPVYYPPVTAS 606
Query: 462 --------DPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNP 499
P+ Q P P PV P P V +SP P
Sbjct: 607 PPPPPVYYTPVIQSPPPPPVYYSPVTQSPPPPPPVYYPPVTQSPPP 652
Score = 42.0 bits (97), Expect = 0.001
Identities = 57/237 (24%), Positives = 78/237 (32%), Gaps = 30/237 (12%)
Query: 283 RSSAQSSKSAVASDDDL-----NLFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSP 337
RSS + S A S D N F P S + + L P P S + T P
Sbjct: 359 RSSKECSSPASRSVDCSKFGCNNFFSPPPPSFKMSPTVRVLPPPPPSSKMSPTFRATPPP 418
Query: 338 RSSFFQPS------PNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIH 391
SS PS P + + +FR PP P + PS + P + P P
Sbjct: 419 PSSKMSPSFRATPPPPSSKMSPSFRATPPPPSSKMSPSVKAYPPPPPPPEYEPSPPPPSS 478
Query: 392 LYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDE 451
+ PP++ P S Y + P P P P P +
Sbjct: 479 EMSPSVRAYPPPPPLSPPP-----------PSPPPPYIYSSPPPPS-PSPPPPYIYSSPP 526
Query: 452 PILANPLHEADPLAQQVQPAPHQQQPVQPE--PEPEQSVSNHSSVRSPNPLVATSEP 506
P++ P P Q P ++Q P E P P ++S P AT P
Sbjct: 527 PVVNCP-----PTTQSPPPPKYEQTPSPREYYPSPSPPYYQYTSSPPPPTYYATQSP 578
>At3g54590 extensin precursor -like protein
Length = 743
Score = 45.1 bits (105), Expect = 1e-04
Identities = 51/204 (25%), Positives = 71/204 (34%), Gaps = 21/204 (10%)
Query: 315 SNPLTPIPESQPAAQTTSPP-----HSPRSSFFQPSPN---EAPLWNNFRTNPPDPLIQP 366
S+P P P SPP SP ++ PSP ++P ++PP P P
Sbjct: 503 SSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPYYSP 562
Query: 367 HPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLE 426
SP H P + + P P + ++ + PP + + S KV
Sbjct: 563 --SPKVHYKSPPPPYVYNSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV---- 616
Query: 427 EYYLTCPSPRRYPGPRPERLVDPDEPILAN----PLHEADPLAQQVQPAPHQQQPVQPEP 482
YY + P P Y P P P + P + P P PH V P P
Sbjct: 617 -YYKSPPPPYVYSSPPPP-YYSPSPKVYYKSPPPPYYSPSPKVYYKSP-PHPHVCVCPPP 673
Query: 483 EPEQSVSNHSSVRSPNPLVATSEP 506
P S S +SP P + P
Sbjct: 674 PPCYSPSPKVVYKSPPPPYVYNSP 697
Score = 38.1 bits (87), Expect = 0.018
Identities = 48/199 (24%), Positives = 66/199 (33%), Gaps = 28/199 (14%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP---S 371
S+P P P + SPP P P P +P + +PP P + P P S
Sbjct: 378 SSPPPPYYSPSPKVEYKSPP-PPYVYSSPPPPTYSPSPKVYYKSPPPPYVYSSPPPPYYS 436
Query: 372 KHQNVWL---PELLIQTLPQPIHLYPS-QLMLLTLLPPITLNPLENSWRSEKEKVSTLEE 427
V+ P + + P P + PS ++ + PP + + S KV
Sbjct: 437 PSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV----- 491
Query: 428 YYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQS 487
YY + P P Y P P P + P P P P P S
Sbjct: 492 YYKSPPPPYVYSSPPP-------------PYYSPSPKVYYKSPPPPYVYSSPPPPYYSPS 538
Query: 488 VSNHSSVRSPNPLVATSEP 506
H +SP P S P
Sbjct: 539 PKVH--YKSPPPPYVYSSP 555
>At3g54580 extensin precursor -like protein
Length = 951
Score = 44.3 bits (103), Expect = 3e-04
Identities = 62/253 (24%), Positives = 83/253 (32%), Gaps = 41/253 (16%)
Query: 316 NPLTPIPESQPAAQTTSPP-----HSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP 370
+P P P SPP SP ++ PSP + +PP P + P P
Sbjct: 545 SPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV------YYKSPPPPYVYSSPPP 598
Query: 371 SKH------QNVWLPELLIQTLPQPIHLYPS-QLMLLTLLPPITLNPLENSWRSEKEKVS 423
H Q P + + P P + PS ++ + PP + + S KV
Sbjct: 599 PYHSPSPKVQYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV- 657
Query: 424 TLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPE 483
YY + P P Y P P P + P P PH V P P
Sbjct: 658 ----YYKSPPPPYVYSSPPP-------------PYYSPSPKVYYKSP-PHPHVCVCPPPP 699
Query: 484 PEQSVSNHSSVRSPNPLVATSE---PHLGASEPHVQTCDIGSPQGASEAHSSNHPASPET 540
P S S +SP P S PH S P V P S + SP+
Sbjct: 700 PCYSPSPKVVYKSPPPPYVYSSPPPPHYSPS-PKVYYKSPPPPYVYSSPPPPYYSPSPKV 758
Query: 541 NLSIIPYTHLRPT 553
+ P + PT
Sbjct: 759 HYKSPPPPYYAPT 771
>At4g33970 extensin-like protein
Length = 699
Score = 43.9 bits (102), Expect = 3e-04
Identities = 54/229 (23%), Positives = 78/229 (33%), Gaps = 43/229 (18%)
Query: 314 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKH 373
SS P P P +P T+P H P P +P+ P P+ +P P P+
Sbjct: 416 SSTPSKPSPVHKPTPVPTTPVHKPTPVPTTPVQKPSPV-------PTTPVQKPSPVPT-- 466
Query: 374 QNVWLPELLIQTLPQPIHLYPSQLMLLTL--LPPITLNPLENSWRSEKEKVSTLEEYYLT 431
P+H PS ++ + P+ P++ + KE + Y +
Sbjct: 467 --------------TPVH-EPSPVLATPVDKPSPVPSRPVQKP-QPPKESPQPDDPYDQS 510
Query: 432 CPSPRRYPGPRPERLVDP---DEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSV 488
+ RR P P P P P P+H P P P P P P
Sbjct: 511 PVTKRRSPPPAPVNSPPPPVYSPPPPPPPVHSPPPPVHSPPPPPVYSPPPPPPP------ 564
Query: 489 SNHSSVRSPNPLVATSEPHLGASEP--HVQTCDIGSPQGASEAHSSNHP 535
V SP P V + P + + P H + SP + HS P
Sbjct: 565 -----VHSPPPPVFSPPPPVYSPPPPVHSPPPPVHSPPPPAPVHSPPPP 608
Score = 42.0 bits (97), Expect = 0.001
Identities = 54/249 (21%), Positives = 79/249 (31%), Gaps = 16/249 (6%)
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQ 365
P+ Q +P+ P +P+ T+P H P P +P+ + P P
Sbjct: 441 PVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPVLATPVDKPSPVPSRPVQKPQPPKES 500
Query: 366 PHPSPSKHQNVWL-----PELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKE 420
P P Q+ P + + P P++ P + PP +P S
Sbjct: 501 PQPDDPYDQSPVTKRRSPPPAPVNSPPPPVYSPPPPPPPVHSPPPPVHSPPPPPVYSPPP 560
Query: 421 KVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQP 480
+ P P + P P P P+H P A P P P P
Sbjct: 561 PPPPVHS-----PPPPVFSPPPPVYSPPPPVHSPPPPVHSPPPPAPVHSPPPPVHSPPPP 615
Query: 481 EPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPA---S 537
P S S +P V S P P + + + SP A PA +
Sbjct: 616 PPVYSPPPPVFSPPPSQSPPVVYSPP---PRPPKINSPPVQSPPPAPVEKKETPPAHAPA 672
Query: 538 PETNLSIIP 546
P + IIP
Sbjct: 673 PSDDEFIIP 681
>At1g23720 unknown protein
Length = 895
Score = 43.9 bits (102), Expect = 3e-04
Identities = 52/209 (24%), Positives = 75/209 (35%), Gaps = 29/209 (13%)
Query: 320 PIPESQPAAQTT---SPP----HSPRSSFFQPSPN---EAPLWNNFRTNPPDPLIQPHPS 369
P P P+ + T SPP SP ++ PSP ++P ++PP P P P
Sbjct: 442 PPPYYSPSPKLTYKSSPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSPSPK 501
Query: 370 PSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLP-------PITLNPLENSWRSEKEKV 422
PS P + P P + PS ++ P P +S + E +
Sbjct: 502 PSYKSP---PPPYVYNSPPPPYYSPSPKVIYKSPPHPHVCVCPPPPPCYSHSPKIEYKSP 558
Query: 423 STLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQP-- 480
T Y+ P P P P+P P + ++P P P P + P P
Sbjct: 559 PTPYVYHSPPPPPYYSPSPKPAYKSSPPPYVYSSP----PPPYYSPAPKPVYKSPPPPYV 614
Query: 481 ---EPEPEQSVSNHSSVRSPNPLVATSEP 506
P P S S + +SP P S P
Sbjct: 615 YNSPPPPYYSPSPKPTYKSPPPPYVYSSP 643
Score = 37.0 bits (84), Expect = 0.040
Identities = 45/195 (23%), Positives = 70/195 (35%), Gaps = 35/195 (17%)
Query: 322 PESQPAAQTTSPPHS---PRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWL 378
P +PA ++ PP+ P ++ PSP P++ + PP P + P P +
Sbjct: 298 PSPKPAYKSPPPPYVYSFPPPPYYSPSPK--PVYKS----PPPPYVYNSPPPPYY----- 346
Query: 379 PELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRY 438
P+P + P + + PP +P S K + Y+ P Y
Sbjct: 347 -----SPSPKPAYKSPPPPYVYSSPPPPYYSP------SPKPTYKSPPPPYVYSSPPPPY 395
Query: 439 PGPRPERLV-DPDEPILAN----PLHEADPLAQQVQPAPH--QQQPVQPEPEPEQSVSNH 491
P P+ + P P + N P + P P P P P P ++
Sbjct: 396 YSPSPKPVYKSPPPPYIYNSPPPPYYSPSPKPSYKSPPPPYVYSSPPPPYYSPSPKLTYK 455
Query: 492 SSVRSPNPLVATSEP 506
S SP P V +S P
Sbjct: 456 S---SPPPYVYSSPP 467
Score = 34.7 bits (78), Expect = 0.20
Identities = 50/209 (23%), Positives = 72/209 (33%), Gaps = 29/209 (13%)
Query: 317 PLTPIPESQPAAQTTSPPH---SPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP--- 370
PL+ P + ++ PP+ SP ++ PSP +PP P + P P
Sbjct: 43 PLSYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVE------YKSPPPPYVYSSPPPPYY 96
Query: 371 ---SKHQNVWLPELLIQTLPQPIHLYPS-QLMLLTLLPPITLNPLENSWRSEKEKV---S 423
K P + + P P + PS + + PP N + S KV S
Sbjct: 97 SPSPKVDYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYNSPPPPYYSPSPKVEYKS 156
Query: 424 TLEEYYLTCPSPRRY-PGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQP-- 480
Y + P P Y P P+ + P + +P P P P + P P
Sbjct: 157 PPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSP----PPPYYSPSPKPTYKSPPPPYI 212
Query: 481 ---EPEPEQSVSNHSSVRSPNPLVATSEP 506
P P S S +SP P S P
Sbjct: 213 YSSPPPPYYSPSPKPVYKSPPPPYVYSSP 241
Score = 33.1 bits (74), Expect = 0.58
Identities = 42/175 (24%), Positives = 59/175 (33%), Gaps = 17/175 (9%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQP-------H 367
S+P P P SPP S P P +P +PP P +
Sbjct: 716 SSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYS 775
Query: 368 PSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPP--ITLNPLENSWRSEKEKV--- 422
PSP P + + P P + PS + PP + +P ++ S KV
Sbjct: 776 PSPKVEYKSPPPPYVYSSPPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYYSPSPKVEYK 835
Query: 423 STLEEYYLTCPSPRRYPGPRPE-RLVDPDEPILAN----PLHEADPLAQQVQPAP 472
S Y P P Y P P+ P P + + P + P A+ P P
Sbjct: 836 SPPPPYVYNSPPPPAYYSPSPKIEYKSPPPPYVYSSPPPPSYSPSPKAEYKSPPP 890
>At1g76930 extensin
Length = 373
Score = 43.1 bits (100), Expect = 6e-04
Identities = 47/205 (22%), Positives = 76/205 (36%), Gaps = 19/205 (9%)
Query: 316 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQN 375
+P P+ P SPP P ++ P P + P P+ + P P K+ +
Sbjct: 177 SPPPPVKHYSPPPVYKSPP--PPVKYYSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKYYS 234
Query: 376 VWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSP 435
P + ++ P P+H P ++ + PP+ +P + S V +Y P P
Sbjct: 235 ---PPPVYKSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPV-----HY--SPPP 284
Query: 436 RRYPGPRPERLVDPDEPILAN---PLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHS 492
Y P P P + + P+H + P P P ++ P P S +
Sbjct: 285 VVYHSPPPPVHYSPPPVVYHSPPPPVHYSPPPVVYHSPPPPKKHYEYKSPPPPVHYSPPT 344
Query: 493 SVRSPNPLV----ATSEPHLGASEP 513
SP P V +P+L S P
Sbjct: 345 VYHSPPPPVHHYSPPHQPYLYKSPP 369
Score = 38.1 bits (87), Expect = 0.018
Identities = 46/197 (23%), Positives = 73/197 (36%), Gaps = 24/197 (12%)
Query: 316 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQN 375
+P P+ P SPP P ++ P P +PP P+ + P P KH
Sbjct: 57 SPPPPVKHYSPPPVYKSPP--PPVKYYSPPP--------VYKSPPPPVYKSPPPPVKH-- 104
Query: 376 VWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVS--TLEEYYLTCP 433
+ P + ++ P P+ Y + + PP+ ++S V + Y + P
Sbjct: 105 -YSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKHYSPPPVYKSPPPPVKHYSPPPVYKSPP 163
Query: 434 SPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAP---HQQQPV-QPEPEPEQSVS 489
P +Y P P P P+ H + P + P P + PV + P P + S
Sbjct: 164 PPVKYYSP-PPVYKSPPPPV----KHYSPPPVYKSPPPPVKYYSPPPVYKSPPPPVKHYS 218
Query: 490 NHSSVRSPNPLVATSEP 506
+SP P V P
Sbjct: 219 PPPVYKSPPPPVKYYSP 235
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 42.4 bits (98), Expect = 0.001
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 6/111 (5%)
Query: 433 PSPRRY-PGPRPERLVDPDEPILANPLHEAD-PLAQQVQPAPHQQQPVQPEPEPEQSVSN 490
PSP PG P D P+ + E D PL+ P P PE + ++
Sbjct: 28 PSPAAVSPGREPST----DSPLSPSSSPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAP 83
Query: 491 HSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETN 541
SS +PL +S P + + +P + + SP S + +N P SP ++
Sbjct: 84 SSSPEVDSPLAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANSPQSPASS 134
Score = 32.7 bits (73), Expect = 0.76
Identities = 44/186 (23%), Positives = 68/186 (35%), Gaps = 22/186 (11%)
Query: 207 SSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQKKQVRIVVKPTRVE 266
+ EES P A S + T P +D P SP ++ + +
Sbjct: 23 AQEESPSP------AAVSPGREPSTDSPLSPSSSPEEDSPLSPSSSPEE------DSPLP 70
Query: 267 PAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIPESQP 326
P++ E + + SS+ S +A + P S+ ++ +PL P S P
Sbjct: 71 PSSSP----EEDSPLAPSSSPEVDSPLAPSSSPEVDSPQPPSSS-PEADSPLPP--SSSP 123
Query: 327 AAQTTSPPHS---PRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLI 383
A + P S P S PSP P ++P P P P+PS + PE
Sbjct: 124 EANSPQSPASSPKPESLADSPSPPPPPPQPESPSSPSYPEPAPVPAPSDDDSDDDPEPET 183
Query: 384 QTLPQP 389
+ P P
Sbjct: 184 EYFPSP 189
Score = 31.2 bits (69), Expect = 2.2
Identities = 35/136 (25%), Positives = 48/136 (34%), Gaps = 12/136 (8%)
Query: 405 PITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEAD-P 463
P T +PL S S E+ S L SP P + D P+ + E D P
Sbjct: 39 PSTDSPLSPS--SSPEEDSPLSP----SSSPEEDSPLPPSSSPEEDSPLAPSSSPEVDSP 92
Query: 464 LAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSP 523
LA P QP PE + + SS + +P S P +P P
Sbjct: 93 LAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANSPQSPASSP-----KPESLADSPSPP 147
Query: 524 QGASEAHSSNHPASPE 539
+ S + P+ PE
Sbjct: 148 PPPPQPESPSSPSYPE 163
>At1g62970 unknown protein
Length = 825
Score = 42.4 bits (98), Expect = 0.001
Identities = 86/378 (22%), Positives = 136/378 (35%), Gaps = 67/378 (17%)
Query: 213 VPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVI 272
+ LN + QS R+ E +P + PS ++ KPT P +E +
Sbjct: 264 INLNNATERVQSSRRFAEPERPPSRSFEGSSHRTPST-----ELTWASKPT---PVSEPV 315
Query: 273 RRVET-------PARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQS----------- 314
R E PAR + S++SS++A L+L ++ S+ Q
Sbjct: 316 RHSELVPWQYSEPARQYQLSSRSSEAA-----QLSLLPSVSDSSHASQPTRSNQSHAVSK 370
Query: 315 ----SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEA--PLWNNFRTNPPDPLIQPHP 368
S P P P SQP PP S QP N P+ + + + P P+ Q
Sbjct: 371 PQPVSKPHPPFPMSQP------PPTSNPFPLSQPPSNSKPFPMSQSSQNSKPFPVSQ--- 421
Query: 369 SPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEY 428
S K + + + + ++ P P+ SQ + NP S S K +
Sbjct: 422 SSQKSKPLLVSQSSQRSKPLPV----SQSLQ-------NSNPFPVSQPSSNSK-----PF 465
Query: 429 YLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPV-QPEPEPEQS 487
++ P P P P + + ++ P A P PA + P+ QP +
Sbjct: 466 PVSQPQPASNPFPVSQPRPNSQPFSMSQPSSTARPFPASQPPAASKSFPISQPPTTSKPF 525
Query: 488 VSNHSSVRSPNPL---VATSEP-HLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLS 543
VS + P P+ TS+P + P Q+ P AS + S P T
Sbjct: 526 VSQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNSTQSF 585
Query: 544 IIPYTHLRPTSLSECINI 561
P P+++ E I
Sbjct: 586 QSPPVSTTPSAVPEASTI 603
Score = 39.3 bits (90), Expect = 0.008
Identities = 53/224 (23%), Positives = 82/224 (35%), Gaps = 9/224 (4%)
Query: 310 LLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPS 369
L+ QSS P+P SQ + P S SS +P P P + P P+ QP P+
Sbjct: 430 LVSQSSQRSKPLPVSQSLQNSNPFPVSQPSSNSKPFPVSQPQ----PASNPFPVSQPRPN 485
Query: 370 PSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYY 429
S+ ++ P + P S+ ++ PP T P + + + + +
Sbjct: 486 -SQPFSMSQPSSTARPFPASQPPAASKSFPIS-QPPTTSKPFVSQPPNTSKPMPVSQPPT 543
Query: 430 LTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVS 489
+ P P P P + P ++ L P+ Q Q PV P S
Sbjct: 544 TSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNSTQ--SFQSPPVSTTPSAVPEAS 601
Query: 490 NHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSN 533
S +P P VA + P QT G+ + SE S+
Sbjct: 602 TIPSPPAPAP-VAQPTHVFNQTPPPEQTPKSGAARTDSEPEVSS 644
>At5g19810 proline-rich protein
Length = 249
Score = 42.0 bits (97), Expect = 0.001
Identities = 46/186 (24%), Positives = 65/186 (34%), Gaps = 21/186 (11%)
Query: 318 LTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQ-PHPSPSKHQNV 376
LT PE P + PP S P N +P +PP P + P P + +
Sbjct: 30 LTSAPEPAPLVDLSPPPPPVNISSPPPPVNLSPPPPPVNLSPPPPPVNLSPPPPPVNLSP 89
Query: 377 WLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSP- 435
P +L+ P P++L P PP+ L+P +S L P P
Sbjct: 90 PPPPVLLSPPPPPVNLSPPP-------PPVNLSP-----PPPPVLLSPPPPPVLLSPPPP 137
Query: 436 --RRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSS 493
P P P L P P+L +P P +P P P P Q+ + +
Sbjct: 138 PVNLSPPPPPVLLSPPPPPVLFSP-----PPPTVTRPPPPPTITRSPPPPRPQAAAYYKK 192
Query: 494 VRSPNP 499
P P
Sbjct: 193 TPPPPP 198
>At4g13340 extensin-like protein
Length = 760
Score = 41.2 bits (95), Expect = 0.002
Identities = 49/185 (26%), Positives = 69/185 (36%), Gaps = 27/185 (14%)
Query: 320 PIPESQPAAQTTSPPHSPRSSFF---QPSPNEAPLWNN-----FRTNPPDPLIQPHPSPS 371
P P S P + PPHSP + P P P+ + + PP P I+P P P
Sbjct: 564 PPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPPPPP 623
Query: 372 --KHQNVWLPELLIQTLPQPIHLY-----PSQLMLLTLLPPITLNPLENSWRSEKEKVST 424
++ P ++ + P P +Y P + + PP P+ S E
Sbjct: 624 CIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPP---PPVHYSSPPPPEV--- 677
Query: 425 LEEYYLTCPSPRRYPGPRPERLVDPDE-PILANPLHEA--DPLAQQVQPAP-HQQQPVQP 480
Y+ PSP Y P P +E P A +H + P+ P P Q P P
Sbjct: 678 --HYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSPPPP 735
Query: 481 EPEPE 485
PE E
Sbjct: 736 SPEYE 740
Score = 38.5 bits (88), Expect = 0.014
Identities = 53/206 (25%), Positives = 64/206 (30%), Gaps = 42/206 (20%)
Query: 306 PISALLKQSSNPLTPIPE---SQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDP 362
P+ L S P P P S P T+ PP SP + P P PP P
Sbjct: 397 PVVTPLPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPP---------PPPP 447
Query: 363 LIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKV 422
+ P P P P + + P P P PP +P S V
Sbjct: 448 VYSPPPPPPPP-----PPPPVYSPPPPPPPPPP--------PPPVYSPPPPSPPPPPPPV 494
Query: 423 STLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQV------QPAPHQQQ 476
+ P P P P P P P+ ++P P V P PH
Sbjct: 495 YS--------PPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPP 546
Query: 477 PVQ---PEPEPEQSVSNHSSVRSPNP 499
P Q P PEP S SP P
Sbjct: 547 PPQFSPPPPEPYYYSSPPPPHSSPPP 572
Score = 38.1 bits (87), Expect = 0.018
Identities = 54/207 (26%), Positives = 68/207 (32%), Gaps = 24/207 (11%)
Query: 314 SSNPLTPIPESQPAAQTTSPP---HSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP 370
SS P P P P T PP HSP F P P E +++ PP PH P
Sbjct: 519 SSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSS--PPPPHSSPPPHSPP 576
Query: 371 SKHQNV--WLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNP------LENSWRSEKEKV 422
H P L P P+ P + PP + P +E S V
Sbjct: 577 PPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVV 636
Query: 423 STLEEYYLTCPSPRRY---PGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQ 479
+Y + P P Y P P P P P P+H + P +V PV
Sbjct: 637 -----HYSSPPPPPVYYSSPPPPPVYYSSPPPP---PPVHYSSPPPPEVHYHSPPPSPVH 688
Query: 480 PEPEPEQSVSNHSSVRSPNPLVATSEP 506
P + P P+V S P
Sbjct: 689 YSSPPPPPSAPCEESPPPAPVVHHSPP 715
Score = 30.4 bits (67), Expect = 3.8
Identities = 26/99 (26%), Positives = 37/99 (37%), Gaps = 6/99 (6%)
Query: 302 FDALPISALLKQSSNPLTPIPESQPAAQTT---SPPHSPRSSFFQPSPNEAPLWNNFRTN 358
+ + P + S P P+ S P SPP SP P P AP +
Sbjct: 648 YSSPPPPPVYYSSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEES---P 704
Query: 359 PPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQL 397
PP P++ P P + P ++ Q+ P P Y L
Sbjct: 705 PPAPVVHHSPPPPMVHHSPPPPVIHQSPPPPSPEYEGPL 743
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,872,927
Number of Sequences: 26719
Number of extensions: 882505
Number of successful extensions: 5882
Number of sequences better than 10.0: 325
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 256
Number of HSP's that attempted gapping in prelim test: 4512
Number of HSP's gapped (non-prelim): 989
length of query: 747
length of database: 11,318,596
effective HSP length: 107
effective length of query: 640
effective length of database: 8,459,663
effective search space: 5414184320
effective search space used: 5414184320
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0234.16


