

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0233.7
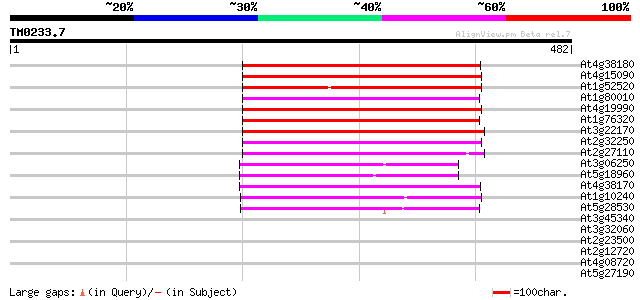
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 182 3e-46
At4g15090 unknown protein 181 1e-45
At1g52520 F6D8.26 178 5e-45
At1g80010 hypothetical protein 170 1e-42
At4g19990 putative protein 169 4e-42
At1g76320 putative phytochrome A signaling protein 168 5e-42
At3g22170 far-red impaired response protein, putative 166 3e-41
At2g32250 Mutator-like transposase 164 8e-41
At2g27110 Mutator-like transposase 156 2e-38
At3g06250 unknown protein 142 5e-34
At5g18960 FAR1 - like protein 141 7e-34
At4g38170 hypothetical protein 138 6e-33
At1g10240 unknown protein 107 1e-23
At5g28530 far-red impaired response protein (FAR1) - like 74 2e-13
At3g45340 putative protein 38 0.014
At3g32060 hypothetical protein 37 0.019
At2g23500 Mutator-like transposase 37 0.019
At2g12720 putative protein 36 0.054
At4g08720 putative protein 35 0.070
At5g27190 putative protein 35 0.092
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 182 bits (463), Expect = 3e-46
Identities = 83/205 (40%), Positives = 131/205 (63%), Gaps = 1/205 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFW D ++ + F D V+FD+TY +NR + F+PF GVNHHGQ ILFGC + + +
Sbjct: 282 VFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEA 341
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
++VWLF + L M P +I T I+AA++ VF + HRFC WHI+KK EKL+ +
Sbjct: 342 SFVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHV 401
Query: 321 -TQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
++ + V + +F+ +++ Y L+++EWL +++SDRR+WVP+Y++
Sbjct: 402 FLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRD 461
Query: 380 IFWAGMSTTQRSESMNAFFDGYVNS 404
F+A MS T RS+S+N++FDGY+N+
Sbjct: 462 TFFADMSLTHRSDSINSYFDGYINA 486
>At4g15090 unknown protein
Length = 768
Score = 181 bits (458), Expect = 1e-45
Identities = 86/206 (41%), Positives = 130/206 (62%), Gaps = 1/206 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
+FW DA+SR Y SF DVVSFD+TY+ + + F+GVNHH Q +L GC L++ + E
Sbjct: 199 LFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESME 258
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNG- 319
T+VWL K+ L M GRAPK I+T Q + + +AV E+ + H F WH+++K+PE +
Sbjct: 259 TFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHV 318
Query: 320 LTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
+ +++ ++ S EFD KM+ + L+ +EWL L R++WVP ++
Sbjct: 319 MKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWLHEHRQKWVPTFMSD 378
Query: 380 IFWAGMSTTQRSESMNAFFDGYVNSK 405
+F AGMST+QRSES+N+FFD Y++ K
Sbjct: 379 VFLAGMSTSQRSESVNSFFDKYIHKK 404
>At1g52520 F6D8.26
Length = 703
Score = 178 bits (452), Expect = 5e-45
Identities = 80/205 (39%), Positives = 129/205 (62%), Gaps = 1/205 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFW DA S+ + F DV+ DS+Y++ + + F GVNHHG++ L CG L+ + E
Sbjct: 280 VFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETME 339
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
+Y WL K L M+ R+P+ I+T +C+ ++AA+ +VF SH RF HIM+K+PEKL GL
Sbjct: 340 SYHWLLKVWLSVMK-RSPQTIVTDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPEKLGGL 398
Query: 321 TQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKSI 380
Y ++K VY++++ EF+ M+ ++ + ENEWL SL+ +R +W P+Y+K
Sbjct: 399 HNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDT 458
Query: 381 FWAGMSTTQRSESMNAFFDGYVNSK 405
F+AG++ E++ FF+ YV+ +
Sbjct: 459 FFAGIAAAHPGETLKPFFERYVHKQ 483
>At1g80010 hypothetical protein
Length = 696
Score = 170 bits (431), Expect = 1e-42
Identities = 84/204 (41%), Positives = 119/204 (58%), Gaps = 1/204 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFW+DAR+RAAY F DV+ FD+T L+N + FVG+NHHG +IL GCGLL+ + E
Sbjct: 275 VFWIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFE 334
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
TYVWLF++ L CM GR P+ IT QC++++ AV EVF +HHR H++ + + + L
Sbjct: 335 TYVWLFRAWLTCMLGRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQL 394
Query: 321 TQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKSI 380
L +VY ++ EF+ +MI + + NE + +F DR W P+Y+K
Sbjct: 395 QDSDLFPMALNRVVYGCLKVEEFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDT 454
Query: 381 FWAGMSTTQRSESMNAF-FDGYVN 403
F AG T F F GYV+
Sbjct: 455 FLAGALTFPLGNVAAPFIFSGYVH 478
>At4g19990 putative protein
Length = 672
Score = 169 bits (427), Expect = 4e-42
Identities = 80/207 (38%), Positives = 128/207 (61%), Gaps = 2/207 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLL-SRKDK 259
+FWVDA+ R Y F DVVS D+T++ N + F GVNHHGQ +L G GLL + + K
Sbjct: 189 IFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESK 248
Query: 260 ETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNG 319
+VWLF++ L+ M G P+ I+T + ++ AV+EVF S H F W + ++PEKL
Sbjct: 249 SGFVWLFRAWLKAMHGCRPRVILTKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGH 308
Query: 320 LTQY-KEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVK 378
+ + K++ + +Y S ++ +F+ ++++ + +++N WL SL+ DR WVP+Y+K
Sbjct: 309 VIRLEKKLVDEINDAIYGSCQSEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMK 368
Query: 379 SIFWAGMSTTQRSESMNAFFDGYVNSK 405
+ AGM T QRS+S+N+ D Y+ K
Sbjct: 369 DVSLAGMCTAQRSDSVNSGLDKYIQRK 395
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 168 bits (426), Expect = 5e-42
Identities = 82/204 (40%), Positives = 127/204 (62%), Gaps = 1/204 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFWVDA+ Y SF DVVSF+++Y ++ + FVGVNHH Q +L GCGLL+
Sbjct: 195 VFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGVNHHVQPVLLGCGLLADDTVY 254
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
TYVWL +S L M G+ PK ++T Q +I+AA+ V E+ H +C WH++ ++P L+
Sbjct: 255 TYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPETRHCYCLWHVLDQLPRNLDYW 314
Query: 321 TQYKE-IKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
+ +++ K L +Y S EFD K+I+ + L++ W+ SL+ +R+ W P +++
Sbjct: 315 SMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDVPWMRSLYEERKFWAPTFMRG 374
Query: 380 IFWAGMSTTQRSESMNAFFDGYVN 403
I +AG+S RSES+N+ FD YV+
Sbjct: 375 ITFAGLSMRCRSESVNSLFDRYVH 398
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 166 bits (419), Expect = 3e-41
Identities = 81/209 (38%), Positives = 127/209 (60%), Gaps = 1/209 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFWVDA+SR Y SF DVVS D+TY+ N+ M + FVGVN H Q ++ GC L+S +
Sbjct: 267 VFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAA 326
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
TY WL ++ L + G+APK +IT + + V E+F + H WH++ KV E L +
Sbjct: 327 TYSWLMETWLRAIGGQAPKVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQV 386
Query: 321 T-QYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
Q+ + +Y S + +F +K + + L++++W+ SL+ DR++W P Y+
Sbjct: 387 VKQHDNFMPKFEKCIYKSGKDEDFARKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTD 446
Query: 380 IFWAGMSTTQRSESMNAFFDGYVNSKNLI 408
+ AGMST+QR++S+NAFFD Y++ K +
Sbjct: 447 VLLAGMSTSQRADSINAFFDKYMHKKTSV 475
>At2g32250 Mutator-like transposase
Length = 684
Score = 164 bits (416), Expect = 8e-41
Identities = 83/206 (40%), Positives = 123/206 (59%), Gaps = 1/206 (0%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFW+DA+++ Y SF DVV FD+ Y+ N + F+PF+GV+HH Q +L GC L+ +
Sbjct: 202 VFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHHRQYVLLGCALIGEVSES 261
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNG- 319
TY WLF++ L+ + G+AP +IT Q + + VVEVF + H FC W ++ K+ E LN
Sbjct: 262 TYSWLFRTWLKAVGGQAPGVMITDQDKLLSDIVVEVFPDVRHIFCLWSVLSKISEMLNPF 321
Query: 320 LTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
++Q ++ V S F+ MI + L ENEW+ LF DR++WVP Y
Sbjct: 322 VSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNENEWVQLLFRDRKKWVPHYFHG 381
Query: 380 IFWAGMSTTQRSESMNAFFDGYVNSK 405
I AG+S +RS S+ + FD Y+NS+
Sbjct: 382 ICLAGLSGPERSGSIASHFDKYMNSE 407
>At2g27110 Mutator-like transposase
Length = 851
Score = 156 bits (395), Expect = 2e-38
Identities = 79/209 (37%), Positives = 123/209 (58%), Gaps = 3/209 (1%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
VFW D+RSR AY F D V+ D+ Y N+ + F+PF GVNHHGQ+ILFGC L+ +
Sbjct: 229 VFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTGVNHHGQAILFGCALILDESDT 288
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKL-NG 319
+++WLFK+ L MR + P +++T Q R+IQ A +VF + H W ++++ EKL +
Sbjct: 289 SFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPGARHCINKWDVLREGQEKLAHV 348
Query: 320 LTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYVKS 379
Y + L + + EF+ +I+ Y L +EWL+SL++ R +WVP+Y +
Sbjct: 349 CLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRHEWLNSLYNARAQWVPVYFRD 408
Query: 380 IFWAGMSTTQRSESMNAFFDGYVNSKNLI 408
F+A + +Q +FFDGYVN + +
Sbjct: 409 SFFAAVFPSQGYS--GSFFDGYVNQQTTL 435
>At3g06250 unknown protein
Length = 764
Score = 142 bits (357), Expect = 5e-34
Identities = 69/188 (36%), Positives = 104/188 (54%), Gaps = 2/188 (1%)
Query: 198 CEMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRK 257
C +FW D+RSR A F D V FD++Y ++ F+ F+G NHH Q +L G L++ +
Sbjct: 355 CMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSVPFATFIGFNHHRQPVLLGGALVADE 414
Query: 258 DKETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKL 317
KE + WLF++ L M GR P++++ Q IQ AV +VF +HHRF W I K E L
Sbjct: 415 SKEAFSWLFQTWLRAMSGRRPRSMVADQDLPIQQAVAQVFPGTHHRFSAWQIRSKERENL 474
Query: 318 NGLTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYV 377
E K + +Y S EFD ++ Y L++N WL ++ R +WVP Y+
Sbjct: 475 RSFP--NEFKYEYEKCLYQSQTTVEFDTMWSSLVNKYGLRDNMWLREIYEKREKWVPAYL 532
Query: 378 KSIFWAGM 385
++ F+ G+
Sbjct: 533 RASFFGGI 540
>At5g18960 FAR1 - like protein
Length = 788
Score = 141 bits (356), Expect = 7e-34
Identities = 67/188 (35%), Positives = 109/188 (57%), Gaps = 2/188 (1%)
Query: 198 CEMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRK 257
C +FW D+R+R A F D V FD++Y ++ F+ +G NHH Q +L GC +++ +
Sbjct: 379 CMSIFWADSRARFACSQFGDSVVFDTSYRKGSYSVPFATIIGFNHHRQPVLLGCAMVADE 438
Query: 258 DKETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKL 317
KE ++WLF++ L M GR P++I+ Q IQ A+V+VF +HHR+ W I +K E+
Sbjct: 439 SKEAFLWLFQTWLRAMSGRRPRSIVADQDLPIQQALVQVFPGAHHRYSAWQIREK--ERE 496
Query: 318 NGLTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIYV 377
N + E K + +Y + EFD +I Y L+++ WL ++ R WVP Y+
Sbjct: 497 NLIPFPSEFKYEYEKCIYQTQTIVEFDSVWSALINKYGLRDDVWLREIYEQRENWVPAYL 556
Query: 378 KSIFWAGM 385
++ F+AG+
Sbjct: 557 RASFFAGI 564
>At4g38170 hypothetical protein
Length = 531
Score = 138 bits (348), Expect = 6e-33
Identities = 70/209 (33%), Positives = 118/209 (55%), Gaps = 2/209 (0%)
Query: 198 CEMVFWVDARSRAAYDSFEDVVSFDSTYLTN-R*NMSFSPFVGVNHHGQSILFGCGLLSR 256
C VFW D R Y F D + FD+TY R + F+ F G NHHGQ +LFGC L+
Sbjct: 29 CGNVFWADPTCRLNYTYFGDTLVFDTTYRRGKRYQVPFAAFTGFNHHGQPVLFGCALILN 88
Query: 257 KDKETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEK 316
+ + ++ WLF++ L+ M P +I R IQ AV VFS++ RF I ++ EK
Sbjct: 89 ESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQVAVSRVFSQTRLRFSQPLIFEETEEK 148
Query: 317 LNGLTQ-YKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPI 375
L + Q + + V ++ A+EF+ ++ Y +++N+WL S+++ R++WV +
Sbjct: 149 LAHVFQAHPTFESEFINCVTETETAAEFEASWDSIVRRYYMEDNDWLQSIYNARQQWVRV 208
Query: 376 YVKSIFWAGMSTTQRSESMNAFFDGYVNS 404
+++ F+ +ST + S +N+FF G+V++
Sbjct: 209 FIRDTFYGELSTNEGSSILNSFFQGFVDA 237
>At1g10240 unknown protein
Length = 680
Score = 107 bits (267), Expect = 1e-23
Identities = 64/210 (30%), Positives = 112/210 (52%), Gaps = 5/210 (2%)
Query: 199 EMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKD 258
E + W A S +Y+ F D V FD+T+ + M +VGVN++G FGC LL ++
Sbjct: 258 ENIAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDEN 317
Query: 259 KETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLN 318
++ W ++ M G+AP+ I+T ++ A+ + H C W ++ K P N
Sbjct: 318 LRSWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFN 377
Query: 319 -GL-TQYKEIKKTLKTLVY-DSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPI 375
GL +Y + K L + +S+E EF+ G M+ + L N +++L++ R W
Sbjct: 378 AGLGERYNDWKAEFYRLYHLESVE--EFELGWRDMVNSFGLHTNRHINNLYASRSLWSLP 435
Query: 376 YVKSIFWAGMSTTQRSESMNAFFDGYVNSK 405
Y++S F AGM+ T RS+++NAF +++++
Sbjct: 436 YLRSHFLAGMTLTGRSKAINAFIQRFLSAQ 465
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 73.9 bits (180), Expect = 2e-13
Identities = 49/207 (23%), Positives = 97/207 (46%), Gaps = 3/207 (1%)
Query: 199 EMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKD 258
E + W S Y F DVV FD++Y + + F G++++G+++L GC LL +
Sbjct: 288 ENIAWAYGDSVRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDES 347
Query: 259 KETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLN 318
++ W ++ + MRGR P+ I+T ++ A+ ++H HI+ K+ +
Sbjct: 348 CRSFTWALQTFVRFMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFS 407
Query: 319 GL--TQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQENEWLSSLFSDRRRWVPIY 376
+ Y+E + L EF+ ++ + L + + L+S R W+P
Sbjct: 408 QTLGSHYEEFRAGFDMLCRAG-NVDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCC 466
Query: 377 VKSIFWAGMSTTQRSESMNAFFDGYVN 403
++ F A T++ + S+++F V+
Sbjct: 467 IREHFVAQTMTSEFNLSIDSFLKRVVD 493
>At3g45340 putative protein
Length = 693
Score = 37.7 bits (86), Expect = 0.014
Identities = 38/163 (23%), Positives = 68/163 (41%), Gaps = 18/163 (11%)
Query: 241 NHHGQSILFGCGLLSRKDKETYVWLFKS*LECMRGRAPKAIITYQCR-SIQAAVVEVFSE 299
NHH I FG ++ ++ ++ W F+ L + P+ + SI V +VF +
Sbjct: 411 NHHHYPIAFG--VIDSENHASWNWFFRK-LNSIIPDDPELVFIRDIHGSIIKGVADVFPK 467
Query: 300 SHHRFCFWHIMKKVPEKLNGLTQYKEIKKTLKTLVYDSIEASEFDDG*HKMIEDYRLQEN 359
+ H C WH+ + + + L G + K +Y A+EF+ I +
Sbjct: 468 ASHGHCVWHLSQNIKKMLAGDKEAGMAKFMELAHIY---TATEFN------IRYAEFRRK 518
Query: 360 EWLSSLFSDR----RRWVPIYVKSIFWAGMSTTQRSESMNAFF 398
+ + DR +W Y + + + T+ +ESMNA F
Sbjct: 519 HPKVATYLDRAIGIEKWARCYFQGDKY-NIDTSNSAESMNAVF 560
>At3g32060 hypothetical protein
Length = 487
Score = 37.4 bits (85), Expect = 0.019
Identities = 26/117 (22%), Positives = 51/117 (43%), Gaps = 4/117 (3%)
Query: 199 EMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*N--MSFSPFVGVNHHGQSILFGCGLLSR 256
+ VF S + V+ D+T+L N + F+ N H I F +L
Sbjct: 247 QYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFA--VLDG 304
Query: 257 KDKETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKV 313
++ ++ W F+ + + +T + S+ A+ V++ +HH +C WH+ + V
Sbjct: 305 ENDASWEWFFEKLKTVVPDTSELVFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNV 361
>At2g23500 Mutator-like transposase
Length = 784
Score = 37.4 bits (85), Expect = 0.019
Identities = 26/117 (22%), Positives = 51/117 (43%), Gaps = 4/117 (3%)
Query: 199 EMVFWVDARSRAAYDSFEDVVSFDSTYLTNR*N--MSFSPFVGVNHHGQSILFGCGLLSR 256
+ VF S + V+ D+T+L N + F+ N H I F +L
Sbjct: 377 QYVFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFA--VLDG 434
Query: 257 KDKETYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKV 313
++ ++ W F+ + + +T + S+ A+ V++ +HH +C WH+ + V
Sbjct: 435 ENDASWEWFFEKLKTVVPDTSELVFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNV 491
>At2g12720 putative protein
Length = 819
Score = 35.8 bits (81), Expect = 0.054
Identities = 28/131 (21%), Positives = 55/131 (41%)
Query: 201 VFWVDARSRAAYDSFEDVVSFDSTYLTNR*NMSFSPFVGVNHHGQSILFGCGLLSRKDKE 260
+F V S A + VV D T+L + + + + Q G++ ++ +
Sbjct: 432 MFIVFGASIAGFHYMRRVVVVDGTFLHGSYKGTLLTALAQDGNFQIFPLAFGVVDTENDD 491
Query: 261 TYVWLFKS*LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKVPEKLNGL 320
++ WLF + AII+ + +SI A+ EV+ + C +H+ K + K
Sbjct: 492 SWRWLFTQLKVVIPDATDLAIISDRHKSIGKAIGEVYPLAARGICTYHLYKNILVKFKRK 551
Query: 321 TQYKEIKKTLK 331
+ +KK +
Sbjct: 552 DLFPLVKKAAR 562
>At4g08720 putative protein
Length = 848
Score = 35.4 bits (80), Expect = 0.070
Identities = 20/104 (19%), Positives = 46/104 (44%), Gaps = 4/104 (3%)
Query: 212 YDSFEDVVSFDSTYLTNR*N--MSFSPFVGVNHHGQSILFGCGLLSRKDKETYVWLFKS* 269
+ + V+ D+T+L N + F+ N H + G+L ++ ++ W F+
Sbjct: 384 FQAMRKVIIVDATHLKNGYGGVLVFASARDPNRH--HYIIAVGVLDGENDASWGWFFEKL 441
Query: 270 LECMRGRAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKV 313
L + ++ + S+ + ++ +HH +C WH+ + V
Sbjct: 442 LSVVPDTPELVFMSDRNSSLIKGIRNAYTAAHHGYCVWHLSQNV 485
>At5g27190 putative protein
Length = 779
Score = 35.0 bits (79), Expect = 0.092
Identities = 21/98 (21%), Positives = 45/98 (45%), Gaps = 4/98 (4%)
Query: 218 VVSFDSTYLTNR*N--MSFSPFVGVNHHGQSILFGCGLLSRKDKETYVWLFKS*LECMRG 275
VV D+T+L + F+ NHH + + ++ R++ ++ W F +
Sbjct: 393 VVIVDATFLKTVYGGMLVFATAQDPNHH--NYIIASAVIDRENDASWSWFFNKLKTVIPD 450
Query: 276 RAPKAIITYQCRSIQAAVVEVFSESHHRFCFWHIMKKV 313
++ + +SI ++++VF + H C WH+ + V
Sbjct: 451 VPGLVFVSDRHQSIIKSIMQVFPNARHGHCVWHLSQNV 488
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.358 0.160 0.581
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,046,142
Number of Sequences: 26719
Number of extensions: 326719
Number of successful extensions: 1713
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1674
Number of HSP's gapped (non-prelim): 34
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0233.7


