

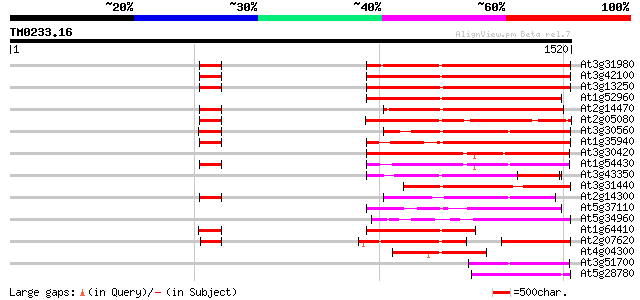
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0233.16
(1520 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g31980 hypothetical protein 521 e-148
At3g42100 putative protein 520 e-147
At3g13250 hypothetical protein 516 e-146
At1g52960 hypothetical protein 503 e-142
At2g14470 pseudogene 463 e-130
At2g05080 putative helicase 458 e-128
At3g30560 hypothetical protein 452 e-127
At1g35940 hypothetical protein 446 e-125
At3g30420 hypothetical protein 441 e-123
At1g54430 hypothetical protein 424 e-118
At3g43350 putative protein 405 e-113
At3g31440 hypothetical protein 390 e-108
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 385 e-107
At5g37110 putative helicase 342 7e-94
At5g34960 putative protein 330 3e-90
At1g64410 unknown protein 285 1e-76
At2g07620 putative helicase 264 3e-70
At4g04300 hypothetical protein 223 9e-58
At3g51700 unknown protein 198 2e-50
At5g28780 putative protein 186 9e-47
>At3g31980 hypothetical protein
Length = 1099
Score = 521 bits (1343), Expect = e-148
Identities = 274/556 (49%), Positives = 376/556 (67%), Gaps = 9/556 (1%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVE 1026
DLV+ + ++ L+EIE +L+ NG +L+DF MP T++ N + E N+++ +
Sbjct: 548 DLVLTDAERRNYALLEIEDMLLCNGSTLQDFKDMPKPTKEG-TDHSNRFITEEKNYNIEK 606
Query: 1027 MSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQK 1086
+ HD+ F+K+ Q IY EII A+ N+GG FFVYG+GGT KTF+WKTL+ +R +
Sbjct: 607 LREDHDDWFNKMTSEQKGIYDEIIKAVLENSGGIFFVYGFGGTSKTFMWKTLSAAVRMRG 666
Query: 1087 QIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDE 1146
I +NVASSGIASLLL GGRTAHS F IP+ ++ + C I S A +L+ ASLIIWDE
Sbjct: 667 LISVNVASSGIASLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDE 726
Query: 1147 APMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVM 1206
APM+SRY FE+LDR+L D+ I N KPFGGKVVV GGDFRQ+LPVI RAEIV+
Sbjct: 727 APMMSRYCFESLDRSLNDV----IGNIDGKPFGGKVVVFGGDFRQVLPVIHGAGRAEIVL 782
Query: 1207 STINSSRLWRFCKVLNLTQNMRLTTSTCDGDN-EEVKHFAKWVLDMGDGKLGDYNDGESN 1265
+ +NSS LW CKVL LT+NMRL ++ D D EE+K F+ W+L +GDG++ + NDGE
Sbjct: 783 AALNSSYLWEHCKVLTLTKNMRLMSNDLDKDEAEEIKEFSNWLLAVGDGRVSEPNDGEVL 842
Query: 1266 VQIPKDLLIYQSSNPIADIVHLMYPDI--VQNVGCVKYYSDKAILAPTLDAVDSINQYAL 1323
+ IP++LLI +++PI I +Y D+ +Q K++ +AIL P V++IN L
Sbjct: 843 IDIPEELLIKDANDPIEAITKAVYGDLDLLQPNNDPKFFQQRAILCPRNTDVNTINDIML 902
Query: 1324 SLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMR 1383
G TYLS+DS+ D + LT +FLN +K SG+P+H L LK G P+ML+R
Sbjct: 903 DKLNGELVTYLSADSIDPQDA-ASLNNPVLTPDFLNSIKLSGLPNHNLTLKIGTPVMLLR 961
Query: 1384 NLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQ 1443
N+D GLCNGTRL V +G +++ A V++G +G V I + + PSD +P + +RRQ
Sbjct: 962 NIDPKGGLCNGTRLQVTQMGNHILEARVITGDRVGDKVIIIKSQISPSDTKLPFRMRRRQ 1021
Query: 1444 FPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTSQ 1503
FP+ +FAMTINKSQGQSL +VG+YLPKPVFSHGQLYVA+SRV S+ GLK+LI +++ +
Sbjct: 1022 FPIAVAFAMTINKSQGQSLKEVGIYLPKPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNT 1081
Query: 1504 LDVTKNIVFQEVFQKI 1519
T N+VF+E+FQ I
Sbjct: 1082 QSQTMNVVFKEIFQNI 1097
Score = 72.8 bits (177), Expect = 1e-12
Identities = 32/60 (53%), Positives = 45/60 (74%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YT+EFQKRGLPHAHILL++ K + ID++I AE+PD ++ P+L+E V + M+HGP
Sbjct: 266 YTVEFQKRGLPHAHILLFMDKSCKFPTSDDIDNIISAEIPDKSKDPKLYEVVKDCMIHGP 325
>At3g42100 putative protein
Length = 1752
Score = 520 bits (1340), Expect = e-147
Identities = 281/557 (50%), Positives = 378/557 (67%), Gaps = 10/557 (1%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDV-V 1025
DL + +++ L EIEKI++ NG +LK+ P + + N L+++EL +++
Sbjct: 1201 DLKLTLAEIRNYTLQEIEKIMLRNGATLKEIQDFPQPSREGI-DNSNRLVVDELRYNIDS 1259
Query: 1026 EMSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQ 1085
+ HDE F LN Q IY EI A+ + GG FF+YG+GGTGKTF+WKTL +RS+
Sbjct: 1260 NLKEKHDEWFQMLNTEQRGIYDEITGAVFNDLGGVFFIYGFGGTGKTFIWKTLAAAVRSR 1319
Query: 1086 KQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWD 1145
QI+LNVASSGIASLLL GGRTAHS F+IPL +E S C I S A L++ ASLIIWD
Sbjct: 1320 GQIVLNVASSGIASLLLEGGRTAHSRFAIPLNPDEFSVCKITPKSDLANLIKEASLIIWD 1379
Query: 1146 EAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIV 1205
EAPM+S++ FE+LD++ DI+ +N +K FGGKVVV GGDFRQ+LPVI R EIV
Sbjct: 1380 EAPMMSKFCFESLDKSFYDIL----NNKDNKVFGGKVVVFGGDFRQVLPVINGAGRVEIV 1435
Query: 1206 MSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDN-EEVKHFAKWVLDMGDGKLGDYNDGES 1264
MS++N+S LW CKVL LT+NMRL + + +E++ F+ W+L +GDG++ + NDGE+
Sbjct: 1436 MSSLNASYLWDHCKVLKLTKNMRLLSGGLSSEEAKEIQQFSDWLLAVGDGRINEPNDGEA 1495
Query: 1265 NVQIPKDLLIYQSSNPIADIVHLMY--PDIVQNVGCVKYYSDKAILAPTLDAVDSINQYA 1322
+ IP++LLI ++ NPI I +Y P + + K++ +AILAPT + V++INQY
Sbjct: 1496 LIDIPEELLIKEAGNPIEAISKEIYGDPSELHMINDPKFFQRRAILAPTNEDVNTINQYM 1555
Query: 1323 LSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLM 1382
L E+ YLS+DS+ D D + +T +FLN ++ +GMP H L LK GAP+ML+
Sbjct: 1556 LEHLKSEERIYLSADSIDPTDSD-SLANPVITPDFLNSIQLTGMPHHALRLKVGAPVMLL 1614
Query: 1383 RNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRR 1442
RNLD GLCNGTRL + L V+ A V++ IG +V I +NL PSD +P K +RR
Sbjct: 1615 RNLDPKGGLCNGTRLQITQLAKQVVQAKVITRDRIGDIVLIPLINLTPSDTKLPFKMRRR 1674
Query: 1443 QFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTS 1502
QFPL +FAMTINKSQGQSL QVG+YLPKPVFSHGQLYVA+SRV S+ GLKILI ++D +
Sbjct: 1675 QFPLSVAFAMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGN 1734
Query: 1503 QLDVTKNIVFQEVFQKI 1519
T N+VF+EVFQ I
Sbjct: 1735 MQKQTTNVVFKEVFQNI 1751
Score = 76.6 bits (187), Expect = 1e-13
Identities = 33/60 (55%), Positives = 46/60 (76%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
+T+EFQKRGLPHAHILL++ ++KL + ID +I AE+PD + PEL+E + N M+HGP
Sbjct: 779 HTVEFQKRGLPHAHILLFMDAKSKLPTADDIDKIISAEIPDKDKEPELYEVIKNSMIHGP 838
>At3g13250 hypothetical protein
Length = 1419
Score = 516 bits (1330), Expect = e-146
Identities = 279/557 (50%), Positives = 378/557 (67%), Gaps = 10/557 (1%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFD-VV 1025
+L + E ++++ L EIEKI++ NG +L+D P + + N L+++EL ++
Sbjct: 869 ELTLTEAQIQNYTLQEIEKIMLFNGATLEDIEHFPKPSREGI-DNSNRLIIDELRYNNQS 927
Query: 1026 EMSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQ 1085
++ H + KL Q IY +I A+ + GG FFVYG+GGTGKTF+WKTL +RS+
Sbjct: 928 DLKKKHSDWIQKLTPEQRGIYDQITNAVFNDLGGVFFVYGFGGTGKTFIWKTLAAAVRSK 987
Query: 1086 KQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWD 1145
QI LNVASSGIASLLL GGRTAHS FSIPL +E S C I S A L++ ASLIIWD
Sbjct: 988 GQICLNVASSGIASLLLEGGRTAHSRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWD 1047
Query: 1146 EAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIV 1205
EAPM+S++ FEALD++ DI++ +K FGGKV+V GGDFRQ+LPVI RAEIV
Sbjct: 1048 EAPMMSKFCFEALDKSFSDIIK----RVDNKVFGGKVMVFGGDFRQVLPVINGAGRAEIV 1103
Query: 1206 MSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDN-EEVKHFAKWVLDMGDGKLGDYNDGES 1264
MS++N+S LW CKVL LT+NMRL + D +E++ F+ W+L +GDG++ + NDGE
Sbjct: 1104 MSSLNASYLWDHCKVLRLTKNMRLLNNDLSVDEAKEIQEFSDWLLAVGDGRVNEPNDGEV 1163
Query: 1265 NVQIPKDLLIYQSSNPIADIVHLMY--PDIVQNVGCVKYYSDKAILAPTLDAVDSINQYA 1322
+ IP++LLI ++ NPI I +Y P + + K++ +AILAP + V++INQY
Sbjct: 1164 IIDIPEELLIQEADNPIEAISREIYGDPTKLHEISDPKFFQRRAILAPKNEDVNTINQYM 1223
Query: 1323 LSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLM 1382
L E+ YLS+DS+ D D ++ +T +FLN +K SGMP H L LK GAP+ML+
Sbjct: 1224 LEHLDSEERIYLSADSIDPSDSD-SLKNPVITPDFLNSIKVSGMPHHSLRLKVGAPVMLL 1282
Query: 1383 RNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRR 1442
RNLD GLCNGTRL + L +++ A V++G IG++VYI +N+ PSD +P K +RR
Sbjct: 1283 RNLDPKGGLCNGTRLQITQLCSHIVEAKVITGDRIGQIVYIPLINITPSDTKLPFKMRRR 1342
Query: 1443 QFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTS 1502
QFPL +F MTINKSQGQSL QVG+YLPKPVFSHGQLYVA+SRV S+ GLKILI +++
Sbjct: 1343 QFPLSVAFVMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKTGLKILILDKEGK 1402
Query: 1503 QLDVTKNIVFQEVFQKI 1519
T N+VF+EVFQ I
Sbjct: 1403 IQKQTTNVVFKEVFQNI 1419
Score = 75.9 bits (185), Expect = 2e-13
Identities = 33/60 (55%), Positives = 46/60 (76%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YT+EFQKRGLPHAHILL+++ +KL + ID +I AE+P+ + PEL+E + N M+HGP
Sbjct: 447 YTVEFQKRGLPHAHILLFMAANSKLPTADDIDKIISAEIPNKDKEPELYEVIKNSMMHGP 506
>At1g52960 hypothetical protein
Length = 924
Score = 503 bits (1295), Expect = e-142
Identities = 257/527 (48%), Positives = 353/527 (66%), Gaps = 6/527 (1%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVE 1026
DL + + + CL EI ++L NG SL + MP + ++ E N +L+E +D
Sbjct: 402 DLQLSDEERQQYCLQEIARLLTKNGVSLSKWKQMPQI-SDEHVEKCNHFILDERKYDRAY 460
Query: 1027 MSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQK 1086
+ H+E + + Q KIY EI+ A+ + GG FFVYG+GGTGKTFLWK L+ +RS+
Sbjct: 461 LIEKHEEWLTMVTSEQKKIYDEIMDAVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKG 520
Query: 1087 QIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDE 1146
I LNVASSGIA+LLL GGRT HS F IP+ NE S C I +GS +L++ A+LIIWDE
Sbjct: 521 DISLNVASSGIAALLLDGGRTTHSRFGIPINPNESSTCNISRGSDLGELVKEANLIIWDE 580
Query: 1147 APMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVM 1206
PM+S++ FE+LDRTLRDIM +N DKPFGGK +V GGDFRQ+LPVI R EIV
Sbjct: 581 TPMMSKHCFESLDRTLRDIM----NNPGDKPFGGKGIVFGGDFRQVLPVINGAGREEIVF 636
Query: 1207 STINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNV 1266
+ +NSS +W CKVL LT+NMRL + + + ++++F+KW+LD+GDGK+ NDG + +
Sbjct: 637 AALNSSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGIALI 696
Query: 1267 QIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPTLDAVDSINQYALSLF 1326
IP++ LI ++P+ I+ +Y + K++ +AIL PT + V+SIN++ +S+
Sbjct: 697 DIPEEFLINGDNDPVESIIEAVYGNTFMEEKDPKFFQGRAILCPTNEDVNSINEHMMSML 756
Query: 1327 PGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMRNLD 1386
G E+ YLSSDS+ D D + +FLN ++ SG+P+H L LK G P+ML+RN+D
Sbjct: 757 DGEERIYLSSDSIDPADTS-SANNDAYSADFLNSVRVSGLPNHCLRLKVGCPVMLLRNMD 815
Query: 1387 ISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQFPL 1446
+ GLCNGTRL V + VI A ++G +GK+V I RM + PSD +P K +RRQFPL
Sbjct: 816 PNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTRLPFKMRRRQFPL 875
Query: 1447 IASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLK 1493
+FAMTINKSQGQ+L VG+YLP+PVFSHGQLYVAISRV S+ G K
Sbjct: 876 SVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 922
Score = 35.0 bits (79), Expect = 0.33
Identities = 15/38 (39%), Positives = 23/38 (60%)
Query: 537 NKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
+KL + D +I AE+PD + P L+ V + M+HGP
Sbjct: 5 SKLSTAEDTDKIITAEIPDKKKKPGLYAVVKDCMIHGP 42
>At2g14470 pseudogene
Length = 1265
Score = 463 bits (1191), Expect = e-130
Identities = 248/494 (50%), Positives = 336/494 (67%), Gaps = 13/494 (2%)
Query: 1013 NVLLLNELNFDVVEMSNL---HDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGT 1069
N L++ EL ++ SNL H+E L Q +Y EI A+ N GG FFVYG+GGT
Sbjct: 776 NRLIVEELRYN--RESNLKEKHEEWKQMLTPEQRGVYNEITEAVFNNLGGVFFVYGFGGT 833
Query: 1070 GKTFLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQG 1129
GKTF+WKTL+ +R + QI+LNVASSGIASLLL GGRTAHS F IPL +E S C I
Sbjct: 834 GKTFIWKTLSATIRYRDQIVLNVASSGIASLLLEGGRTAHSRFGIPLNPDEFSVCKIKPK 893
Query: 1130 SPKAKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDF 1189
S A L++ ASL+IWDEAPM+SR+ FEALD++ DI++ N+ + FGGKVVV GGDF
Sbjct: 894 SDLANLVKKASLVIWDEAPMMSRFCFEALDKSFSDIIK----NTDNTVFGGKVVVFGGDF 949
Query: 1190 RQILPVIPKGRRAEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTC-DGDNEEVKHFAKWV 1248
RQ+ PVI RAEIVMS++N+S LW CKVL LT+N RL + + + +E++ F+ W+
Sbjct: 950 RQVFPVINGAGRAEIVMSSLNASYLWDNCKVLKLTKNTRLLANNLSETEAKEIQEFSDWL 1009
Query: 1249 LDMGDGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMY--PDIVQNVGCVKYYSDKA 1306
L +GDG++ + NDG + + IP+DLLI + PI I + +Y P I+ + K++ +A
Sbjct: 1010 LAVGDGRINESNDGVAIIDIPEDLLITNADKPIESITNEIYGDPKILHEITDPKFFQGRA 1069
Query: 1307 ILAPTLDAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGM 1366
ILA + V++IN+Y L E+ YLS+DS+ D D + +T +FLN +K G+
Sbjct: 1070 ILASKNEDVNTINEYLLDQLHAEERIYLSADSIDPTDSD-SLSNPVITPDFLNSIKLPGL 1128
Query: 1367 PDHKLVLKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRM 1426
P+H L LK GAP++L+RNLD GLCNGTRL + L ++ A V++G IG ++ I +
Sbjct: 1129 PNHSLRLKVGAPVLLLRNLDPKGGLCNGTRLQITQLCTQIVEAKVITGDRIGHIILIPTV 1188
Query: 1427 NLMPSDESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRV 1486
NL P++ +P K +RRQFPL +F MTINKS+GQSL VG+YLPKPVFSHGQLYVA+SRV
Sbjct: 1189 NLTPTNTKLPFKMRRRQFPLSVAFVMTINKSEGQSLEHVGLYLPKPVFSHGQLYVALSRV 1248
Query: 1487 KSRAGLKILICNED 1500
S+ GLKILI ++D
Sbjct: 1249 TSKKGLKILILDKD 1262
Score = 77.4 bits (189), Expect = 6e-14
Identities = 34/60 (56%), Positives = 46/60 (76%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YT+EFQKRGLPHAHILL++ ++KL + ID +I AE+PD + PEL+E + N M+HGP
Sbjct: 448 YTVEFQKRGLPHAHILLFMHAKSKLPTSDDIDKLISAEIPDKEKEPELYEVIKNSMIHGP 507
>At2g05080 putative helicase
Length = 1219
Score = 458 bits (1178), Expect = e-128
Identities = 264/560 (47%), Positives = 352/560 (62%), Gaps = 39/560 (6%)
Query: 964 LSLDLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFD 1023
L DL++ + K L EI+ IL NG SL + MP V + + NVL+L+E +D
Sbjct: 674 LFTDLILSDEEKKVYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDT-NVLILDEKGYD 732
Query: 1024 VVEMSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLR 1083
+ H + KL Q +Y II A+N N GG FFVYG+GGTGKTFLWKTL+ LR
Sbjct: 733 RESETKKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFGGTGKTFLWKTLSAALR 792
Query: 1084 SQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLII 1143
S+ I+LNVASSGIASLLL GGRTAHS IPL NE + C + GS +A L++ ASLII
Sbjct: 793 SKGDIVLNVASSGIASLLLEGGRTAHSRSGIPLNPNEFTTCNMKAGSDRANLVKEASLII 852
Query: 1144 WDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAE 1203
WDEAPM+SR+ FE+LDR+L DI N +KPFGGKVVV GGDFRQ+LPVIP A+
Sbjct: 853 WDEAPMMSRHCFESLDRSLSDI----CGNCDNKPFGGKVVVFGGDFRQVLPVIPGADTAD 908
Query: 1204 IVMSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGE 1263
IVM+ +NSS LW CKVL LT+NM L + +W+L +GDG++G+ NDGE
Sbjct: 909 IVMAALNSSYLWSHCKVLTLTKNMCLFSE-------------EWILAVGDGRIGEPNDGE 955
Query: 1264 SNVQIPKDLLIYQSSNPIADIVHLMYPDI--VQNVGCVKYYSDKAILAPTLDAVDSINQY 1321
+ + IP + LI ++ +PI I +Y DI + ++ ++AIL PT + V+ IN+
Sbjct: 956 ALIDIPSEFLITKAKDPIQAICTEIYGDITKIHEQKDPVFFQERAILCPTNEDVNQINET 1015
Query: 1322 ALSLFPGNEKTYLSSDSVWRVDEDVGIEAD-WLTTEFLNGLKCSGMPDHKLVLKEGAPIM 1380
L G E T+LSSDS+ D+G + LT EFLN +K G+ +HKL LK G+P+M
Sbjct: 1016 MLDNLQGEELTFLSSDSL--DTADIGSRNNPVLTPEFLNNVKVLGLSNHKLRLKIGSPVM 1073
Query: 1381 LMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQ 1440
L+RN+D GL NGTRL + + P ++ A++L+G +D +P + +
Sbjct: 1074 LLRNIDPIGGLMNGTRLQIMQMSPFILQAMILTGDR--------------ADTKLPFRMR 1119
Query: 1441 RRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNED 1500
R Q PL FAMTINKSQGQSL +VG++LP+P FSH QLYVAISRV S++GLKILI N++
Sbjct: 1120 RTQLPLAVCFAMTINKSQGQSLKRVGIFLPRPCFSHSQLYVAISRVTSKSGLKILIVNDE 1179
Query: 1501 TSQLDVTKNIVFQEVFQKIY 1520
TK F + F +I+
Sbjct: 1180 GKPQKQTKK--FTKKFLRIF 1197
Score = 79.7 bits (195), Expect = 1e-14
Identities = 37/60 (61%), Positives = 42/60 (69%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YTIEFQKRGLPHAHIL+WL + KL ID I AE+PD + PELFE + MVHGP
Sbjct: 265 YTIEFQKRGLPHAHILIWLDSKCKLTRAEHIDKAISAEIPDKLKDPELFEVIKEMMVHGP 324
>At3g30560 hypothetical protein
Length = 1473
Score = 452 bits (1164), Expect = e-127
Identities = 244/507 (48%), Positives = 322/507 (63%), Gaps = 32/507 (6%)
Query: 1013 NVLLLNELNFDVVEMSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKT 1072
N L+L+E N++ + +HD+ L Q K+Y +I+ A+ N GG
Sbjct: 999 NQLILDERNYNRETLKTIHDDWLKMLTTEQKKVYDKIMDAVLNNKGG------------- 1045
Query: 1073 FLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPK 1132
I LNVASSGIASLLL GGRTAHS F IPL +E S C + +GS
Sbjct: 1046 --------------DICLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDL 1091
Query: 1133 AKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQI 1192
A+L+ A LIIWDEAPM+S+Y FE+LD++L+DI+ D PFGGK+++ GGDFRQI
Sbjct: 1092 AELVTAAKLIIWDEAPMMSKYCFESLDKSLKDILS----TPEDMPFGGKLIIFGGDFRQI 1147
Query: 1193 LPVIPKGRRAEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMG 1252
LPVI R IV S++NSS LW++CKV LT+NMRL + E++ F+KW+L +G
Sbjct: 1148 LPVILAAGRELIVKSSLNSSHLWQYCKVFKLTKNMRLLQDIDINEAREIEDFSKWILAVG 1207
Query: 1253 DGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPTL 1312
+GKL NDG + +QI D+LI + NPI I+ +Y K++ D+AIL PT
Sbjct: 1208 EGKLNQPNDGVTQIQIRDDILIPEGDNPIESIIKAVYGTSFDEERDPKFFQDRAILCPTN 1267
Query: 1313 DAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLV 1372
D V+SIN + LS G EK Y SSDS+ D + T +FLN +K SG+P+H L
Sbjct: 1268 DDVNSINDHMLSKLTGEEKIYRSSDSIDPSDTRADKNPVY-TPDFLNKIKISGLPNHLLW 1326
Query: 1373 LKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSD 1432
LK G P+ML+RNLD GL NGTRL + LG ++ +L+GT +GK+V I RM L PSD
Sbjct: 1327 LKVGCPVMLLRNLDSHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKLVIIPRMPLTPSD 1386
Query: 1433 ESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGL 1492
+P K +RRQFPL +FAMTINKSQGQSL VG+YLPKPVFSHGQLYVA+SRVKS+ GL
Sbjct: 1387 RRLPFKMKRRQFPLSVAFAMTINKSQGQSLGNVGIYLPKPVFSHGQLYVAMSRVKSKGGL 1446
Query: 1493 KILICNEDTSQLDVTKNIVFQEVFQKI 1519
K+LI + Q + T N+VF+E+F+ +
Sbjct: 1447 KVLITDSKGKQKNETTNVVFKEIFRNL 1473
Score = 75.9 bits (185), Expect = 2e-13
Identities = 33/63 (52%), Positives = 43/63 (67%)
Query: 512 LGTYTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMV 571
L TYTIEFQKRGLPHAHI++W+ K +D +I AE+PD P+L++ VS M+
Sbjct: 551 LATYTIEFQKRGLPHAHIIVWMDPRYKFPIADHVDKIIFAEIPDKENDPKLYQAVSECMI 610
Query: 572 HGP 574
HGP
Sbjct: 611 HGP 613
>At1g35940 hypothetical protein
Length = 1678
Score = 446 bits (1146), Expect = e-125
Identities = 251/556 (45%), Positives = 336/556 (60%), Gaps = 67/556 (12%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVE 1026
DL + E +K+ L EIEKI++ NG +L+D P
Sbjct: 1186 DLTLTETEIKNYTLQEIEKIMLSNGATLEDIDEFP------------------------- 1220
Query: 1027 MSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQK 1086
DE L Q +Y I A+ N GG FFVYG+GGTGKTF+WKTL+ +R +
Sbjct: 1221 -KPTRDEWKQMLTPEQRGVYNAITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSAAIRCRG 1279
Query: 1087 QIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDE 1146
QI+LNVASSGIASLLL GGRTAHS F IPL +E S
Sbjct: 1280 QIVLNVASSGIASLLLEGGRTAHSRFGIPLNHDEFSV----------------------- 1316
Query: 1147 APMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVM 1206
+LD++ DI++ N+++K FGGKVVV GGDFRQ+LPVI RAEIVM
Sbjct: 1317 ----------SLDKSFSDIIK----NTNNKVFGGKVVVFGGDFRQVLPVINGAGRAEIVM 1362
Query: 1207 STINSSRLWRFCKVLNLTQNMRLTTSTCDG-DNEEVKHFAKWVLDMGDGKLGDYNDGESN 1265
S++N+S LW CKVL LT+NMRL + + +E++ F+ W+L + DG++ + NDG +
Sbjct: 1363 SSLNASYLWDHCKVLKLTKNMRLLANNLSATEAKEIQEFSDWLLAVSDGRINEPNDGVAT 1422
Query: 1266 VQIPKDLLIYQSSNPIADIVHLMY--PDIVQNVGCVKYYSDKAILAPTLDAVDSINQYAL 1323
+ IP+DLLI + PI I + +Y P I+ + K++ +AILAP + V++IN+Y L
Sbjct: 1423 IDIPEDLLITNADKPIETITNEIYGDPKILHEITDPKFFQGRAILAPKNEDVNTINEYLL 1482
Query: 1324 SLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMR 1383
E+ YLS+DS+ D D + +T +FLN +K G+P+H L LK GAP+ML+R
Sbjct: 1483 EQLDAEERIYLSADSIDPTDSD-SLNNPVITPDFLNSIKLPGLPNHSLCLKVGAPVMLLR 1541
Query: 1384 NLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQ 1443
NLD GLCNGTRL + L ++ A V++G IG +V I +NL P+D +P K +RRQ
Sbjct: 1542 NLDPKGGLCNGTRLQITQLCTQIVEAKVITGDRIGNIVLIPTVNLTPTDTKLPFKMRRRQ 1601
Query: 1444 FPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTSQ 1503
FPL +FAMTINKSQGQSL +G+YLPKPVFSHGQLYVA+SRV S+ GLKILI ++D
Sbjct: 1602 FPLSVAFAMTINKSQGQSLEHIGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGKL 1661
Query: 1504 LDVTKNIVFQEVFQKI 1519
T N+VF+EVFQ I
Sbjct: 1662 QKQTTNVVFKEVFQNI 1677
Score = 76.3 bits (186), Expect = 1e-13
Identities = 33/60 (55%), Positives = 46/60 (76%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YT+EFQKRGLPHAHILL++ ++KL + ID +I AE+PD + P+L+E + N M+HGP
Sbjct: 789 YTVEFQKRGLPHAHILLFMHAKSKLPTADDIDKLISAEIPDKEKEPDLYEVIKNSMIHGP 848
>At3g30420 hypothetical protein
Length = 837
Score = 441 bits (1133), Expect = e-123
Identities = 255/560 (45%), Positives = 339/560 (60%), Gaps = 23/560 (4%)
Query: 968 LVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVEM 1027
L ++ L+ L+EIE +L + KSL D+ MP E + L E N LL E ++ +
Sbjct: 279 LELKAKELEKYTLIEIETLLRQHEKSLSDYPEMPQPEKSILEEVNNSLLRQEFQINIDKE 338
Query: 1028 SNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQ 1087
H FSKLN+ Q IY +++ ++ G FF+YG GGTGKTFL+KT+ LRS +
Sbjct: 339 RETHANLFSKLNEQQRIIYDDVLKSVTNKEGKLFFLYGDGGTGKTFLYKTIISALRSNGK 398
Query: 1088 IILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDEA 1147
++ VASS IA+LLLPGGRTAHS F IP+ ++ED C I GS A +L LIIWDEA
Sbjct: 399 NVMPVASSAIAALLLPGGRTAHSWFKIPINVHEDFICDIKIGSMLANVLSKVDLIIWDEA 458
Query: 1148 PMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVMS 1207
PM R+ FEA+DRTLRDI+ + K GGK V+LGGDFRQILPVIP+ R E V +
Sbjct: 459 PMAHRHTFEAVDRTLRDILSVGDEKALTKTLGGKTVLLGGDFRQILPVIPQRTRQETVSA 518
Query: 1208 TINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLG------DYND 1261
IN S LW C L+QNMR+ EE+K FA+W+L +GDG+ D +
Sbjct: 519 AINRSYLWESCHKYLLSQNMRV-------QPEEIK-FAEWILQIGDGEAPRKTHGIDDDQ 570
Query: 1262 GESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPTLDAVDSINQY 1321
E N+ I K+LL+ ++ NP+ + + PD ++ A+L P + VD IN Y
Sbjct: 571 EEDNIIIDKNLLLPETENPLEVLCQSVSPDFTNTFQDLENLKGTAVLTPRNETVDEINDY 630
Query: 1322 ALSLFPGNEKTYLSSDSVWRVDEDVGIEADWL----TTEFLNGLKCSGMPDHKLVLKEGA 1377
LS PG K Y S+DS +D+D + + E+LN L+ G+P H+L LK G
Sbjct: 631 LLSKVPGLAKEYFSADS---IDQDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVGV 687
Query: 1378 PIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSG-THIGKVVYISRMNLMPSDESMP 1436
PIML+RNL+ GLCNGTRL V +LG V+ A +LS T K V I R+ L P D P
Sbjct: 688 PIMLLRNLNQKEGLCNGTRLTVTHLGDKVLKAEILSDTTKKRKKVLIPRIILSPQDSKHP 747
Query: 1437 VKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILI 1496
+RRQFP+ +AMT+NKSQGQ+L++V +YLPKPVFSHGQLYVA+SRV S GL +L
Sbjct: 748 FTLRRRQFPVRMCYAMTVNKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLD 807
Query: 1497 CNEDTSQLDVTKNIVFQEVF 1516
++ VT NIV++EVF
Sbjct: 808 TSKKKEGKYVT-NIVYREVF 826
>At1g54430 hypothetical protein
Length = 1639
Score = 424 bits (1091), Expect = e-118
Identities = 252/558 (45%), Positives = 333/558 (59%), Gaps = 50/558 (8%)
Query: 968 LVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVEM 1027
L ++ L+ L+EIE +L + KSL D+ MP E
Sbjct: 1112 LELKAEELEKYTLIEIETLLRQHEKSLSDYPEMPQPE----------------------- 1148
Query: 1028 SNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQ 1087
+KLN+ Q IY +++ ++ G FF+YG GGTGKTFL+KT+ LRS +
Sbjct: 1149 --------NKLNEQQRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGK 1200
Query: 1088 IILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDEA 1147
++ VASS IA+LLLPGGRTAHS F IP+ ++EDS C I GS A +L LIIWDEA
Sbjct: 1201 NVMPVASSAIAALLLPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVLSKVDLIIWDEA 1260
Query: 1148 PMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVMS 1207
PM R+ FEA+DRTLRDI+ + K FGGK V+LGGDFRQILPVIP+G R E V +
Sbjct: 1261 PMAHRHTFEAVDRTLRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVIPQGTRQETVSA 1320
Query: 1208 TINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLG------DYND 1261
IN S LW C L+QNMR+ EE+K FA+W+L +GDG+ D +
Sbjct: 1321 AINRSYLWESCHKYLLSQNMRV-------QPEEIK-FAEWILQVGDGEAPRKTHGIDDDQ 1372
Query: 1262 GESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPTLDAVDSINQY 1321
E N+ I K+LL+ ++ NP+ + ++PD ++ A+L P + VD IN Y
Sbjct: 1373 EEDNIIIDKNLLLPETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDY 1432
Query: 1322 ALSLFPGNEKTYLSSDSVWRVD--EDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPI 1379
LS PG K Y S+DS+ R + + G E + E+LN L+ G+P H+L LK G PI
Sbjct: 1433 LLSKVPGLAKEYFSADSIDRDEALTEEGFEMSY-PMEYLNSLEFPGLPAHRLCLKVGVPI 1491
Query: 1380 MLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSG-THIGKVVYISRMNLMPSDESMPVK 1438
ML+RNL+ GLCNGTRLIV +LG V+ A +LS T K V I R+ L P D P
Sbjct: 1492 MLLRNLNQKEGLCNGTRLIVTHLGDKVLKAEILSDTTKERKKVLIPRIILSPQDSKHPFT 1551
Query: 1439 FQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICN 1498
+RRQFP+ +AMTINKSQGQ+L++V +YLPKPVFSHGQLYVA+SRV S GL +L +
Sbjct: 1552 LRRRQFPVRMCYAMTINKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVLDTS 1611
Query: 1499 EDTSQLDVTKNIVFQEVF 1516
+ VT NIV++EVF
Sbjct: 1612 KKKEGKYVT-NIVYREVF 1628
Score = 68.9 bits (167), Expect = 2e-11
Identities = 34/60 (56%), Positives = 40/60 (66%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YTIEFQKRGLPHAHILLWL + K +P ID I AE+P + PE V +M+HGP
Sbjct: 700 YTIEFQKRGLPHAHILLWLQGDLKKPTPNDIDKYISAEIPVKDKDPEGHTLVEQHMMHGP 759
>At3g43350 putative protein
Length = 830
Score = 405 bits (1042), Expect = e-113
Identities = 227/527 (43%), Positives = 309/527 (58%), Gaps = 71/527 (13%)
Query: 968 LVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVEM 1027
L + + + CL EI ++L NG SL ++ + NT+ +YG+
Sbjct: 132 LQLSDEERQQYCLQEIARLLTKNGVSLSKWNRCHKFQMNTMVDYGDF------------- 178
Query: 1028 SNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQ 1087
KIY EI+ + + GG FFVYG+GGTGKTFLWK L+ +RS+
Sbjct: 179 -------------RAKKIYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAVRSKGD 225
Query: 1088 IILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDEA 1147
I LNVASSGIA+L L GGRTAHS F IP+ NE S C I +GS +L++ A LIIWDEA
Sbjct: 226 ISLNVASSGIAALRLDGGRTAHSRFDIPINPNESSTCNISRGSDLGELVKEAKLIIWDEA 285
Query: 1148 PMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVMS 1207
PM+S++ FE+LDRTL+DI ++N DKP GGKV+V GGDFRQ+LPVI R EIV +
Sbjct: 286 PMMSKHCFESLDRTLKDI----VNNPGDKPLGGKVIVFGGDFRQVLPVINGAGREEIVFA 341
Query: 1208 TINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNVQ 1267
+NSS +W KVL LT+NMRL + + +++ F+KW+LD+GDGK+ NDG + +
Sbjct: 342 ALNSSYIWEHSKVLELTKNMRLLADISEHEKRDIEDFSKWILDVGDGKISQPNDGIALID 401
Query: 1268 IPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVK-----YYSDKAILAPTLDAVDSINQYA 1322
IP++ LI ++P+ I+ +Y + K Y +AIL PT + V+SIN++
Sbjct: 402 IPEEFLINGDNDPVESIIEAVYGNTFMEEKDPKKTDYPQYQGRAILCPTNEDVNSINEHM 461
Query: 1323 LSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLM 1382
+ + G E+ YLSSDS+ D A +L +FLN ++ G+P+H L LK G P+ML+
Sbjct: 462 MRMLDGEERIYLSSDSIDPADISSANNAAYL-ADFLNNVRVYGLPNHCLRLKVGCPVMLL 520
Query: 1383 RNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRR 1442
RN+D + GLCNGTRL V + +I
Sbjct: 521 RNMDPNKGLCNGTRLQVTQMTDTII----------------------------------- 545
Query: 1443 QFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSR 1489
Q I +FAMTINKSQGQ+L VG+YLP+PVFSHGQLYVAISRV S+
Sbjct: 546 QARFITAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 592
Score = 142 bits (359), Expect = 1e-33
Identities = 69/118 (58%), Positives = 87/118 (73%)
Query: 1376 GAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESM 1435
G P+ML+RN+D + GLCNGTRL V + VI A ++G +GK+V I RM + P D +
Sbjct: 711 GCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPLDTRL 770
Query: 1436 PVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLK 1493
P K +R+QF L +FAMTINKSQGQ+L VG+YLP+PVFSHGQLYVAISRV S+ G K
Sbjct: 771 PFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 828
Score = 141 bits (355), Expect = 3e-33
Identities = 68/114 (59%), Positives = 86/114 (74%)
Query: 1376 GAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESM 1435
G P+ML+RN+D + GLCNGTRL V + VI A ++G +GK+V I RM + PSD +
Sbjct: 595 GCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTRL 654
Query: 1436 PVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSR 1489
P K +R+QF L +FAMTINKSQGQ+L VG+YLP+PVFSHGQLYVAISRV S+
Sbjct: 655 PFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSK 708
>At3g31440 hypothetical protein
Length = 536
Score = 390 bits (1001), Expect = e-108
Identities = 220/455 (48%), Positives = 293/455 (64%), Gaps = 32/455 (7%)
Query: 1068 GTGKTFLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIP 1127
GT KT +WKTL +R + QI+LN+ASSGIASLLL GGRTAHS F I L +E S C I
Sbjct: 110 GTWKTIIWKTLFAAIRRRDQIVLNMASSGIASLLLEGGRTAHSRFGIRLNPDEFSVCKIK 169
Query: 1128 QGSPKAKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGG 1187
S A L++ ASL+I D+APM+SR+ FEALD++ DI++ N+ +K FGGKVVV G
Sbjct: 170 PKSDLANLVKEASLVICDKAPMMSRFCFEALDKSFSDIIK----NTYNKVFGGKVVVFSG 225
Query: 1188 DFRQILPVIPKGRRAEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTC-DGDNEEVKHFAK 1246
DFRQ+LPVI RAEIVMS++N+S LW CKVL LT+NMRL + + + +E+ F+
Sbjct: 226 DFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANNLSETEAKEIHEFSD 285
Query: 1247 WVLDMGDGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMY--PDIVQNVGCVKYYSD 1304
W+L +GDG++ + ND + + IPKDLLI + PI I + +Y P I+ + K++
Sbjct: 286 WLLAVGDGRINEPNDDVAIIDIPKDLLITNADKPIEWITNEIYGDPKILHEITDPKFFQG 345
Query: 1305 KAILAPTLDAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCS 1364
+AILAP + V++IN+Y L E+ YLS+DS+ D D + +T +FLN +K
Sbjct: 346 RAILAPKNEDVNTINEYLLEQLHAEERIYLSADSIDPTDSD-SLNNPVITPDFLNSIKLP 404
Query: 1365 GMPDHKLVLKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYIS 1424
G GLCNG RL + L ++ A V++G IG +V I
Sbjct: 405 G------------------------GLCNGARLQITQLFTEIVEAKVITGDRIGHIVLIP 440
Query: 1425 RMNLMPSDESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAIS 1484
+NL P+D +P K +RRQFPL +FAMTINKSQGQSL VG+YLPKPVFSHGQLYVA+S
Sbjct: 441 TVNLTPTDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEHVGLYLPKPVFSHGQLYVALS 500
Query: 1485 RVKSRAGLKILICNEDTSQLDVTKNIVFQEVFQKI 1519
RV S+ GLKILI +++ T NIVF+EVFQ I
Sbjct: 501 RVTSKKGLKILILDKNGKLQKQTTNIVFKEVFQNI 535
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 385 bits (990), Expect = e-107
Identities = 214/466 (45%), Positives = 281/466 (59%), Gaps = 30/466 (6%)
Query: 1013 NVLLLNELNFDVVEMSNLHDECFSKLNDGQLKIYQ-EIITAINANNGGFFFVYGYGGTGK 1071
N L+++E N++ + HD L K+Y EI+ + + GG FF+Y +GGTGK
Sbjct: 794 NPLIIDERNYNRESLKKKHDNWLKTLTPEHKKVYHDEIMDDVLNDKGGVFFLYAFGGTGK 853
Query: 1072 TFLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSP 1131
TFLWK L+ +R + LNVASS IASLLL GGRTAHS F IPL +E S C + +GS
Sbjct: 854 TFLWKVLSAAIRCKGDTCLNVASSSIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSD 913
Query: 1132 KAKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQ 1191
A+L+ A LIIWDE D PFG KV++ GGDFRQ
Sbjct: 914 LAELVTAAKLIIWDE----------------------------DMPFGRKVILFGGDFRQ 945
Query: 1192 ILPVIPKGRRAEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDM 1251
IL VIP R IV S++NSS LW+ CKVL LT+NMRL + E++ F KW+L +
Sbjct: 946 ILHVIPAAGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQDIDINEAREIEDFLKWILTV 1005
Query: 1252 GDGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPT 1311
G+GKL + +DG +++QIP D+LI + NPI I+ +Y K++ KAIL PT
Sbjct: 1006 GEGKLNEPSDGVTHIQIPDDILIPEGDNPIESIIKAVYGTTFAQKRDPKFFQHKAILCPT 1065
Query: 1312 LDAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKL 1371
D V+SIN + LS G E+ Y SS+S+ D + T +FLN +K SG+ +H L
Sbjct: 1066 NDDVNSINDHMLSKLTGEERIYRSSNSIDPSDTRADKNPIY-TPDFLNKIKISGLANHLL 1124
Query: 1372 VLKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPS 1431
LK G P+ML+RN GL NGTRL + LG ++ +L+GT +GK+V I RM+L PS
Sbjct: 1125 RLKVGCPVMLLRNFYPHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKLVIIPRMSLTPS 1184
Query: 1432 DESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHG 1477
D +P K +RR FPL +FAMTINKSQGQSL VG+YLPK VFSHG
Sbjct: 1185 DRRLPFKMKRRHFPLSVAFAMTINKSQGQSLGNVGMYLPKAVFSHG 1230
Score = 77.8 bits (190), Expect = 4e-14
Identities = 31/60 (51%), Positives = 44/60 (72%)
Query: 515 YTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
YT+EFQKRGLPHAHI++W+ K + +D +I AE+PD ++PEL++ VS M+HGP
Sbjct: 406 YTVEFQKRGLPHAHIIVWMDPRYKFHTADHVDKIIFAEIPDKEKHPELYQAVSECMIHGP 465
>At5g37110 putative helicase
Length = 1307
Score = 342 bits (878), Expect = 7e-94
Identities = 213/530 (40%), Positives = 290/530 (54%), Gaps = 86/530 (16%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVE 1026
DL++ + K L EI+ IL NG SL + MP V + + NVL+L+E +D
Sbjct: 847 DLILSDEEKKLYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDT-NVLILDEKGYDRDN 905
Query: 1027 MSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQK 1086
++ H + L Q IY +II A+N N G FVYG+GGT
Sbjct: 906 LTEKHAKWIKMLTPEQKSIYDDIIGAVNENVGVVVFVYGFGGT----------------- 948
Query: 1087 QIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDE 1146
GGRTAHS F IPL NE + C + GS +A L++ ASLIIWDE
Sbjct: 949 ----------------EGGRTAHSRFGIPLNPNEFTTCNMKVGSDRANLVKEASLIIWDE 992
Query: 1147 APMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVM 1206
APM+SRY FE+LDR+L DI N +KPFGGKVVV GG
Sbjct: 993 APMMSRYCFESLDRSLSDICG----NGDNKPFGGKVVVFGG------------------- 1029
Query: 1207 STINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNV 1266
L+ S + +++K F++W+L +GDG++ + NDGE+ +
Sbjct: 1030 ----------------------LSVS----EAKDIKEFSEWILAVGDGRIVEPNDGEALI 1063
Query: 1267 QIPKDLLIYQSSNPIADIVHLMYPDI--VQNVGCVKYYSDKAILAPTLDAVDSINQYALS 1324
IP + LI ++ +PI I +Y DI + ++ +KAIL PT + V+ IN+ L
Sbjct: 1064 VIPSEFLITKAKDPIEAICTEIYGDITKIHEQNDPIFFQEKAILCPTNEDVNQINETMLD 1123
Query: 1325 LFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMRN 1384
G E T+LSSDS+ D G LT +FLN +K S +P+HKL LK G P+ML+RN
Sbjct: 1124 NLQGEEFTFLSSDSLDPADIG-GKNNPALTPDFLNSVKVSRLPNHKLRLKIGCPVMLLRN 1182
Query: 1385 LDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQF 1444
+D GL NGTRL + +GP ++ A++L+G G +V I R+ L PSD +P + +R Q
Sbjct: 1183 IDPIGGLMNGTRLRITQMGPFILQAMILTGDRAGHLVLIPRLKLAPSDTKLPFRMRRTQL 1242
Query: 1445 PLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKI 1494
PL FAMTINKSQGQSL +VG++L +P FSHGQLYVAISRV S+ LKI
Sbjct: 1243 PLAVCFAMTINKSQGQSLKRVGIFLLRPCFSHGQLYVAISRVTSKTRLKI 1292
Score = 42.4 bits (98), Expect = 0.002
Identities = 20/37 (54%), Positives = 23/37 (62%)
Query: 538 KLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
KL ID VI AE+PD + PELFE + MVHGP
Sbjct: 520 KLTKAEHIDKVISAEIPDKLKDPELFEVIKESMVHGP 556
>At5g34960 putative protein
Length = 1033
Score = 330 bits (847), Expect = 3e-90
Identities = 216/544 (39%), Positives = 289/544 (52%), Gaps = 108/544 (19%)
Query: 981 MEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVEMSNL---HDECFSK 1037
+ EKI++ NG +L+D P + + + N L++ EL ++ SNL H+E
Sbjct: 592 VSFEKIMLSNGATLEDIDEFPKPTRDGI-DNSNRLIVEELRYN--RESNLKEKHEEWKQM 648
Query: 1038 LNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQIILNVASSGI 1097
L Q +Y EI A+ N QI+LNVASSGI
Sbjct: 649 LTPEQRGVYNEITEAVFNN----------------------------LDQIVLNVASSGI 680
Query: 1098 ASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDEAPMVSRYAFEA 1157
ASLLL GGRTAHS F I L +E S C I S A L++ ASL+IWDEAPM+SR
Sbjct: 681 ASLLLEGGRTAHSRFGISLNPDEFSVCKIKPKSDLANLVKEASLVIWDEAPMMSR----- 735
Query: 1158 LDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVMSTINSSRLWRF 1217
Q+L VI RAEIVMS++N+S LW
Sbjct: 736 ---------------------------------QVLLVINGAGRAEIVMSSLNASYLWDH 762
Query: 1218 CKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDGESNVQIPKDLLIYQS 1277
CKVL K+ + NDG + + IP+DLLI +
Sbjct: 763 CKVL---------------------------------KINEPNDGVAIIDIPEDLLITNT 789
Query: 1278 SNPIADIVHLMY--PDIVQNVGCVKYYSDKAILAPTLDAVDSINQYALSLFPGNEKTYLS 1335
PI I + +Y P I+ + K++ +AILAPT + V++IN+Y L E+ YLS
Sbjct: 790 DKPIESITNEIYGDPKILHEITDPKFFQGRAILAPTNEDVNTINEYLLEQLHAEERIYLS 849
Query: 1336 SDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMRNLDISTGLCNGT 1395
+DS+ D + + +T +FLN +K +G+P+H L LK AP+ML+RNLD GLCNGT
Sbjct: 850 ADSIDPTDSN-SLNNPVITPDFLNSIKLAGLPNHSLRLKVSAPVMLLRNLDPKGGLCNGT 908
Query: 1396 RLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQFPLIASFAMTIN 1455
RL + L ++ A V++G IG +V I +NL P+D +P K +RRQFPL +FAMTIN
Sbjct: 909 RLQITQLCTQIVEAKVITGDIIGHIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTIN 968
Query: 1456 KSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTSQLDVTKNIVFQEV 1515
SQGQSL VG+YLPK VFSHGQLYVA+SRV S+ GLK LI ++D T N+VF+EV
Sbjct: 969 TSQGQSLEHVGLYLPKAVFSHGQLYVALSRVTSKKGLKFLILDKDGKLQKQTTNVVFKEV 1028
Query: 1516 FQKI 1519
FQ I
Sbjct: 1029 FQNI 1032
>At1g64410 unknown protein
Length = 753
Score = 285 bits (730), Expect = 1e-76
Identities = 146/296 (49%), Positives = 199/296 (66%), Gaps = 5/296 (1%)
Query: 967 DLVIEEHRLKDLCLMEIEKILMVNGKSLKDFSGMPHVEANTLTEYGNVLLLNELNFDVVE 1026
DL + + + CL EI ++L NG SL + MP + ++ E N +L+E +D
Sbjct: 452 DLQLSDEERQQYCLQEIARLLTKNGVSLSKWKQMPQI-SDEHVEKCNHFILDERKYDRAY 510
Query: 1027 MSNLHDECFSKLNDGQLKIYQEIITAINANNGGFFFVYGYGGTGKTFLWKTLTYKLRSQK 1086
++ H+E + + Q KIY EI+ + + GG FFVYG+GGTGKTFLWK L+ +RS+
Sbjct: 511 LTEKHEEWLTMVTLEQKKIYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKG 570
Query: 1087 QIILNVASSGIASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKAKLLQLASLIIWDE 1146
I LNVASSGIA+LLL GGRTAHS F IP+ NE S C I +G +L++ A+LIIWDE
Sbjct: 571 DISLNVASSGIAALLLDGGRTAHSRFGIPINPNESSTCNISRGLDFGELVKEANLIIWDE 630
Query: 1147 APMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRRAEIVM 1206
A M+S++ FE+LDRTLRDIM +N DKPFGGKV+V GGDFRQ+L VI R EIV
Sbjct: 631 AHMMSKHCFESLDRTLRDIM----NNPGDKPFGGKVIVFGGDFRQVLSVINGAGREEIVF 686
Query: 1207 STINSSRLWRFCKVLNLTQNMRLTTSTCDGDNEEVKHFAKWVLDMGDGKLGDYNDG 1262
+ +NSS +W CKVL LT+NMRL + + + ++++F+KW+LD+GDGK+ NDG
Sbjct: 687 AALNSSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDG 742
Score = 74.7 bits (182), Expect = 4e-13
Identities = 35/62 (56%), Positives = 44/62 (70%)
Query: 513 GTYTIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVH 572
G YTIEFQKRGLPHAHILL++ +KL + D VI AE+PD + PEL+ V + M+H
Sbjct: 178 GMYTIEFQKRGLPHAHILLFMHPTSKLSTAEDTDKVITAEIPDKKKKPELYAVVKDCMIH 237
Query: 573 GP 574
GP
Sbjct: 238 GP 239
>At2g07620 putative helicase
Length = 1241
Score = 264 bits (674), Expect = 3e-70
Identities = 147/302 (48%), Positives = 194/302 (63%), Gaps = 12/302 (3%)
Query: 944 FMFLSIIF-IY*TC------LISKAFKLSLDLVIEEHRLKDLCLMEIEKILMVNGKSLKD 996
F FL IIF IY C L+ ++ + LV+ + + L+EIE +L+ NG +L+D
Sbjct: 747 FFFLGIIFRIYFLCCFWINVLLDQSMCGKMLLVLSDAEKINYALLEIEDMLLCNGSTLED 806
Query: 997 FSGMPHVEANTLTEYGNVLLLNELNFDVVEMSNLHDECFSKLNDGQLKIYQEIITAINAN 1056
F MP T++ N + E N++V ++ HD+ F+K+ Q +IY EI+ A+ N
Sbjct: 807 FKHMPK-PTKEGTDHSNRFITEEKNYNVEKLKEDHDDWFNKMTSEQKEIYDEIMKAVLEN 865
Query: 1057 NGGFFFVYGYGGTGKTFLWKTLTYKLRSQKQIILNVASSGIASLLLPGGRTAHSLFSIPL 1116
+GG FFVYG+GGTGKTF+WKTL+ +R + I +NVASSGIA LLL GGRTAHS F IP+
Sbjct: 866 SGGIFFVYGFGGTGKTFMWKTLSAAVRMKGLISVNVASSGIAFLLLQGGRTAHSRFGIPI 925
Query: 1117 CLNEDSCCGIPQGSPKAKLLQLASLIIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDK 1176
++ + C I S A +L+ ASLIIWDEAPM+SRY FE+LDR+L D+ I N K
Sbjct: 926 NPDDFTTCHIVPNSDLANMLKEASLIIWDEAPMMSRYCFESLDRSLNDV----IGNVDGK 981
Query: 1177 PFGGKVVVLGGDFRQILPVIPKGRRAEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTCDG 1236
PFGGKVVV GGDFRQ+L VI RAEIV++ +NSS LW C VL LT+NM L C
Sbjct: 982 PFGGKVVVFGGDFRQVLHVIHGAGRAEIVLAALNSSYLWEHCNVLTLTKNMSLILFCCKN 1041
Query: 1237 DN 1238
N
Sbjct: 1042 SN 1043
Score = 181 bits (460), Expect = 2e-45
Identities = 90/188 (47%), Positives = 128/188 (67%), Gaps = 1/188 (0%)
Query: 1332 TYLSSDSVWRVDEDVGIEADWLTTEFLNGLKCSGMPDHKLVLKEGAPIMLMRNLDISTGL 1391
TYLS+DS+ D + T FLN +K SG+ +H L LK G P+ML++N+D GL
Sbjct: 1053 TYLSADSIDPQDP-ASLNNPVFTPYFLNSIKLSGLSNHNLTLKIGTPVMLLKNIDPKGGL 1111
Query: 1392 CNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMPSDESMPVKFQRRQFPLIASFA 1451
CNGTRL V +G +++ A V++G + V I + + PSD +P + +RRQFP+ +FA
Sbjct: 1112 CNGTRLQVTQMGNHILEARVITGDRVRDKVIIIKAQISPSDTKLPFRMRRRQFPIAVAFA 1171
Query: 1452 MTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRAGLKILICNEDTSQLDVTKNIV 1511
M I KSQGQSL +V +YLP+PVFSHGQLYVA+SRV S+ GLK+LI +++ + T N+V
Sbjct: 1172 MRIKKSQGQSLKEVEIYLPRPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQTMNVV 1231
Query: 1512 FQEVFQKI 1519
F+E+FQ +
Sbjct: 1232 FKEIFQNL 1239
Score = 66.6 bits (161), Expect = 1e-10
Identities = 29/59 (49%), Positives = 43/59 (72%)
Query: 516 TIEFQKRGLPHAHILLWLSMENKLRSPAMIDSVICAELPDPTQYPELFECVSNYMVHGP 574
T+ FQKRGLPHAHILL++ K + ID+++ AE+PD + P+L+E V++ M+HGP
Sbjct: 406 TVMFQKRGLPHAHILLFMDKSCKFPTSDDIDNILSAEIPDKAKDPKLYEVVNDCMIHGP 464
>At4g04300 hypothetical protein
Length = 286
Score = 223 bits (567), Expect = 9e-58
Identities = 131/271 (48%), Positives = 169/271 (62%), Gaps = 21/271 (7%)
Query: 1038 LNDGQLKIYQEIITAINANNGG-FFFVYGYGGTGKTFLWKTLTYKLRSQKQIILNVASSG 1096
L Q IY +I A+ + GG FFFVYG GG GKTF+WKTL RS+ Q LN+ASSG
Sbjct: 2 LTPEQRGIYDQITNAVFNDMGGVFFFVYGSGGIGKTFIWKTLAAVGRSKGQTCLNIASSG 61
Query: 1097 IASLLLPGGRTAHSLFSIPLCLNEDSCCGIPQGSPKA---------------KLLQLASL 1141
IASLLL GGR AH FSIPL +E S C I S A L++ ASL
Sbjct: 62 IASLLLEGGRIAHYRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDKLVDLIKKASL 121
Query: 1142 IIWDEAPMVSRYAFEALDRTLRDIMRFKIHNSSDKPFGGKVVVLGGDFRQILPVIPKGRR 1201
IIWD+APM S++ FEALD++ DI I +K F GKV+V GGDFRQ+LPVI R
Sbjct: 122 IIWDKAPMKSKFCFEALDKSFSDI----IKRVDNKVFCGKVMVFGGDFRQVLPVINGAGR 177
Query: 1202 AEIVMSTINSSRLWRFCKVLNLTQNMRLTTSTCDGDN-EEVKHFAKWVLDMGDGKLGDYN 1260
AE VMS++N+ +W CKVL LT+NMRL + D +E++ F W+L +GDG++ + N
Sbjct: 178 AETVMSSLNAVYIWDHCKVLKLTKNMRLLNNDLSVDEAKEIQEFFDWLLVVGDGRVNEPN 237
Query: 1261 DGESNVQIPKDLLIYQSSNPIADIVHLMYPD 1291
DGE+ + IP++LLI ++ PI I +Y D
Sbjct: 238 DGEALIDIPEELLIQEADIPIEAISREIYGD 268
>At3g51700 unknown protein
Length = 344
Score = 198 bits (504), Expect = 2e-50
Identities = 112/274 (40%), Positives = 166/274 (59%), Gaps = 2/274 (0%)
Query: 1244 FAKWVLDMGDGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYS 1303
F KW+ ++G + NDGE+ + I +DLLI + +PI IV +Y + +Y
Sbjct: 60 FTKWITNIGGENINKPNDGETKIDIHEDLLITECKDPIKTIVDEVYGESFTESYNPDFYQ 119
Query: 1304 DKAILAPTLDAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLTTEFLNGLKC 1363
++AIL T D D IN Y LS G E +D+++ + + EFLN +K
Sbjct: 120 ERAILCHTNDVADEINDYMLSQLQGEETKCYGADTIYPTHASPNDKMLY-PLEFLNSIKI 178
Query: 1364 SGMPDHKLVLKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYI 1423
G PD KL LK GAP+ML+R+L L GTRL + + V+ A++++G + G+ V I
Sbjct: 179 PGFPDFKLRLKVGAPVMLLRDLAPYGWLRKGTRLQITRVETFVLEAMIITGNNHGEKVLI 238
Query: 1424 SRMNLMPSDESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAI 1483
R+ + P+K +RRQFP+ +FAMTI++SQ Q+LS+VG+YLP+ + HGQ YVAI
Sbjct: 239 PRIPSDLREAKFPIKMRRRQFPVKLAFAMTIDESQRQTLSKVGIYLPRQLLFHGQRYVAI 298
Query: 1484 SRVKSRAGLKILICNED-TSQLDVTKNIVFQEVF 1516
S+VKSRAGLK+LI ++D + TKN+VF+E+F
Sbjct: 299 SKVKSRAGLKVLITDKDGKPDQEETKNVVFKELF 332
>At5g28780 putative protein
Length = 337
Score = 186 bits (472), Expect = 9e-47
Identities = 111/269 (41%), Positives = 156/269 (57%), Gaps = 5/269 (1%)
Query: 1252 GDGKLGDYNDGESNVQIPKDLLIYQSSNPIADIVHLMYPDIVQNVGCVKYYSDKAILAPT 1311
G+ + +DGE V++ + + N + + Y + +Y +++ IL P
Sbjct: 66 GNAPTCETDDGEV-VEMDTSFFLKHNGNRLQQVTKGAYVQFSVSQPNFQYLTERGILTPH 124
Query: 1312 LDAVDSINQYALSLFPGNEKTYLSSDSVWRVDEDVGIEADWLT-TEFLNGLKCSGMPDHK 1370
+ VD IN Y LS G+ K YLSS S+ + D +G + + L ++LN L+ +P HK
Sbjct: 125 NEYVDEINAYMLSQVGGDSKEYLSSYSIGKADT-IGADYEALYHVKYLNSLEFPSLPKHK 183
Query: 1371 LVLKEGAPIMLMRNLDISTGLCNGTRLIVDYLGPNVIGAIVLSGTHIGKVVYISRMNLMP 1430
+ LK+G PIM MRN + GLCNGTRLIV LG VI A +++GTH GK+V I R L P
Sbjct: 184 ISLKKGVPIMQMRNFNQKEGLCNGTRLIVTNLGEQVIEAQIVTGTHAGKMVSIPRFILSP 243
Query: 1431 SDESMPVKFQRRQFPLIASFAMTINKSQGQSLSQVGVYLPKPVFSHGQLYVAISRVKSRA 1490
P +R+QFP+ +AMTI K+QGQSL +YLP PVFSH QLYVA+SRV S
Sbjct: 244 PQSEHPFTLRRQQFPMRVCYAMTIIKNQGQSLKSDVLYLPNPVFSHVQLYVALSRVTSPI 303
Query: 1491 GLKILICNEDTSQLDVTKNIVFQEVFQKI 1519
GL IL + D + D KNIV++E + +
Sbjct: 304 GLTIL--HGDDQKNDEVKNIVYKEFYNDL 330
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.359 0.162 0.586
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,826,593
Number of Sequences: 26719
Number of extensions: 1121800
Number of successful extensions: 5532
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 5339
Number of HSP's gapped (non-prelim): 103
length of query: 1520
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1408
effective length of database: 8,326,068
effective search space: 11723103744
effective search space used: 11723103744
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0233.16


