

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.9
(273 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
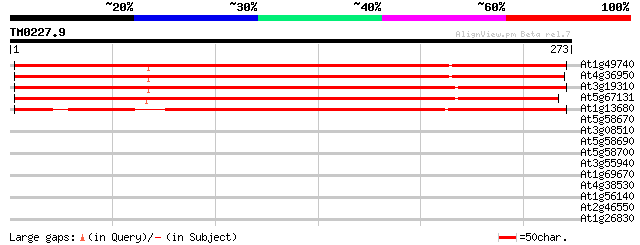
Score E
Sequences producing significant alignments: (bits) Value
At1g49740 unknown protein 338 2e-93
At4g36950 MAP3K-like protein kinase 325 2e-89
At3g19310 unknown protein 322 2e-88
At5g67131 unknown protein 302 1e-82
At1g13680 hypothetical protein 248 3e-66
At5g58670 phosphoinositide specific phospholipase C (AtPLC1) 37 0.009
At3g08510 phosphoinositide specific phospholipase C (AtPLC2) 37 0.011
At5g58690 phosphoinositide-specific phospholipase C-line (MZN1.13) 35 0.056
At5g58700 phosphoinositide-specific phospholipase C4 (PLC4) 34 0.096
At3g55940 phosphoinositide-specific phospholipase C - like protein 33 0.21
At1g69670 putative cullin 30 1.8
At4g38530 phosphoinositide-specific phospholipase C 29 2.4
At1g56140 receptor-like protein kinase, putative 28 4.0
At2g46550 unknown protein 27 9.0
At1g26830 cullin 3-like protein 27 9.0
>At1g49740 unknown protein
Length = 359
Score = 338 bits (867), Expect = 2e-93
Identities = 161/272 (59%), Positives = 209/272 (76%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+YSWLTTHNSFA G TGS +LA TNQ+DSIT QL NGVRG MLDM+D+++ I
Sbjct: 73 LPFNKYSWLTTHNSFARLGEVSRTGSAILAPTNQQDSITSQLNNGVRGFMLDMYDFQNDI 132
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C +T FQPA+N+L+E +VFL + E++TI I+D+V S G+ KVFD A
Sbjct: 133 WLCHSFDGTCFNFTAFQPAINILREFQVFLEKNKEEVVTIIIEDYVKSPKGLTKVFDAAG 192
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
LRKF FPVS+MPKNG DWP + M+RKN RL+VFTS++ +EA+EGIAY+W Y+VE+Q+GN
Sbjct: 193 LRKFMFPVSRMPKNGGDWPRLDDMVRKNQRLLVFTSDSHKEATEGIAYQWKYMVENQYGN 252
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K G C NR +S PM++ +KSLVL+N+F + + AC+ NS+ L+ + C++AAG
Sbjct: 253 GGLKVGVCPNRAQSAPMSDKSKSLVLVNHFPDAAD-VIVACKQNSASLLESIKTCYQAAG 311
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
RWPNFIAVDFYKR DGGGAP+A+D+AN NLI
Sbjct: 312 QRWPNFIAVDFYKRSDGGGAPQAVDVANGNLI 343
>At4g36950 MAP3K-like protein kinase
Length = 799
Score = 325 bits (833), Expect = 2e-89
Identities = 154/271 (56%), Positives = 202/271 (73%), Gaps = 4/271 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+YSWLTTHNS+A G N +TGS +++ NQEDSIT+QLKNGVRG+MLD +D+++ I
Sbjct: 383 LPFNKYSWLTTHNSYAITGANSATGSFLVSPKNQEDSITNQLKNGVRGIMLDTYDFQNDI 442
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C +T FQPA+N LKE+ FL ++ +EI+TI ++D+V S G+ VF+ +
Sbjct: 443 WLCHSTGGTCFNFTAFQPAINALKEINDFLESNLSEIVTIILEDYVKSQMGLTNVFNASG 502
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L KF P+S+MPK+G+DWPTV M+++N RL+VFTS +EASEG+AY+WNY+VE+Q+GN
Sbjct: 503 LSKFLLPISRMPKDGTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQWNYMVENQYGN 562
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K GSC +R ES ++ ++SLV NYF N +AC DNSSPLI MM C AAG
Sbjct: 563 DGMKDGSCSSRSESSSLDTMSRSLVFQNYFETSPN-STQACADNSSPLIEMMRTCHEAAG 621
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNL 270
RWPNFIAVDFY+R D GGA EA+D AN L
Sbjct: 622 KRWPNFIAVDFYQRSDSGGAAEAVDEANGRL 652
>At3g19310 unknown protein
Length = 413
Score = 322 bits (824), Expect = 2e-88
Identities = 150/272 (55%), Positives = 204/272 (74%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LP+N+YSWLTTHNSFA G TGS +LA +NQ+DSIT QL NGVRG MLD++D+++ I
Sbjct: 73 LPYNKYSWLTTHNSFARMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDI 132
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C YT FQPA+N+LKE +VFL + ++T+ ++D+V S NG+ +VFD +
Sbjct: 133 WLCHSYGGNCFNYTAFQPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASG 192
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
LR F FPVS+MPKNG DWPT+ MI +N RL+VFTSN +EASEGIA+ W Y++E+Q+G+
Sbjct: 193 LRNFMFPVSRMPKNGEDWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGD 252
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K G C NRPES+ M + ++SL+L+NYF + + +C+ NS+PL+ + C A+G
Sbjct: 253 GGMKAGVCTNRPESVAMGDRSRSLILVNYFPDTADVIG-SCKQNSAPLLDTVKNCQEASG 311
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
RWPNFIAVDFYKR DGGGAP+A+D+AN + +
Sbjct: 312 KRWPNFIAVDFYKRSDGGGAPKAVDVANGHAV 343
>At5g67131 unknown protein
Length = 426
Score = 302 bits (774), Expect = 1e-82
Identities = 142/268 (52%), Positives = 192/268 (70%), Gaps = 4/268 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+Y+WL THN+F+ G + F NQED+IT+QL+NGVRGLMLDM+D+ + I
Sbjct: 80 LPFNKYTWLMTHNAFSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDI 139
Query: 63 WLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC RG C +T FQPA+N+L+EV FL +PTEI+TI I+D+V G++ +F A
Sbjct: 140 WLCHSLRGQCFNFTAFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAG 199
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+WFPVSKMP+ G DWPTV M+++N+RL+VFTS A++E EG+AY+W Y+VE++ G+
Sbjct: 200 LDKYWFPVSKMPRKGEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGD 259
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K GSC NR ES P+N+ + SL LMNYF D AC+++S+PL M+ C ++ G
Sbjct: 260 PGVKRGSCPNRKESQPLNSKSSSLFLMNYFPTYPVEKD-ACKEHSAPLAEMVGTCLKSGG 318
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLAN 267
NR PNF+AV+FY R DGGG E LD N
Sbjct: 319 NRMPNFLAVNFYMRSDGGGVFEILDRMN 346
>At1g13680 hypothetical protein
Length = 300
Score = 248 bits (632), Expect = 3e-66
Identities = 123/269 (45%), Positives = 173/269 (63%), Gaps = 22/269 (8%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
+PFN+Y++LTTHNS+A +G L QED+I QL +GVR LMLD +DYE
Sbjct: 45 MPFNKYAFLTTHNSYAIEG-------KALHVATQEDTIVQQLNSGVRALMLDTYDYEGD- 96
Query: 63 WLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARLRK 122
A++ KE+ FL +P+EI+T+ ++D+V S NG+ KVF + L+K
Sbjct: 97 -------------NRAIDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSGLKK 143
Query: 123 FWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNVGI 182
FWFPV MP G DWP VK M+ N+RLIVFTS S++ +EGIAY+WNY+VE+Q+G+ G+
Sbjct: 144 FWFPVQNMPIGGQDWPLVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGDDGV 203
Query: 183 KGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGNRW 242
K C NR +S + + TK+LV +N+F+ V C +NS L+ M+ C+ AAGNRW
Sbjct: 204 KPDECSNRADSALLTDKTKALVSVNHFKTV-PVKILTCEENSEQLLDMIKTCYVAAGNRW 262
Query: 243 PNFIAVDFYKRGDGGGAPEALDLANRNLI 271
NF+AV+FYKR +GGG +A+D N L+
Sbjct: 263 ANFVAVNFYKRSNGGGTFQAIDKLNGELL 291
>At5g58670 phosphoinositide specific phospholipase C (AtPLC1)
Length = 561
Score = 37.4 bits (85), Expect = 0.009
Identities = 30/109 (27%), Positives = 49/109 (44%), Gaps = 14/109 (12%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMW---D 57
M P + Y T HNS+ G ++ S + + I L+NGVR + LD+W
Sbjct: 107 MNQPLSHYFLYTGHNSYLT-GNQLNSNSSI-------EPIVKALRNGVRVIELDLWPNSS 158
Query: 58 YEDTIWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVT 106
++ G T Q LNV+KE + +P + + ++DH+T
Sbjct: 159 GKEAEVRHGGTLTSREDLQKCLNVVKENAFQVSAYP---VVLTLEDHLT 204
>At3g08510 phosphoinositide specific phospholipase C (AtPLC2)
Length = 581
Score = 37.0 bits (84), Expect = 0.011
Identities = 32/108 (29%), Positives = 48/108 (43%), Gaps = 12/108 (11%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMW--DY 58
M+ P + Y T HNS+ TG+ L+ E I D LK GVR + LD+W
Sbjct: 105 MDAPISHYFIFTGHNSYL-------TGNQ-LSSDCSEVPIIDALKKGVRVIELDIWPNSN 156
Query: 59 EDTIWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVT 106
+D I + G TT + LK +R + + ++DH+T
Sbjct: 157 KDDIDVLHG--MTLTTPVGLIKCLKAIRAHAFDVSDYPVVVTLEDHLT 202
>At5g58690 phosphoinositide-specific phospholipase C-line (MZN1.13)
Length = 578
Score = 34.7 bits (78), Expect = 0.056
Identities = 33/112 (29%), Positives = 49/112 (43%), Gaps = 20/112 (17%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDS---ITDQLKNGVRGLMLDMW- 56
M P + Y T+HNS+ TG+ + N E S + LK GVR L LDMW
Sbjct: 114 MASPLSHYFIYTSHNSYL-------TGNQI----NSECSDVPLIKALKRGVRALELDMWP 162
Query: 57 -DYEDTIWLCRG-PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVT 106
+D I + G T L +KE + +P + + ++DH+T
Sbjct: 163 NSTKDDILVLHGWAWTPPVELVKCLRSIKEHAFYASAYP---VILTLEDHLT 211
>At5g58700 phosphoinositide-specific phospholipase C4 (PLC4)
Length = 597
Score = 33.9 bits (76), Expect = 0.096
Identities = 31/108 (28%), Positives = 48/108 (43%), Gaps = 13/108 (12%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDY-E 59
M+ P + Y T HNS+ TG+ L+ E I D L+ GVR + LD+W
Sbjct: 116 MDAPLSHYFIFTGHNSYL-------TGNQ-LSSNCSELPIADALRRGVRVVELDLWPRGT 167
Query: 60 DTIWLCRG-PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVT 106
D + + G TK L +K + +P + I ++DH+T
Sbjct: 168 DDVCVKHGRTLTKEVKLGKCLESIKANAFAISKYP---VIITLEDHLT 212
>At3g55940 phosphoinositide-specific phospholipase C - like protein
Length = 584
Score = 32.7 bits (73), Expect = 0.21
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 14/109 (12%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMW--DY 58
M+ P + Y T HNS+ TG+ L+ E I + LK GVR + LD+W
Sbjct: 105 MDAPLSHYFIYTGHNSYL-------TGNQ-LSSDCSELPIIEALKKGVRVIELDIWPNSD 156
Query: 59 EDTIWLCRG-PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVT 106
ED I + G T L ++E + +P + + ++DH+T
Sbjct: 157 EDGIDVLHGRTLTSPVELIKCLRAIREHAFDVSDYP---VVVTLEDHLT 202
>At1g69670 putative cullin
Length = 732
Score = 29.6 bits (65), Expect = 1.8
Identities = 16/35 (45%), Positives = 20/35 (56%)
Query: 74 TFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSG 108
TFQ ALN E V L T E I++F+DD + G
Sbjct: 356 TFQNALNSSFEYFVNLNTRSPEFISLFVDDKLRKG 390
>At4g38530 phosphoinositide-specific phospholipase C
Length = 526
Score = 29.3 bits (64), Expect = 2.4
Identities = 27/108 (25%), Positives = 49/108 (45%), Gaps = 14/108 (12%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYE- 59
M+ P + Y T HNS+ TG+ V + ++ E I L+ GV+ + LD+W
Sbjct: 70 MKAPLSHYFVYTGHNSYL-------TGNQVNSRSSVEP-IVQALRKGVKVIELDLWPNPS 121
Query: 60 -DTIWLCRG-PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHV 105
+ + G T + Q L +K+ + +P + I ++DH+
Sbjct: 122 GNAAEVRHGRTLTSHEDLQKCLTAIKDNAFHVSDYP---VIITLEDHL 166
>At1g56140 receptor-like protein kinase, putative
Length = 2083
Score = 28.5 bits (62), Expect = 4.0
Identities = 33/161 (20%), Positives = 67/161 (41%), Gaps = 17/161 (10%)
Query: 61 TIW-LCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
T+W LC F P V + R THP E ++ +++TS N ++++ D
Sbjct: 1028 TVWFLCN--------FGPVYVVRAQNRTGATTHPDEGLSSSSLNNLTSSNSIDRIEDCVA 1079
Query: 120 LRKFWFPVSKMPKNGSDW--PTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVE-SQ 176
F + +G++ +T++R+ YR ++S + ++ +
Sbjct: 1080 SASFELNFRGLEDSGAEGMEHQRRTLLRRRYRRQCSRLKPCLQSSHQMRLQFRKLHNLPH 1139
Query: 177 FGNVGIKGGSCQNRPES-----LPMNNATKSLVLMNYFRNV 212
+ + G+ GSC+ + +P + T S + FRN+
Sbjct: 1140 YQHQGLCDGSCRIYTSTTLDLGIPDKSHTFSQFKITSFRNL 1180
>At2g46550 unknown protein
Length = 397
Score = 27.3 bits (59), Expect = 9.0
Identities = 14/36 (38%), Positives = 21/36 (57%)
Query: 170 NYVVESQFGNVGIKGGSCQNRPESLPMNNATKSLVL 205
+Y V Q KG SC+NRPE+ ++ +KS +L
Sbjct: 232 DYSVSGQTIKGTSKGSSCKNRPEASSESDLSKSELL 267
>At1g26830 cullin 3-like protein
Length = 732
Score = 27.3 bits (59), Expect = 9.0
Identities = 14/35 (40%), Positives = 19/35 (54%)
Query: 74 TFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSG 108
TFQ ALN E + L E I++F+DD + G
Sbjct: 356 TFQNALNSSFEYFINLNARSPEFISLFVDDKLRKG 390
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,620,302
Number of Sequences: 26719
Number of extensions: 280707
Number of successful extensions: 540
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 521
Number of HSP's gapped (non-prelim): 15
length of query: 273
length of database: 11,318,596
effective HSP length: 98
effective length of query: 175
effective length of database: 8,700,134
effective search space: 1522523450
effective search space used: 1522523450
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0227.9


