

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0229.10
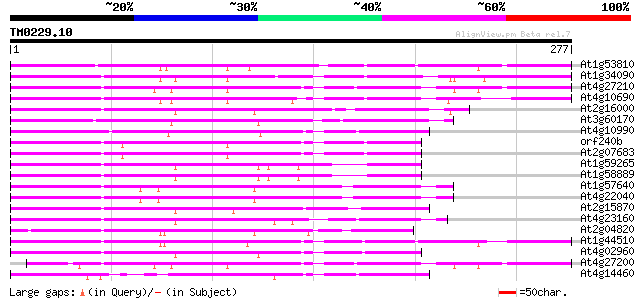
(277 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g53810 84 6e-17
At1g34090 putative protein 83 1e-16
At4g27210 putative protein 83 2e-16
At4g10690 retrotransposon like protein 79 3e-15
At2g16000 putative retroelement pol polyprotein 76 2e-14
At3g60170 putative protein 76 2e-14
At4g10990 putative retrotransposon polyprotein 74 9e-14
orf240b -mitochondrial genome- 71 6e-13
At2g07683 putative protein 71 6e-13
At1g59265 polyprotein, putative 70 9e-13
At1g58889 polyprotein, putative 70 9e-13
At1g57640 70 9e-13
At4g22040 LTR retrotransposon like protein 70 1e-12
At2g15870 putative retroelement pol polyprotein 69 2e-12
At4g23160 putative protein 68 5e-12
At2g04820 putative retroelement pol polyprotein 68 6e-12
At1g44510 polyprotein, putative 67 1e-11
At4g02960 putative polyprotein of LTR transposon 67 1e-11
At4g27200 putative protein 64 9e-11
At4g14460 retrovirus-related like polyprotein 62 3e-10
>At1g53810
Length = 1522
Score = 84.3 bits (207), Expect = 6e-17
Identities = 87/306 (28%), Positives = 147/306 (47%), Gaps = 38/306 (12%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D++ITG++S + + ++ LN F +K +G + +FLGI++++ DG L ++Q +Y DL
Sbjct: 1112 DMVITGNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQIQTY-DGGLFMSQQKYAEDLLI 1170
Query: 61 KVNISYGEKLQIP--------HNR-----GCSFYGSIAGSLQ*LTITRPEQSYSVKKVC- 106
+++ + P N+ +++ S+AG LQ LT+TRP+ ++V VC
Sbjct: 1171 TASMANCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQ 1230
Query: 107 ----PFMSSLHDSHR-------TAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*L 155
P +S + R T NS + S++++ ++ + ++ S C
Sbjct: 1231 KMHQPSVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYESDYDLSAYSDSDYANC-- 1288
Query: 156 GHGS*RSSFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITW 215
R S C ++ + I I W S KQ VS SS+EAEYRSL + +K SI
Sbjct: 1289 --KETRRSVGGYCTFMGQNI-ISWSSKKQPTVSRSSTEAEYRSLS-ETASEIKWMSSILR 1344
Query: 216 LKVVNLNTAPSIF*Q----VYHHFT*L*HYPTC*YYMELDILFFRKIVINRQLTAQHIPA 271
V+L P +F VY H T + ++D + R+ V + L +HIP
Sbjct: 1345 EIGVSLPDTPELFCDNLSAVYLTANPAFHART--KHFDVDHHYIRERVALKTLVVKHIPG 1402
Query: 272 DLQLAN 277
LQLA+
Sbjct: 1403 HLQLAD 1408
>At1g34090 putative protein
Length = 761
Score = 83.2 bits (204), Expect = 1e-16
Identities = 92/303 (30%), Positives = 139/303 (45%), Gaps = 41/303 (13%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D+ ITG+SS ++N + +LN F +K +G L +FLGI+ S G L L+Q +Y+ DL
Sbjct: 333 DMAITGNSSTALSNLLDELNKQFRMKDMGKLHYFLGIQAHYHSGG-LFLSQQKYVEDLLI 391
Query: 61 KVNISYGEKLQIP--------HNRGCSF-----YGSIAGSLQ*LTITRPEQSYSVKKVC- 106
+++ + P N+ F + S+AG LQ LT+TRP+ ++V VC
Sbjct: 392 TASMADCAPMPTPLPLQLNKVPNQDEQFSDQRYFRSLAGKLQYLTLTRPDIQFAVNYVCQ 451
Query: 107 ----PFMSSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
P +S H R T + +S A C R S + G R
Sbjct: 452 KMHSPTVSDYHLLKRILRYVKGTTTMGIS-FAKDTDCTVRAYSDSDYG-----GCKRTRR 505
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWL-KV--- 218
S C + I I W S KQ V+ SS+EAEYR+L ++ ITWL KV
Sbjct: 506 STAGFCTFFGSNI-ISWCSQKQETVARSSTEAEYRALSDAAA-------EITWLCKVLKE 557
Query: 219 --VNLNTAPSIF*QVYH--HFT*L*HYPTC*YYMELDILFFRKIVINRQLTAQHIPADLQ 274
+ L+TAP ++ + T + + E + R+ V LT +HIP+ +Q
Sbjct: 558 LQIPLHTAPELYADNLSSIYLTANPSFHKRSKHFETHYHYVRERVALGSLTVKHIPSHVQ 617
Query: 275 LAN 277
LA+
Sbjct: 618 LAD 620
>At4g27210 putative protein
Length = 1318
Score = 82.8 bits (203), Expect = 2e-16
Identities = 86/305 (28%), Positives = 134/305 (43%), Gaps = 45/305 (14%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D++ITG+SS + + ++ LN F + +G L +FLGI+V+ +G L ++Q +Y DL
Sbjct: 910 DMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQVQRQQNG-LFMSQQKYAEDLLI 968
Query: 61 KVNISYGEKL---------QIPHNRGC----SFYGSIAGSLQ*LTITRPEQSYSVKKVC- 106
++ + L ++PH +++ SIAG LQ LT+TRP+ ++V VC
Sbjct: 969 AASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQ 1028
Query: 107 ----PFMSSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
P +S H R T + +S S S S W R
Sbjct: 1029 KMHQPTISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYS-DSDWG-----NCKQTRR 1082
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKV---- 218
S C + + L+ W S K VS SS+EAEY+SL ++ I WL
Sbjct: 1083 SVGGLCTF-MGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAA-------SEILWLSTLLRE 1134
Query: 219 --VNLNTAPSIF*Q----VYHHFT*L*HYPTC*YYMELDILFFRKIVINRQLTAQHIPAD 272
+ L P +F VY H T + ++D F R+ V + L +HIP
Sbjct: 1135 LRIPLPDTPELFCDNLSAVYLTANPAFHART--KHFDIDFHFVRERVALKALVVKHIPGS 1192
Query: 273 LQLAN 277
Q+A+
Sbjct: 1193 EQIAD 1197
>At4g10690 retrotransposon like protein
Length = 1515
Score = 79.0 bits (193), Expect = 3e-15
Identities = 84/318 (26%), Positives = 138/318 (42%), Gaps = 73/318 (22%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D+I+TG++ L+ ++ L+ F +K +G L +FLGI+ +DG L L+Q +Y DL
Sbjct: 1116 DMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHYHNDG-LFLSQEKYTSDLLV 1174
Query: 61 KVNISYGEKLQIP--------HNRGC---SFYGSIAGSLQ*LTITRPEQSYSVKKVC--- 106
+S + P +N+ +++ +AG LQ LT+TRP+ ++V VC
Sbjct: 1175 NAGMSDCSSMPTPLQLDLLQGNNKPFPEPTYFRRLAGKLQYLTLTRPDIQFAVNFVCQKM 1234
Query: 107 --PFMSSLHDSHRTAINASSTNSKAMSLLASSQA---CFSRLSFISSWV**C*LGHGS*R 161
P MS H R T + ++L +++ + C+S S W G R
Sbjct: 1235 HAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYSD----SDWA-----GCKDTR 1285
Query: 162 SSFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITW------ 215
S C +L I I W + + VS SS+EAEYR+L ++ ++W
Sbjct: 1286 RSTGGFCTFLGYNI-ISWSAKRHPTVSKSSTEAEYRTLSFAA-------SEVSWIGFLLQ 1337
Query: 216 ----------------LKVVNLNTAPSIF*QVYHHFT*L*HYPTC*YYMELDILFFRKIV 259
L V L+ P++ + H ++D + R+ V
Sbjct: 1338 EIGLPQQQIPEMYCDNLSAVYLSANPALHSRSKH--------------FQVDYYYVRERV 1383
Query: 260 INRQLTAQHIPADLQLAN 277
LT +HIPA QLA+
Sbjct: 1384 ALGALTVKHIPASQQLAD 1401
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 76.3 bits (186), Expect = 2e-14
Identities = 72/245 (29%), Positives = 112/245 (45%), Gaps = 31/245 (12%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DI+I +S A +L+ F L+ LG L++FLG+EV + G + + Q +Y +L
Sbjct: 1140 DIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAG-ISICQRKYALELLQ 1198
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ + + +P Y I G L LTITRP+ +++V K+C
Sbjct: 1199 STGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQ 1258
Query: 108 FMSSLHDSHRTA---INASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSSF 164
F S+ +H TA + + L S+ + + F S C S +SF
Sbjct: 1259 FSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRS-TTSF 1317
Query: 165 YLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWL--KVVNLN 222
+ + LI WRS KQ VS SS+EAEYR+L L++C V WL +V+L
Sbjct: 1318 TM----FVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMV-------WLFTLLVSLQ 1366
Query: 223 TAPSI 227
+P +
Sbjct: 1367 ASPPV 1371
>At3g60170 putative protein
Length = 1339
Score = 75.9 bits (185), Expect = 2e-14
Identities = 70/237 (29%), Positives = 105/237 (43%), Gaps = 31/237 (13%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D+I TGS A+ + F + L F + LG ++HFLGIEVK SDG + + Q RY R++ A
Sbjct: 1023 DLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQ-SDGGIFICQRRYAREVLA 1081
Query: 61 KVNISYGEKLQIPHNRGC-------------SFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ + ++ P G + + + GSL LT+TRP+ Y V +
Sbjct: 1082 RFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTRPDLMYGVCLISR 1141
Query: 108 FMSSLHDSHRTAIN-----ASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
FMS+ SH A T + ++F S G + R
Sbjct: 1142 FMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLKLMAFTDSDY----AGDLNDRR 1197
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKVV 219
S V+L+ I W S KQ VV+ S++EAEY + +C+ V WL+ V
Sbjct: 1198 S-TSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCV-------WLRKV 1246
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 73.9 bits (180), Expect = 9e-14
Identities = 66/225 (29%), Positives = 104/225 (45%), Gaps = 26/225 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D++I + S+ + N L F +K LG FLG+E+ S+G V Q +Y ++L
Sbjct: 725 DLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVC-QRKYAQNLLE 783
Query: 61 KVNISYGEKLQIPHNRG-------------CSFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
V +S + IP + + Y + G L L ITRP+ +++V +
Sbjct: 784 DVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQ 843
Query: 108 FMSSLHDSHRTAINA-----SSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
F+S+ D H A + + + ASS+ C + S + W R
Sbjct: 844 FLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSD-ADWG-----TCKDSRR 897
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYV 207
S C+YL LI W+S KQ+VVS SS+E+EYRSL ++C +
Sbjct: 898 SVTGFCIYL-GTSLITWKSKKQSVVSRSSTESEYRSLAQATCEII 941
>orf240b -mitochondrial genome-
Length = 240
Score = 71.2 bits (173), Expect = 6e-13
Identities = 66/220 (30%), Positives = 103/220 (46%), Gaps = 25/220 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRY------ 54
DI++TGSS+ L+N I +L+ F +K LG + +FLGI++K+ G L L+Q +Y
Sbjct: 9 DILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSG-LFLSQTKYAEQILN 67
Query: 55 ------IRDL*AKVNISYGEKLQIPHNRGCSFYGSIAGSLQ*LTITRPEQSYSVKKVC-- 106
+ + + + + S + SI G+LQ LT+TRP+ SY+V VC
Sbjct: 68 NAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNIVCQR 127
Query: 107 ---PFMSSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSS 163
P ++ R T + + +S+ S W G S R S
Sbjct: 128 MHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFC-DSDWA-----GCTSTRRS 181
Query: 164 FYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSS 203
C +L I I W + +Q VS SS+E EYR+L L++
Sbjct: 182 TTGFCTFLGCNI-ISWSAKRQPTVSRSSTETEYRALALTA 220
>At2g07683 putative protein
Length = 385
Score = 71.2 bits (173), Expect = 6e-13
Identities = 66/220 (30%), Positives = 103/220 (46%), Gaps = 25/220 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRY------ 54
DI++TGSS+ L+N I +L+ F +K LG + +FLGI++K+ G L L+Q +Y
Sbjct: 154 DILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSG-LFLSQTKYAEQILN 212
Query: 55 ------IRDL*AKVNISYGEKLQIPHNRGCSFYGSIAGSLQ*LTITRPEQSYSVKKVC-- 106
+ + + + + S + SI G+LQ LT+TRP+ SY+V VC
Sbjct: 213 NAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNIVCQR 272
Query: 107 ---PFMSSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSS 163
P ++ R T + + +S+ S W G S R S
Sbjct: 273 MHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFC-DSDWA-----GCTSTRRS 326
Query: 164 FYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSS 203
C +L I I W + +Q VS SS+E EYR+L L++
Sbjct: 327 TTGFCTFLGCNI-ISWSAKRQPTVSRSSTETEYRALALTA 365
>At1g59265 polyprotein, putative
Length = 1466
Score = 70.5 bits (171), Expect = 9e-13
Identities = 72/231 (31%), Positives = 103/231 (44%), Gaps = 46/231 (19%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DI+ITG+ L++N + L+ F +K L +FLGIE K + G L L+Q RYI DL A
Sbjct: 1153 DILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLA 1211
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ N+ + + P Y I GSLQ L TRP+ SY+V ++
Sbjct: 1212 RTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQ 1271
Query: 108 FMSSLHDSHRTAIN------ASSTN-------SKAMSLLASSQACFS--RLSFISSWV** 152
FM + H A+ A + N +SL A S A ++ + ++S+
Sbjct: 1272 FMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYI 1331
Query: 153 C*LGHGS*RSSFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSS 203
LGH I W S KQ V SS+EAEYRS+ +S
Sbjct: 1332 VYLGHHP-----------------ISWSSKKQKGVVRSSTEAEYRSVANTS 1365
>At1g58889 polyprotein, putative
Length = 1466
Score = 70.5 bits (171), Expect = 9e-13
Identities = 72/231 (31%), Positives = 103/231 (44%), Gaps = 46/231 (19%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DI+ITG+ L++N + L+ F +K L +FLGIE K + G L L+Q RYI DL A
Sbjct: 1153 DILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLA 1211
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ N+ + + P Y I GSLQ L TRP+ SY+V ++
Sbjct: 1212 RTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQ 1271
Query: 108 FMSSLHDSHRTAIN------ASSTN-------SKAMSLLASSQACFS--RLSFISSWV** 152
FM + H A+ A + N +SL A S A ++ + ++S+
Sbjct: 1272 FMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYI 1331
Query: 153 C*LGHGS*RSSFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSS 203
LGH I W S KQ V SS+EAEYRS+ +S
Sbjct: 1332 VYLGHHP-----------------ISWSSKKQKGVVRSSTEAEYRSVANTS 1365
>At1g57640
Length = 1444
Score = 70.5 bits (171), Expect = 9e-13
Identities = 70/235 (29%), Positives = 103/235 (43%), Gaps = 29/235 (12%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D+II+GS + F S L F +K LG L++FLGIEV + G L+Q +Y+ D+ +
Sbjct: 1130 DLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQG-FYLSQRKYVLDIIS 1188
Query: 61 KVN----------ISYGEKLQI---PHNRGCSFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
++ + KL + P S Y + G L L +TRPE SYSV +
Sbjct: 1189 EMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQ 1248
Query: 108 FMSSLHDSHRTA---INASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSSF 164
FM + H A + ++ +L SS + + S C L S F
Sbjct: 1249 FMQNPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYF 1308
Query: 165 YLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKVV 219
L I W++ KQ VS SS+EAEYR++ + + WLK V
Sbjct: 1309 -----VQLGDTPISWKTKKQPTVSRSSAEAEYRAMAFLT-------QELMWLKRV 1351
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 70.1 bits (170), Expect = 1e-12
Identities = 69/235 (29%), Positives = 103/235 (43%), Gaps = 29/235 (12%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
D+II+GS + F S L F +K LG L++FLGIEV + G L+Q +Y+ D+ +
Sbjct: 795 DLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQG-FYLSQRKYVLDIIS 853
Query: 61 KVN----------ISYGEKLQI---PHNRGCSFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
++ + KL + P S Y + G L L +TRPE SYSV +
Sbjct: 854 EMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQ 913
Query: 108 FMSSLHDSHRTA---INASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSSF 164
FM + H A + ++ +L SS + + S C L S F
Sbjct: 914 FMQNPRQDHWNAAIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYF 973
Query: 165 YLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKVV 219
L I W++ KQ +S SS+EAEYR++ + + WLK V
Sbjct: 974 -----VQLGDTPISWKTKKQPTISRSSAEAEYRAMAFLT-------QELMWLKRV 1016
>At2g15870 putative retroelement pol polyprotein
Length = 1264
Score = 69.3 bits (168), Expect = 2e-12
Identities = 65/225 (28%), Positives = 100/225 (43%), Gaps = 26/225 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DI+I ++ A L +F L+ LG L++FLG+E+ G + L Q +Y +L A
Sbjct: 950 DIVIASTNEAAAVQLSQDLQNLFKLRDLGDLKYFLGLEITRTEAG-ISLCQRKYALELLA 1008
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ + + +P Y I G+L LTITRP+ +++V K+C
Sbjct: 1009 STGMINCKLVSVPMVPNLKLMKVDGELLEDREQYRRIVGTLMYLTITRPDITFAVNKLCQ 1068
Query: 108 FM-----SSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
F + L +HR T + + ASS + S W H +
Sbjct: 1069 FSYAPTTAHLQAAHRVLQYIKGTVGQGLFYSASSDLTLKGFAD-SDWASCPDSRHSTTGF 1127
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYV 207
+ ++ LI RS KQ VVS SS+EAEYR+L L++C V
Sbjct: 1128 TMFVG------DSLISLRSKKQHVVSRSSAEAEYRALALATCELV 1166
>At4g23160 putative protein
Length = 1240
Score = 68.2 bits (165), Expect = 5e-12
Identities = 71/237 (29%), Positives = 108/237 (44%), Gaps = 39/237 (16%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DIII ++ A ++ S+L F L+ LG L++FLG+E+ + G + + Q +Y DL
Sbjct: 286 DIIICSNNDAAVDELKSQLKSCFKLRDLGPLKYFLGLEIARSAAG-INICQRKYALDLLD 344
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ + + +P + +F Y + G L L ITR + S++V K+
Sbjct: 345 ETGLLGCKPSSVPMDPSVTFSAHSGGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQ 404
Query: 108 FMSSLHDSHRTAINASSTNSKA---MSLLASSQA-----CFSRLSFISSWV**C*LGHGS 159
F + +H+ A+ K L SSQA FS SF S
Sbjct: 405 FSEAPRLAHQQAVMKILHYIKGTVGQGLFYSSQAEMQLQVFSDASFQSC---------KD 455
Query: 160 *RSSFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWL 216
R S C++L LI W+S KQ VVS SS+EAEYR+L ++ D + WL
Sbjct: 456 TRRSTNGYCMFL-GTSLISWKSKKQQVVSKSSAEAEYRALSFAT-------DEMMWL 504
>At2g04820 putative retroelement pol polyprotein
Length = 387
Score = 67.8 bits (164), Expect = 6e-12
Identities = 72/216 (33%), Positives = 100/216 (45%), Gaps = 25/216 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DIIITG +S L+ NF L F LK LG L +FLGIE G L L Q +YI DL A
Sbjct: 75 DIIITGDAS-LVKNFNVSLADRFSLKDLGALSYFLGIEATRTKKG-LHLMQRKYITDLLA 132
Query: 61 KVNISYGEKLQIP---------HN----RGCSFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
K ++ + + P H+ Y I GSLQ L+ TRP+ ++ V ++
Sbjct: 133 KTHMLDAKPISTPMATTPKLSLHSGTILDDAKEYRMIIGSLQYLSFTRPDIAFVVNRLSQ 192
Query: 108 FMSSLHDSHRTA---INASSTNSKAMSLLASSQACFSRLSFI-SSWV**C*LGHGS*RSS 163
FM D H A + +K+ + S A + SF + W LG G ++
Sbjct: 193 FMHKPTDEHWQAAKRVLRYLAGTKSHGIFLRSDAPLTVHSFSDADWA--GDLGDGLSTNA 250
Query: 164 FYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSL 199
+ + I W S KQ S SS+EAEYR++
Sbjct: 251 Y----IVYFGGSPISWSSKKQRRFSRSSTEAEYRAV 282
>At1g44510 polyprotein, putative
Length = 1459
Score = 67.0 bits (162), Expect = 1e-11
Identities = 85/306 (27%), Positives = 131/306 (42%), Gaps = 47/306 (15%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DII+TGS ++ +S L F +K L +FLGIE + G L L Q +Y+ DL A
Sbjct: 1146 DIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYFLGIEATRTNTG-LHLMQRKYMTDLLA 1204
Query: 61 KVNISYGEKLQIP---------HN----RGCSFYGSIAGSLQ*LTITRPEQSYSVKKVCP 107
K N+ + + P H S Y S+ GSLQ L TRP+ +++V ++
Sbjct: 1205 KHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIAFAVNRLSQ 1264
Query: 108 FMSSLHDSH-----RTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
FM H R + T + + L +SS S + W G +
Sbjct: 1265 FMHQPTSDHWQAAKRVLRYLAGTTTHGIFLNSSSPIHLHAFS-DADWA-----GDSADYV 1318
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKVVNLN 222
S +YL R I W S KQ VS SS+E+EYR++ ++ ++ S+ + L
Sbjct: 1319 STNAYVIYLGRN-PISWSSKKQRGVSRSSTESEYRAVA-NAASEIRWLCSLLTELHIRLP 1376
Query: 223 TAPSIF*Q-----------VYHHFT*L*HYPTC*YYMELDILFFRKIVINRQLTAQHIPA 271
P+IF V+H ++ LD F R ++ +R L H+
Sbjct: 1377 HGPTIFCDNIGATYICANPVFHSRM---------KHIALDYHFVRGMIQSRALRVSHVST 1427
Query: 272 DLQLAN 277
+ QLA+
Sbjct: 1428 NDQLAD 1433
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 66.6 bits (161), Expect = 1e-11
Identities = 68/221 (30%), Positives = 97/221 (43%), Gaps = 26/221 (11%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGIEVKSLSDGTLVLTQFRYIRDL*A 60
DI+ITG+ + L+ + + L+ F +K+ L +FLGIE K + G L L+Q RY DL A
Sbjct: 1136 DILITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQG-LHLSQRRYTLDLLA 1194
Query: 61 KVNISYGEKLQIPHNRGCSF-------------YGSIAGSLQ*LTITRPEQSYSVKKVCP 107
+ N+ + + P Y I GSLQ L TRP+ SY+V ++
Sbjct: 1195 RTNMLTAKPVATPMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQ 1254
Query: 108 FMSSLHDSHRTAIN-----ASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RS 162
+M D H A+ + T + L + S + W G
Sbjct: 1255 YMHMPTDDHWNALKRVLRYLAGTPDHGIFLKKGNTLSLHAYS-DADWA-----GDTDDYV 1308
Query: 163 SFYLRCVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSS 203
S VYL I W S KQ V SS+EAEYRS+ +S
Sbjct: 1309 STNGYIVYLGHH-PISWSSKKQKGVVRSSTEAEYRSVANTS 1348
>At4g27200 putative protein
Length = 819
Score = 63.9 bits (154), Expect = 9e-11
Identities = 81/300 (27%), Positives = 126/300 (42%), Gaps = 50/300 (16%)
Query: 9 SALMNNFISKLNLVFLLKQLG*LEH---FLGIEVKSLSDGTLVLTQFRYIRDL*AKVNIS 65
S + + ++ LN F + +G +H FLGI+V+ +G L ++Q +Y DL ++
Sbjct: 418 SQTLTSLLAALNKEFRMTDMG--QHSLTFLGIQVQRQQNG-LFMSQQKYAEDLLIAASME 474
Query: 66 YGEKL---------QIPHNRGC----SFYGSIAGSLQ*LTITRPEQSYSVKKVC-----P 107
+ L ++PH +++ SIAG LQ LT+TRP+ ++V VC P
Sbjct: 475 HCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMHQP 534
Query: 108 FMSSLHDSHRTAINASSTNSKAMSLLASSQACFSRLSFISSWV**C*LGHGS*RSSFYLR 167
+S H R T + +S S S S W R S
Sbjct: 535 TISDFHLLKRILRYIKGTITMGISYSRDSPTLLQAYS-DSDWG-----NCKQTRRSVGGL 588
Query: 168 CVYLLRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYVKDPDSITWLKV------VNL 221
C + + L+ W S K VS SS+EAEY+SL ++ I WL + L
Sbjct: 589 CTF-MGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAA-------SEILWLSTLLRELRIPL 640
Query: 222 NTAPSIF*Q----VYHHFT*L*HYPTC*YYMELDILFFRKIVINRQLTAQHIPADLQLAN 277
P +F VY H T + ++D F R+ V + L +HIP Q+A+
Sbjct: 641 PDTPELFCDNLSAVYLTANPAFHART--KHFDIDFHFVRERVALKALVVKHIPGSEQIAD 698
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 62.0 bits (149), Expect = 3e-10
Identities = 66/216 (30%), Positives = 99/216 (45%), Gaps = 34/216 (15%)
Query: 1 DIIITGSSSALMNNFISKLNLVFLLKQLG*LEHFLGI---EVKSLS-DGTLVLTQFRYIR 56
DI+I ++ A + N + L F +K LG FLG+ + S+ D TL L +R
Sbjct: 1196 DIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLGLLGCKPSSIPMDPTLHL-----VR 1250
Query: 57 DL*AKVNISYGEKLQIPHNRGCSFYGSIAGSLQ*LTITRPEQSYSVKKVCPFMSSLHDSH 116
D+ G L P + Y + G L LTITRP+ +Y+V ++ F+S+ D H
Sbjct: 1251 DM--------GTPLPNP-----TAYRKLIGRLLYLTITRPDITYAVHQLSQFISAPSDIH 1297
Query: 117 RTAINASSTNSKA-----MSLLASSQACFSRLSFISSWV**C*LGHGS*RSSFYLRCVYL 171
A + KA + A + C + S + W R S C+YL
Sbjct: 1298 LQAAHKVLRYIKANPGQGLMYSADYEICLNGFSD-ADWA-----ACKDTRRSISGFCIYL 1351
Query: 172 LRPILIFWRS*KQTVVSGSSSEAEYRSLGLSSCRYV 207
LI W+S KQ V S SS+E+EYRS+ ++C +
Sbjct: 1352 -GTSLISWKSKKQAVASRSSTESEYRSMAQATCEII 1386
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.152 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,062,002
Number of Sequences: 26719
Number of extensions: 173538
Number of successful extensions: 805
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 625
Number of HSP's gapped (non-prelim): 126
length of query: 277
length of database: 11,318,596
effective HSP length: 98
effective length of query: 179
effective length of database: 8,700,134
effective search space: 1557323986
effective search space used: 1557323986
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0229.10


