

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.5
(406 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
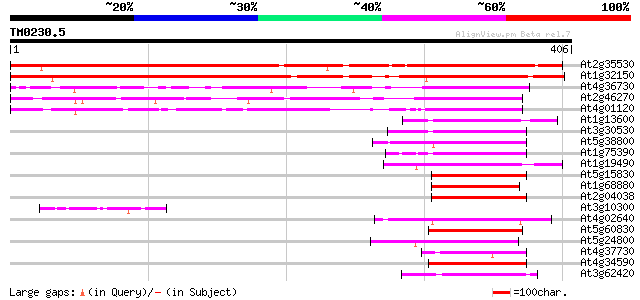
Score E
Sequences producing significant alignments: (bits) Value
At2g35530 bZIP transcription factor AtbZip16 490 e-139
At1g32150 putative G-Box binding protein 459 e-130
At4g36730 G-box binding bZip transcription factor GBF1 / AtbZip41 181 6e-46
At2g46270 G-box binding bZIP transcription factor GBF3 / AtbZip55 165 4e-41
At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54 122 3e-28
At1g13600 bZip transcription factor AtbZip58 64 1e-10
At3g30530 bZip transcription factor AtbZip42 63 3e-10
At5g38800 bZIP transcription factor AtbZip43 62 4e-10
At1g75390 bZip transcription factor AtbZip44 62 4e-10
At1g19490 bZip transcription factor AtbZip62 61 1e-09
At5g15830 bZIP transcription factor AtbZip3 59 4e-09
At1g68880 bZip transcription factor AtbZip8 59 4e-09
At2g04038 bZip transcription factor AtbZip48 59 5e-09
At3g10300 unknown protein 59 6e-09
At4g02640 bZIP protein BZO2H1 58 8e-09
At5g60830 bZip transcription factor AtbZip70 58 1e-08
At5g24800 bZIP protein BZO2H2 57 2e-08
At4g37730 bZip transcription factor AtbZip7 56 4e-08
At4g34590 bZIP transcription factor ATB2/ Atbzip11 55 5e-08
At3g62420 bZip transcription factor AtbZip53 55 9e-08
>At2g35530 bZIP transcription factor AtbZip16
Length = 409
Score = 490 bits (1261), Expect = e-139
Identities = 252/410 (61%), Positives = 302/410 (73%), Gaps = 21/410 (5%)
Query: 1 MGSSEMDKTPKEKESKTPPLT----PQEQPPTTSTGT-VNPPDWSSFQAYSPMPP-HGFL 54
M S+EM+K+ KEKE KTPP + P Q P+++ + PDWS FQAYSPMPP HG++
Sbjct: 1 MASNEMEKSSKEKEPKTPPPSSTAPPSSQEPSSAVSAGMATPDWSGFQAYSPMPPPHGYV 60
Query: 55 ASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
AS+PQ HPYMWGVQH+MPPYGTPPHPYVAMYP GG+YAHPS+PPGSYP+SP+AMPSPNG+
Sbjct: 61 ASSPQPHPYMWGVQHMMPPYGTPPHPYVAMYPPGGMYAHPSMPPGSYPYSPYAMPSPNGM 120
Query: 115 VEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
E SGNT G + D K EVKEKLPIKRS+GSLGSLNMIT KNNE GK G SANGA+SK
Sbjct: 121 TEVSGNTTGGTDGDAKQSEVKEKLPIKRSRGSLGSLNMITGKNNEPGKNSGASANGAYSK 180
Query: 175 SGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQ 230
SGES S+G+SEGSD NSQNDS G + S+NG + + QNG TP Q
Sbjct: 181 SGESASDGSSEGSDGNSQNDS---GSGLDGKDAEAASENGGSANGPQNGSAGTPILPVSQ 237
Query: 231 TMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
T+ ++P++A G VPGP TNLNIGMDYWG PTS+ IP +HG V ST V G +A GSR
Sbjct: 238 TVPIMPMTAAG----VPGPPTNLNIGMDYWGAPTSAGIPGMHGKV-STPVPGVVAP-GSR 291
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
DG SQ WLQD+RELKRQRRKQSNRESARRSRLRKQAECDELAQRAE L EEN +LR+E+
Sbjct: 292 DGGHSQPWLQDDRELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNEENTNLRAEI 351
Query: 351 SRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTE 400
++++S E+L TEN +LK++L P + + + H D Q+G E
Sbjct: 352 NKLKSQCEELTTENTSLKDQLSLFPPLEGI--SMDNDHQEPDTNQTGAAE 399
>At1g32150 putative G-Box binding protein
Length = 389
Score = 459 bits (1182), Expect = e-130
Identities = 244/414 (58%), Positives = 289/414 (68%), Gaps = 38/414 (9%)
Query: 1 MGSSEMDKTPKEKESKT-PPLTPQEQPPTT---------STGTVNPPDWSSFQAYSPMPP 50
MGSSEM+K+ KEKE KT PP T P T S G DWS FQAYSPMPP
Sbjct: 1 MGSSEMEKSGKEKEPKTTPPSTSSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAYSPMPP 60
Query: 51 HGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPS 110
HG++AS+PQ HPYMWGVQH+MPPYGTPPHPYV MYP GG+YAHPS+PPGSYP+SP+AMPS
Sbjct: 61 HGYVASSPQPHPYMWGVQHMMPPYGTPPHPYVTMYPPGGMYAHPSLPPGSYPYSPYAMPS 120
Query: 111 PNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANG 170
PNG+ EASGNT +E DGKP + KEKLPIKRSKGSLGSLNMI KNNE GK G SANG
Sbjct: 121 PNGMAEASGNTGSVIEGDGKPSDGKEKLPIKRSKGSLGSLNMIIGKNNEAGKNSGASANG 180
Query: 171 AHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP-H 229
A SKS ESGS+G+S+GSDANSQNDS + G+ + S++G + H G N P +
Sbjct: 181 ACSKSAESGSDGSSDGSDANSQNDSGSRHNGKDG---ETASESGGSAHGPPRNGSNLPVN 237
Query: 230 QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGS 289
QT++++P+SA G VPGP TNLNIGMDYW HGNV G G
Sbjct: 238 QTVAIMPVSATG----VPGPPTNLNIGMDYWSG---------HGNV------SGAVPGVV 278
Query: 290 RDGVQSQHWLQ--DERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLR 347
DG QSQ WLQ DERE+KRQRRKQSNRESARRSRLRKQAECDELAQRAE L EN+SLR
Sbjct: 279 VDGSQSQPWLQVSDEREIKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNGENSSLR 338
Query: 348 SEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEA 401
+E+++++S YE+LL EN++LK + P L G D++ + + + Q A
Sbjct: 339 AEINKLKSQYEELLAENSSLKNKFSSAP---SLEGGDLDKNEQEPQRSTRQDVA 389
>At4g36730 G-box binding bZip transcription factor GBF1 / AtbZip41
Length = 315
Score = 181 bits (459), Expect = 6e-46
Identities = 143/390 (36%), Positives = 186/390 (47%), Gaps = 110/390 (28%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDW-SSFQAY-----SPMPPHGFL 54
MG+SE DK P + + P + QE PPT PDW +S QAY +P P
Sbjct: 1 MGTSE-DKMPFK--TTKPTSSAQEVPPTPY------PDWQNSMQAYYGGGGTPNPFFPSP 51
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
+P HPYMWG QH +MPPYGTP PY AMYP G +YAHPS+P MP +G
Sbjct: 52 VGSPSPHPYMWGAQHHMMPPYGTPV-PYPAMYPPGAVYAHPSMP----------MPPNSG 100
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHG-KTPGTSANGAH 172
+ P +A GK +SKG+ ++K E G K S N
Sbjct: 101 ---PTNKEPAKDQASGK-----------KSKGN-------SKKKAEGGDKALSGSGNDGA 139
Query: 173 SKSGESGSEGTSEGSD--ANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQ 230
S S ES + G+S+ +D AN Q ++ D SQ+ T
Sbjct: 140 SHSDESVTAGSSDENDENANQQEQGSIRKPSFGQMLADASSQSTT--------------- 184
Query: 231 TMSMVPISAGGAPGAVP----GPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMAT 286
G G+VP P TNLNIGMD W + VP
Sbjct: 185 ---------GEIQGSVPMKPVAPGTNLNIGMDLWSS---------QAGVP---------- 216
Query: 287 GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASL 346
++DERELKRQ+RKQSNRESARRSRLRKQAEC++L QR E+L EN SL
Sbjct: 217 ------------VKDERELKRQKRKQSNRESARRSRLRKQAECEQLQQRVESLSNENQSL 264
Query: 347 RSEVSRIRSDYEQLLTENAALKERLGELPG 376
R E+ R+ S+ ++L +EN ++++ L + G
Sbjct: 265 RDELQRLSSECDKLKSENNSIQDELQRVLG 294
>At2g46270 G-box binding bZIP transcription factor GBF3 / AtbZip55
Length = 382
Score = 165 bits (418), Expect = 4e-41
Identities = 130/386 (33%), Positives = 181/386 (46%), Gaps = 75/386 (19%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS----PMPPH---GF 53
MG+S + P K K PP T PDW++ QAY MPP+
Sbjct: 1 MGNSSEEPKPPTKSDKP------SSPPVDQTNVHVYPDWAAMQAYYGPRVAMPPYYNSAM 54
Query: 54 LASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGG-IYAHPSIPPGSYPFS----PFAM 108
AS PYMW QH+M PYG P Y A+YPHGG +YAHP IP GS P P
Sbjct: 55 AASGHPPPPYMWNPQHMMSPYGAP---YAAVYPHGGGVYAHPGIPMGSLPQGQKDPPLTT 111
Query: 109 PSPNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSA 168
P ++ + G+ + +G ++KE + S G+ G
Sbjct: 112 PGTLLSIDTPTKSTGNTD-NGLMKKLKEFDGLAMSLGN------------------GNPE 152
Query: 169 NGA--HSKSGESG-SEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGL 225
NGA H +S S ++G+++GSD N+ E K + S E P+++G + +
Sbjct: 153 NGADEHKRSRNSSETDGSTDGSDGNTTGADEPKL---KRSREGTPTKDGKQLVQASSFHS 209
Query: 226 NTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMA 285
+P + V + G PG + N N P S A+
Sbjct: 210 VSPSSGDTGVKLIQGSGAILSPGVSANSN--------PFMSQSLAM-------------- 247
Query: 286 TGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENAS 345
V + WLQ+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ EAL EN +
Sbjct: 248 -------VPPETWLQNERELKRERRKQSNRESARRSRLRKQAETEELARKVEALTAENMA 300
Query: 346 LRSEVSRIRSDYEQLLTENAALKERL 371
LRSE++++ ++L NA L ++L
Sbjct: 301 LRSELNQLNEKSDKLRGANATLLDKL 326
>At4g01120 G-box binding bZip transcription factor GBF2 / AtbZip54
Length = 360
Score = 122 bits (307), Expect = 3e-28
Identities = 112/378 (29%), Positives = 170/378 (44%), Gaps = 69/378 (18%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS------PMPPHGFL 54
MGS+E + + P++ S V DW++ QAY P + L
Sbjct: 1 MGSNEEGNPTNNSDKPSQAAAPEQ-----SNVHVYHHDWAAMQAYYGPRVGIPQYYNSNL 55
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
A PYMW +M PYG P P+ P GG+YAHP + GS P P + S +G
Sbjct: 56 APGHAPPPYMWASPSPMMAPYGAPYPPFC---PPGGVYAHPGVQMGSQPQGPVSQ-SASG 111
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
+ TP +++A + +K+ K G I+ NN+ G +S+ H
Sbjct: 112 VT-----TPLTIDAPANSAGNSDHGFMKKLKEFDGLAMSIS--NNKVGSAEHSSSE--HR 162
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
S S ++G+S GSD N+ + + R+ + + G P TM
Sbjct: 163 SSQSSENDGSSNGSDGNTTGGEQSRRKRRQQRSPSTGERPSSQNSLPLRGENEKPDVTM- 221
Query: 234 MVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGV 293
GTP V TA++ + G +GV
Sbjct: 222 ---------------------------GTP-----------VMPTAMSFQNSAG--MNGV 241
Query: 294 QSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRI 353
Q W +E+E+KR++RKQSNRESARRSRLRKQAE ++L+ + +AL EN SLRS++ ++
Sbjct: 242 P-QPW--NEKEVKREKRKQSNRESARRSRLRKQAETEQLSVKVDALVAENMSLRSKLGQL 298
Query: 354 RSDYEQLLTENAALKERL 371
++ E+L EN A+ ++L
Sbjct: 299 NNESEKLRLENEAILDQL 316
>At1g13600 bZip transcription factor AtbZip58
Length = 196
Score = 63.9 bits (154), Expect = 1e-10
Identities = 42/112 (37%), Positives = 59/112 (52%), Gaps = 9/112 (8%)
Query: 285 ATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENA 344
+ + D Q + DER +QRR SNRESARRSR+RKQ DEL + L+ +N
Sbjct: 68 SNNSTSDEDHQQSMVIDER---KQRRMISNRESARRSRMRKQRHLDELWSQVIRLRTDNH 124
Query: 345 SLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQS 396
L +++R+ +E L ENA LKE DLR S+ +H++ S
Sbjct: 125 CLMDKLNRVSESHELALKENAKLKEE------TSDLRQLISEIKSHNEDDNS 170
>At3g30530 bZip transcription factor AtbZip42
Length = 173
Score = 62.8 bits (151), Expect = 3e-10
Identities = 38/101 (37%), Positives = 59/101 (57%), Gaps = 3/101 (2%)
Query: 274 NVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELA 333
N S +++ +T + Q+ + + +ER +QRR SNRESARRSR+RKQ DEL
Sbjct: 52 NPQSMSLSSNNSTSDEAEEQQTNNNIINER---KQRRMISNRESARRSRMRKQRHLDELW 108
Query: 334 QRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
+ L+ EN L +++ + ++++L ENA LKE EL
Sbjct: 109 SQVMWLRIENHQLLDKLNNLSESHDKVLQENAQLKEETFEL 149
>At5g38800 bZIP transcription factor AtbZip43
Length = 165
Score = 62.4 bits (150), Expect = 4e-10
Identities = 43/120 (35%), Positives = 63/120 (51%), Gaps = 9/120 (7%)
Query: 263 PTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDEREL--------KRQRRKQSN 314
PTSS + N P + G+ T + S + DE E ++Q+RK SN
Sbjct: 22 PTSSPFCGQNPN-PFFSFETGVNTSQFMSLISSNNSTSDEAEENHKEIINERKQKRKISN 80
Query: 315 RESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
RESARRSR+RKQ + DEL + L++EN L +++ + E+++ EN LKE EL
Sbjct: 81 RESARRSRMRKQRQVDELWSQVMWLRDENHQLLRKLNCVLESQEKVIEENVQLKEETTEL 140
>At1g75390 bZip transcription factor AtbZip44
Length = 173
Score = 62.4 bits (150), Expect = 4e-10
Identities = 40/102 (39%), Positives = 60/102 (58%), Gaps = 5/102 (4%)
Query: 273 GNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDEL 332
GN S + G +A GS ++ Q L DER +++RKQSNRESARRSR+RKQ D+L
Sbjct: 13 GNCSSVSTTG-LANSGSESDLR-QRDLIDER---KRKRKQSNRESARRSRMRKQKHLDDL 67
Query: 333 AQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGEL 374
+ L++ENA + + ++ Y + EN L+ ++ EL
Sbjct: 68 TAQVTHLRKENAQIVAGIAVTTQHYVTIEAENDILRAQVLEL 109
>At1g19490 bZip transcription factor AtbZip62
Length = 471
Score = 60.8 bits (146), Expect = 1e-09
Identities = 42/132 (31%), Positives = 69/132 (51%), Gaps = 10/132 (7%)
Query: 271 LHGNVPSTAVAGGMATGGSRDGV--QSQHWLQDERELKRQRRKQSNRESARRSRLRKQAE 328
++G P +A + +G Q+ + ERE +R RR +NRESAR++ R+QA
Sbjct: 123 INGETPKPILASTLIRCSRSNGCGRSRQNLSEAEREERRIRRILANRESARQTIRRRQAM 182
Query: 329 CDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQH 388
C+EL+++A L EN +LR E +++ L T N LKE++ L+S K D
Sbjct: 183 CEELSKKAADLTYENENLRREKDWALKEFQSLETINKHLKEQV--------LKSVKPDTK 234
Query: 389 THDDAQQSGQTE 400
+++ + Q E
Sbjct: 235 EPEESPKPSQVE 246
>At5g15830 bZIP transcription factor AtbZip3
Length = 186
Score = 59.3 bits (142), Expect = 4e-09
Identities = 30/69 (43%), Positives = 46/69 (66%)
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENA 365
++QRR SNRESARRSR+RKQ DEL + L+ EN L +++++ + + ++ EN+
Sbjct: 74 RKQRRMVSNRESARRSRMRKQRHLDELLSQVAWLRSENHQLLDKLNQVSDNNDLVIQENS 133
Query: 366 ALKERLGEL 374
+LKE EL
Sbjct: 134 SLKEENLEL 142
>At1g68880 bZip transcription factor AtbZip8
Length = 138
Score = 59.3 bits (142), Expect = 4e-09
Identities = 31/64 (48%), Positives = 43/64 (66%)
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENA 365
+++RRK SNRESARRSR+RKQ +EL L +N SL E+S+ R YE+++ EN
Sbjct: 47 RKRRRKVSNRESARRSRMRKQRHMEELWSMLVQLINKNKSLVDELSQARECYEKVIEENM 106
Query: 366 ALKE 369
L+E
Sbjct: 107 KLRE 110
>At2g04038 bZip transcription factor AtbZip48
Length = 166
Score = 58.9 bits (141), Expect = 5e-09
Identities = 31/69 (44%), Positives = 43/69 (61%)
Query: 306 KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENA 365
++QRR SNRESARRSR+RKQ DEL + L+ EN L +++R+ +L EN+
Sbjct: 74 RKQRRMLSNRESARRSRMRKQRHLDELWSQVIRLRNENNCLIDKLNRVSETQNCVLKENS 133
Query: 366 ALKERLGEL 374
LKE +L
Sbjct: 134 KLKEEASDL 142
>At3g10300 unknown protein
Length = 335
Score = 58.5 bits (140), Expect = 6e-09
Identities = 43/103 (41%), Positives = 50/103 (47%), Gaps = 19/103 (18%)
Query: 22 PQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPYGTPPH-P 80
P QPP STG NPP + S +P PP+G AS+P A PY G Q PYG PP P
Sbjct: 18 PPPQPPYGSTGN-NPPPYGS-SGSNPPPPYGSSASSPYAVPY--GAQ--PAPYGAPPSAP 71
Query: 81 YVAM----------YPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
Y ++ PHG Y PS PG Y P + PS G
Sbjct: 72 YASLPGDHNKPHKEKPHGASYGSPS--PGGYGAHPSSGPSDYG 112
>At4g02640 bZIP protein BZO2H1
Length = 417
Score = 58.2 bits (139), Expect = 8e-09
Identities = 44/151 (29%), Positives = 69/151 (45%), Gaps = 26/151 (17%)
Query: 265 SSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERE---------LKRQRRKQSNR 315
+S++PA V T V+ T GS L +E E +K+ RR SNR
Sbjct: 176 TSSLPA---EVKKTGVSMKQVTSGSSREYSDDEDLDEENETTGSLKPEDVKKSRRMLSNR 232
Query: 316 ESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALK------- 368
ESARRSR RKQ + +L + LK E++SL ++S + Y++ N LK
Sbjct: 233 ESARRSRRRKQEQTSDLETQVNDLKGEHSSLLKQLSNMNHKYDEAAVGNRILKADIETLR 292
Query: 369 -------ERLGELPGNDDLRSGKSDQHTHDD 392
E + + G + + G+S H +++
Sbjct: 293 AKVKMAEETVKRVTGMNPMLLGRSSGHNNNN 323
>At5g60830 bZip transcription factor AtbZip70
Length = 188
Score = 57.8 bits (138), Expect = 1e-08
Identities = 31/68 (45%), Positives = 43/68 (62%)
Query: 304 ELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTE 363
E +R RR SNRESARRSR+RK+ + +EL Q+ E L N L +V + Q+L E
Sbjct: 77 EERRARRMVSNRESARRSRMRKKKQIEELQQQVEQLMMLNHHLSEKVINLLESNHQILQE 136
Query: 364 NAALKERL 371
N+ LKE++
Sbjct: 137 NSQLKEKV 144
>At5g24800 bZIP protein BZO2H2
Length = 277
Score = 57.0 bits (136), Expect = 2e-08
Identities = 35/110 (31%), Positives = 50/110 (44%), Gaps = 3/110 (2%)
Query: 262 TPTSSNIPALHGNVPSTAVAGGMATGGSRDG---VQSQHWLQDERELKRQRRKQSNRESA 318
+P S+N P + G V T D D +LKR RR SNRESA
Sbjct: 75 SPVSANKPEVRGGVRRTTSGSSHVNSDDEDAETEAGQSEMTNDPNDLKRIRRMNSNRESA 134
Query: 319 RRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALK 368
+RSR RKQ +L + ++LK +N++L ++ + T N LK
Sbjct: 135 KRSRRRKQEYLVDLETQVDSLKGDNSTLYKQLIDATQQFRSAGTNNRVLK 184
>At4g37730 bZip transcription factor AtbZip7
Length = 305
Score = 55.8 bits (133), Expect = 4e-08
Identities = 33/83 (39%), Positives = 49/83 (58%), Gaps = 10/83 (12%)
Query: 299 LQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRS-------EVS 351
+ DER+ KR +SNRESA+RSR+RKQ+ D L ++ L EN L + ++
Sbjct: 193 MTDERKRKRM---ESNRESAKRSRMRKQSHIDNLREQVNRLDLENRELGNRLRLVLHQLQ 249
Query: 352 RIRSDYEQLLTENAALKERLGEL 374
R+ SD +L+TE L+ RL E+
Sbjct: 250 RVNSDNNRLVTEQEILRLRLSEM 272
>At4g34590 bZIP transcription factor ATB2/ Atbzip11
Length = 159
Score = 55.5 bits (132), Expect = 5e-08
Identities = 29/71 (40%), Positives = 44/71 (61%)
Query: 304 ELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTE 363
E ++++R SNRESARRSR++KQ D+L + LK+EN + + VS Y + E
Sbjct: 25 EQRKRKRMLSNRESARRSRMKKQKLLDDLTAQVNHLKKENTEIVTSVSITTQHYLTVEAE 84
Query: 364 NAALKERLGEL 374
N+ L+ +L EL
Sbjct: 85 NSVLRAQLDEL 95
>At3g62420 bZip transcription factor AtbZip53
Length = 146
Score = 54.7 bits (130), Expect = 9e-08
Identities = 36/99 (36%), Positives = 50/99 (50%), Gaps = 5/99 (5%)
Query: 284 MATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEEN 343
M T D + DER+ KR SNRESARRSR+RKQ + +L LK +N
Sbjct: 6 MQTSPESDNDPRYATVTDERKRKRMI---SNRESARRSRMRKQKQLGDLINEVTLLKNDN 62
Query: 344 ASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRS 382
A + +V Y ++ ++N L+ + EL D LRS
Sbjct: 63 AKITEQVDEASKKYIEMESKNNVLRAQASEL--TDRLRS 99
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,009,420
Number of Sequences: 26719
Number of extensions: 576371
Number of successful extensions: 3330
Number of sequences better than 10.0: 397
Number of HSP's better than 10.0 without gapping: 156
Number of HSP's successfully gapped in prelim test: 243
Number of HSP's that attempted gapping in prelim test: 2061
Number of HSP's gapped (non-prelim): 874
length of query: 406
length of database: 11,318,596
effective HSP length: 102
effective length of query: 304
effective length of database: 8,593,258
effective search space: 2612350432
effective search space used: 2612350432
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0230.5


