

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0231b.3
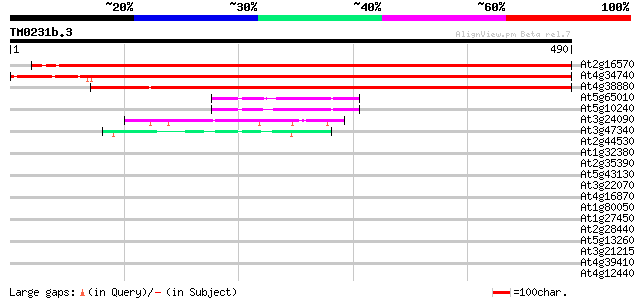
(490 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g16570 amidophosphoribosyltransferase 744 0.0
At4g34740 amidophosphoribosyltransferase 2 precursor 741 0.0
At4g38880 amidophosphoribosyltransferase - like protein 639 0.0
At5g65010 asparagine synthetase (gb|AAC72837.1) 59 6e-09
At5g10240 asparagine synthetase ASN3 56 4e-08
At3g24090 glutamine:fructose-6-phosphate amidotransferase, putative 50 3e-06
At3g47340 glutamine-dependent asparagine synthetase 45 1e-04
At2g44530 phosphoribosyl pyrophosphate synthetase like protein 41 0.002
At1g32380 ribose-phosphate pyrophosphokinase 2 39 0.005
At2g35390 phosphoribosyl pyrophosphate synthetase 38 0.014
At5g43130 unkown protein 34 0.21
At3g22070 unknown protein 34 0.21
At4g16870 retrotransposon like protein 33 0.27
At1g80050 adenine phosphoribosyltransferase 33 0.36
At1g27450 adenine phosphoribosyltransferase 1, APRT 33 0.47
At2g28440 En/Spm-like transposon protein 32 0.80
At5g13260 unknown protein 32 1.0
At3g21215 unknown protein 32 1.0
At4g39410 putative WRKY DNA-binding protein 31 1.4
At4g12440 putative adenine phosphoribosyltransferase 31 1.4
>At2g16570 amidophosphoribosyltransferase
Length = 566
Score = 744 bits (1921), Expect = 0.0
Identities = 378/472 (80%), Positives = 415/472 (87%), Gaps = 5/472 (1%)
Query: 20 KPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNH-NTTFPEPLCNDDEDQKP 78
KPF P L +P S P P +L H + SP + N+ P ND++D+KP
Sbjct: 32 KPFLKPPHL---SLLPSPLSSP-PPSLIHGVSSYFSSPSPSEDNSHTPFDYHNDEDDEKP 87
Query: 79 REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFTE 138
REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTV K+ Q++TGVGLV++VF E
Sbjct: 88 REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVSPEKVLQTITGVGLVSEVFNE 147
Query: 139 AKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEE 198
+KLDQLPG AIGHVRYSTAG SMLKNVQPFVAGYRFGS+GVAHNGNLVNY++LR LEE
Sbjct: 148 SKLDQLPGEFAIGHVRYSTAGASMLKNVQPFVAGYRFGSIGVAHNGNLVNYKTLRAMLEE 207
Query: 199 NGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGF 258
NGSIFNTSSDTEVVLHLIA SK RPF +RI+DACEKL+GAYS VFVTEDKLVAVRDP+GF
Sbjct: 208 NGSIFNTSSDTEVVLHLIAISKARPFFMRIIDACEKLQGAYSMVFVTEDKLVAVRDPYGF 267
Query: 259 RPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQC 318
RPLVMG+RSNGAVVFASETCALDLIEA YEREVYPGEVLVVDK+G++S CL+ EPKQC
Sbjct: 268 RPLVMGRRSNGAVVFASETCALDLIEATYEREVYPGEVLVVDKDGVKSQCLMPKFEPKQC 327
Query: 319 IFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAKAG 378
IFEHIYFSLP+S+VFG+SVYESR +FGEILATESPVECDVVIAVPDSGVVAALGYAAK+G
Sbjct: 328 IFEHIYFSLPNSIVFGRSVYESRHVFGEILATESPVECDVVIAVPDSGVVAALGYAAKSG 387
Query: 379 VPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSK 438
VPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKLSPV+ VLEGKRVVVVDDSIVRGTTSSK
Sbjct: 388 VPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVRGVLEGKRVVVVDDSIVRGTTSSK 447
Query: 439 IVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
IVRL++EAGAKEVHMRIA PPI+ SCYYGVDTPSSEELISNR+SV+EI EFI
Sbjct: 448 IVRLLREAGAKEVHMRIASPPIVASCYYGVDTPSSEELISNRLSVEEINEFI 499
>At4g34740 amidophosphoribosyltransferase 2 precursor
Length = 561
Score = 741 bits (1913), Expect = 0.0
Identities = 390/500 (78%), Positives = 430/500 (86%), Gaps = 15/500 (3%)
Query: 1 MAASNFSTLSSSSSSFSK-SKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCK 59
MAA+ S++SSS S +K +K N ++ L NP F NP++ S + P S
Sbjct: 1 MAAT--SSISSSLSLNAKPNKLSNNNNNNKPHRFLRNP--FLNPSSSSFSPLPASISSSS 56
Query: 60 NHNTTFP----EPLC-----NDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQE 110
+ + +FP PL NDD D+KPREECGVVGIYGD EASRLCYLALHALQHRGQE
Sbjct: 57 S-SPSFPLRVSNPLTLLAADNDDYDEKPREECGVVGIYGDSEASRLCYLALHALQHRGQE 115
Query: 111 GAGIVTVHNNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFV 170
GAGIVTV +K+ Q++TGVGLV++VF+E+KLDQLPG +AIGHVRYSTAG SMLKNVQPFV
Sbjct: 116 GAGIVTVSKDKVLQTITGVGLVSEVFSESKLDQLPGDIAIGHVRYSTAGSSMLKNVQPFV 175
Query: 171 AGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVD 230
AGYRFGSVGVAHNGNLVNY LR LEENGSIFNTSSDTEVVLHLIA SK RPF +RIVD
Sbjct: 176 AGYRFGSVGVAHNGNLVNYTKLRADLEENGSIFNTSSDTEVVLHLIAISKARPFFMRIVD 235
Query: 231 ACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYERE 290
ACEKL+GAYS VFVTEDKLVAVRDP GFRPLVMG+RSNGAVVFASETCALDLIEA YERE
Sbjct: 236 ACEKLQGAYSMVFVTEDKLVAVRDPHGFRPLVMGRRSNGAVVFASETCALDLIEATYERE 295
Query: 291 VYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILAT 350
VYPGEVLVVDK+G++ CL+ HPEPKQCIFEHIYFSLP+S+VFG+SVYESR +FGEILAT
Sbjct: 296 VYPGEVLVVDKDGVKCQCLMPHPEPKQCIFEHIYFSLPNSIVFGRSVYESRHVFGEILAT 355
Query: 351 ESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLK 410
ESPV+CDVVIAVPDSGVVAALGYAAKAGV FQQGLIRSHYVGRTFIEPSQKIRD GVKLK
Sbjct: 356 ESPVDCDVVIAVPDSGVVAALGYAAKAGVAFQQGLIRSHYVGRTFIEPSQKIRDFGVKLK 415
Query: 411 LSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDT 470
LSPV+ VLEGKRVVVVDDSIVRGTTSSKIVRL++EAGAKEVHMRIA PPII SCYYGVDT
Sbjct: 416 LSPVRGVLEGKRVVVVDDSIVRGTTSSKIVRLLREAGAKEVHMRIASPPIIASCYYGVDT 475
Query: 471 PSSEELISNRMSVDEIREFI 490
PSS ELISNRMSVDEIR++I
Sbjct: 476 PSSNELISNRMSVDEIRDYI 495
>At4g38880 amidophosphoribosyltransferase - like protein
Length = 532
Score = 639 bits (1649), Expect = 0.0
Identities = 317/421 (75%), Positives = 372/421 (88%), Gaps = 2/421 (0%)
Query: 71 NDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVG 130
N DED K EECGVVGI+GDPEASRL YLALHALQHRGQEGAGIV + N + +S+TGVG
Sbjct: 66 NVDEDDKLHEECGVVGIHGDPEASRLSYLALHALQHRGQEGAGIVAANQNGL-ESITGVG 124
Query: 131 LVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR 190
LV+DVFTE+KL+ LPG +AIGHVRYST+G SMLKNVQPF+A + GS+ VAHNGN VNY+
Sbjct: 125 LVSDVFTESKLNNLPGDIAIGHVRYSTSGASMLKNVQPFIASCKLGSLAVAHNGNFVNYK 184
Query: 191 SLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLV 250
L+ KLEE GSIF TSSDTE+VLHLIA SK + F+LR++DACEKL+GAYS VFV EDKL+
Sbjct: 185 QLKTKLEEMGSIFITSSDTELVLHLIAKSKAKTFLLRVIDACEKLRGAYSMVFVFEDKLI 244
Query: 251 AVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKN-GLQSLCL 309
AVRDPFGFRPLVMG+RSNGAVVFASETCALDLI+A YEREV PGE++VVD+N G S+ +
Sbjct: 245 AVRDPFGFRPLVMGRRSNGAVVFASETCALDLIDATYEREVCPGEIVVVDRNHGDSSMFM 304
Query: 310 VSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVA 369
+SHPE KQC+FEH YFS P+S+VFG+SVYE+RR++GEILAT +PV+CDVVIAVPDSG VA
Sbjct: 305 ISHPEQKQCVFEHGYFSQPNSIVFGRSVYETRRMYGEILATVAPVDCDVVIAVPDSGTVA 364
Query: 370 ALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDS 429
ALGYAAKAGVPFQ GL+RSHY RTFIEP+Q+IRD VK+KLSPV+AVLEGKRVVVVDDS
Sbjct: 365 ALGYAAKAGVPFQIGLLRSHYAKRTFIEPTQEIRDFAVKVKLSPVRAVLEGKRVVVVDDS 424
Query: 430 IVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREF 489
IVRGTTS KIVR++++AGAKEVHMRIA PP+I SCYYGVDTP S+ELIS++MSV+ I++
Sbjct: 425 IVRGTTSLKIVRMLRDAGAKEVHMRIALPPMIASCYYGVDTPRSQELISSKMSVEAIQKH 484
Query: 490 I 490
I
Sbjct: 485 I 485
>At5g65010 asparagine synthetase (gb|AAC72837.1)
Length = 578
Score = 58.9 bits (141), Expect = 6e-09
Identities = 47/131 (35%), Positives = 67/131 (50%), Gaps = 13/131 (9%)
Query: 177 SVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLK 236
+V V NG + N++ LREKL+ + F T SD EV+ HL + FI + L
Sbjct: 69 TVAVTVNGEIYNHKILREKLKSHQ--FRTGSDCEVIAHLYEEHGEE-FI-------DMLD 118
Query: 237 GAYSFVFV-TEDK-LVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPG 294
G ++FV + T DK +A RD G PL +G +G+V FASE AL + E PG
Sbjct: 119 GMFAFVLLDTRDKSFIAARDAIGITPLYIGWGLDGSVWFASEMKALS-DDCEQFMSFPPG 177
Query: 295 EVLVVDKNGLQ 305
+ + GL+
Sbjct: 178 HIYSSKQGGLR 188
>At5g10240 asparagine synthetase ASN3
Length = 578
Score = 56.2 bits (134), Expect = 4e-08
Identities = 43/131 (32%), Positives = 66/131 (49%), Gaps = 13/131 (9%)
Query: 177 SVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLK 236
++ V NG + N+++LRE L+ + F T SD EV+ HL + + + L
Sbjct: 69 TIAVTVNGEIYNHKALRENLKSHQ--FRTGSDCEVIAHLYEEHGE--------EFVDMLD 118
Query: 237 GAYSFVFV-TEDK-LVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPG 294
G ++FV + T DK +A RD G PL +G +G+V FASE AL + E PG
Sbjct: 119 GMFAFVLLDTRDKSFIAARDAIGITPLYIGWGLDGSVWFASEMKALS-DDCEQFMCFPPG 177
Query: 295 EVLVVDKNGLQ 305
+ + GL+
Sbjct: 178 HIYSSKQGGLR 188
>At3g24090 glutamine:fructose-6-phosphate amidotransferase, putative
Length = 691
Score = 50.1 bits (118), Expect = 3e-06
Identities = 53/216 (24%), Positives = 99/216 (45%), Gaps = 28/216 (12%)
Query: 101 LHALQHRGQEGAGIVTVHNNK-----IFQSVTGVGLVADVFT--EAKLDQLPGTLA-IGH 152
L L++RG + AGI +++ +F+ + + + T + LD++ A I H
Sbjct: 27 LRRLEYRGYDSAGIAIDNSSPSSSPLVFRQAGNIESLVNSITNTDLNLDEVFYFHAGIAH 86
Query: 153 VRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVV 212
R++T G+ +N P +G + V HNG + NY L+E L +G F + +DTEV+
Sbjct: 87 TRWATHGEPAPRNSHPQSSGPGDDFL-VVHNGVITNYEVLKETLVRHGFTFESDTDTEVI 145
Query: 213 LHLI---------ATSKQRPFILRIVDACEKLKGAYSFVFVT---EDKLVAVRDPFGFRP 260
L + F + + L+GAY+ +F + ++L+A + G P
Sbjct: 146 PKLAKFVFDKANEEGGQTVTFCEVVFEVMRHLEGAYALIFKSWHYPNELIACK--LG-SP 202
Query: 261 LVMGKRSNGAVVFASE----TCALDLIEAEYEREVY 292
L++G + + ++ S L+L + E V+
Sbjct: 203 LLLGVKVSNSLSLESRGLLFVSLLELDQGESNSHVF 238
>At3g47340 glutamine-dependent asparagine synthetase
Length = 584
Score = 44.7 bits (104), Expect = 1e-04
Identities = 49/206 (23%), Positives = 83/206 (39%), Gaps = 48/206 (23%)
Query: 82 CGVVGIYG---DPEASRLCYLAL-HALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFT 137
CG++ + G D +A R+ L L L+HRG + +G+ +N + V
Sbjct: 2 CGILAVLGCSDDSQAKRVRVLELSRRLRHRGPDWSGLYQNGDNYLAHQRLAV-------- 53
Query: 138 EAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLE 197
+ ++ Q + + V V NG + N+ LR++L+
Sbjct: 54 ---------------IDPASGDQPLFNEDKTIV---------VTVNGEIYNHEELRKRLK 89
Query: 198 ENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFV--TEDKLVAVRDP 255
+ F T SD EV+ HL VD + L G +SFV + ++ + RD
Sbjct: 90 NHK--FRTGSDCEVIAHLYEEYG--------VDFVDMLDGIFSFVLLDTRDNSFMVARDA 139
Query: 256 FGFRPLVMGKRSNGAVVFASETCALD 281
G L +G +G+V +SE L+
Sbjct: 140 IGVTSLYIGWGLDGSVWISSEMKGLN 165
>At2g44530 phosphoribosyl pyrophosphate synthetase like protein
Length = 394
Score = 40.8 bits (94), Expect = 0.002
Identities = 44/170 (25%), Positives = 69/170 (39%), Gaps = 26/170 (15%)
Query: 324 YFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQ 383
YF +P V+G+ V + LA+++ D+V+ PD G VA AK
Sbjct: 211 YFDIPVDHVYGQPVIL------DYLASKAISSEDLVVVSPDVGGVARARAFAKKLSDAPL 264
Query: 384 GLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLI 443
++ G E I D+ +GK ++VDD I T SK L+
Sbjct: 265 AIVDKRRHGHNVAEVMNLIGDV-------------KGKVAIMVDDMIDTAGTISKGAALL 311
Query: 444 KEAGAKEVHM----RIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREF 489
+ GA+EV+ + PP I G+ E +I+N + + E F
Sbjct: 312 HQEGAREVYACTTHAVFSPPAISRLSSGL---FQEVIITNTIPLSEKNYF 358
>At1g32380 ribose-phosphate pyrophosphokinase 2
Length = 400
Score = 39.3 bits (90), Expect = 0.005
Identities = 64/255 (25%), Positives = 100/255 (39%), Gaps = 60/255 (23%)
Query: 225 ILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIE 284
+L +VDAC + + K+ AV FG+ +++ G A++ A + E
Sbjct: 156 LLIMVDACRR---------ASAKKVTAVIPYFGYARA--DRKTQGRESIAAKLVANLITE 204
Query: 285 AEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLF 344
A +R VL D + QS+ YF +P V+ + V
Sbjct: 205 AGADR------VLACDLHSGQSMG---------------YFDIPVDHVYCQPVIL----- 238
Query: 345 GEILATESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD 404
+ LA++S D+V+ PD G VA AK ++ G E I D
Sbjct: 239 -DYLASKSISSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRHGHNVAEVMNLIGD 297
Query: 405 LGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSC 464
+ +GK V+VDD I T K L+ E GA+EV+ AC C
Sbjct: 298 V-------------KGKVAVMVDDIIDTAGTIVKGAALLHEEGAREVY---AC------C 335
Query: 465 YYGVDTPSSEELISN 479
+ V +P + E +S+
Sbjct: 336 THAVFSPPAIERLSS 350
>At2g35390 phosphoribosyl pyrophosphate synthetase
Length = 403
Score = 37.7 bits (86), Expect = 0.014
Identities = 62/254 (24%), Positives = 98/254 (38%), Gaps = 60/254 (23%)
Query: 225 ILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIE 284
+L +VDAC + + K+ AV FG+ +++ G A++ A + E
Sbjct: 159 LLIMVDACRR---------ASAKKVTAVIPYFGYARA--DRKTQGRESIAAKLVANLITE 207
Query: 285 AEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLF 344
A +R VL D + QS+ YF +P V+ + V
Sbjct: 208 AGADR------VLACDLHSGQSMG---------------YFDIPVDHVYCQPVIL----- 241
Query: 345 GEILATESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD 404
+ LA++S D+V+ PD G VA AK ++ G E I D
Sbjct: 242 -DYLASKSIPSEDLVVVSPDVGGVARARAFAKKLSDAPLAIVDKRRSGHNVAEVMNLIGD 300
Query: 405 LGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSC 464
+ GK ++VDD I T K L+ + GA+EV+ AC C
Sbjct: 301 V-------------RGKVAIMVDDMIDTAGTIVKGAALLHQEGAREVY---AC------C 338
Query: 465 YYGVDTPSSEELIS 478
+ V +P + E +S
Sbjct: 339 THAVFSPPAIERLS 352
>At5g43130 unkown protein
Length = 689
Score = 33.9 bits (76), Expect = 0.21
Identities = 22/85 (25%), Positives = 36/85 (41%)
Query: 10 SSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNHNTTFPEPL 69
SS+S + S P P ++ H P+SFP TT H P P+ + +T P
Sbjct: 176 SSASGTLGSSVPVQGLTKHPQHQMQHPPSSFPMYTTSGSFHSFPGPNTNASGSTLRPHLH 235
Query: 70 CNDDEDQKPREECGVVGIYGDPEAS 94
+ + G G+ G P+++
Sbjct: 236 DSHMRHVAHNQPMGSTGLGGPPQST 260
>At3g22070 unknown protein
Length = 146
Score = 33.9 bits (76), Expect = 0.21
Identities = 29/72 (40%), Positives = 33/72 (45%), Gaps = 8/72 (11%)
Query: 3 ASNFSTLSSSSSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPY--PSPCKN 60
+S+ SSSSSS S + PF TP FP NP PNP S + PP P P
Sbjct: 44 SSSTDPSSSSSSSSSSTSPFI-TP-FPNP----NPNPNPNPPPPSTPNPPPEFSPPPPDL 97
Query: 61 HNTTFPEPLCND 72
TT P P D
Sbjct: 98 DTTTAPPPPSTD 109
>At4g16870 retrotransposon like protein
Length = 1474
Score = 33.5 bits (75), Expect = 0.27
Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 9/75 (12%)
Query: 2 AASNFSTLSSSSS----SFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSP 57
A+ FS L+S +S +F +S TP + L P+ +P T+ H +PP +P
Sbjct: 748 ASFPFSNLTSQNSLPTVTFEQSSSPLVTPILSSSSVL--PSCLSSPCTVLHQQQPPVTTP 805
Query: 58 CKNHN---TTFPEPL 69
H+ TT P PL
Sbjct: 806 NSPHSSQPTTSPAPL 820
>At1g80050 adenine phosphoribosyltransferase
Length = 192
Score = 33.1 bits (74), Expect = 0.36
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query: 414 VKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMR--IACPPIIGSC 464
V+AV +R +++DD + G T S + L++ AGA+ V + P G C
Sbjct: 119 VEAVKSEERALIIDDLVATGGTLSASINLLERAGAEVVECACVVGLPKFKGQC 171
>At1g27450 adenine phosphoribosyltransferase 1, APRT
Length = 243
Score = 32.7 bits (73), Expect = 0.47
Identities = 15/38 (39%), Positives = 23/38 (60%)
Query: 414 VKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEV 451
V AV G+R +++DD I G T + +RL++ G K V
Sbjct: 176 VGAVEPGERAIIIDDLIATGGTLAAAIRLLERVGVKIV 213
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 32.0 bits (71), Expect = 0.80
Identities = 23/74 (31%), Positives = 32/74 (43%), Gaps = 11/74 (14%)
Query: 19 SKPFTNTPCFPGNKTLHN----PTSFPNPTTL--SHAHKPPYPSPCKNHNTTFPEPL--- 69
S P ++P P + N P S P P +L S + PP P P + ++PEP
Sbjct: 109 SSPEADSPLPPSSSPEANSPQSPASSPKPESLADSPSPPPPPPQPESPSSPSYPEPAPVP 168
Query: 70 --CNDDEDQKPREE 81
+DD D P E
Sbjct: 169 APSDDDSDDDPEPE 182
>At5g13260 unknown protein
Length = 537
Score = 31.6 bits (70), Expect = 1.0
Identities = 17/71 (23%), Positives = 31/71 (42%)
Query: 13 SSSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNHNTTFPEPLCND 72
SS+ +K KP ++ P K + + +P A +PP P + TT P P+
Sbjct: 107 SSTIAKPKPVASSAVVPPPKISRSSSPANSPAVSVRASQPPVPPSKLRNQTTNPLPVATP 166
Query: 73 DEDQKPREECG 83
+++ + G
Sbjct: 167 KTEKRVLADIG 177
>At3g21215 unknown protein
Length = 339
Score = 31.6 bits (70), Expect = 1.0
Identities = 16/56 (28%), Positives = 26/56 (45%), Gaps = 2/56 (3%)
Query: 37 PTSFPNPTTLSHAHKPPYPSPCKNHNTTFPEPLCNDDEDQKPREECGVVGIYGDPE 92
P P P + PP+P P +H P P+ D+ ++ P +E + I G P+
Sbjct: 15 PAGAPPPPAAVSSAAPPHPPPIHHHPP--PPPVLVDNHNRPPYDELRTIFIAGLPD 68
>At4g39410 putative WRKY DNA-binding protein
Length = 304
Score = 31.2 bits (69), Expect = 1.4
Identities = 29/80 (36%), Positives = 38/80 (47%), Gaps = 17/80 (21%)
Query: 7 STLSSSSSSFSKSKPFTNTPCFPGNKTL-----HNPTSFPNPTTLSHAHKPPYPSPCKNH 61
STLSSSSSS S S P + F G+ +L +NP+SF +SH P N
Sbjct: 36 STLSSSSSS-SSSSPSSLVSPFLGHNSLNSFLHNNPSSF-----ISH------PQDSINL 83
Query: 62 NTTFPEPLCNDDEDQKPREE 81
T PE L + K R++
Sbjct: 84 MTNLPETLISSLSSSKQRDD 103
>At4g12440 putative adenine phosphoribosyltransferase
Length = 182
Score = 31.2 bits (69), Expect = 1.4
Identities = 16/38 (42%), Positives = 22/38 (57%)
Query: 414 VKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEV 451
V+AV G R +VVDD I G T + L+K GA+ +
Sbjct: 116 VEAVDSGDRALVVDDLIATGGTLCAAMNLLKRVGAEVI 153
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,497,369
Number of Sequences: 26719
Number of extensions: 531571
Number of successful extensions: 1722
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 1669
Number of HSP's gapped (non-prelim): 65
length of query: 490
length of database: 11,318,596
effective HSP length: 103
effective length of query: 387
effective length of database: 8,566,539
effective search space: 3315250593
effective search space used: 3315250593
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0231b.3


