

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
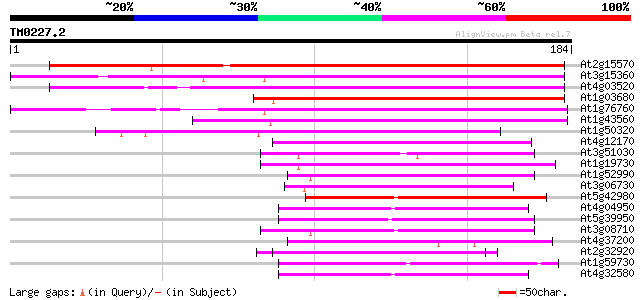
Query= TM0227.2
(184 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g15570 putative thioredoxin M 174 3e-44
At3g15360 thioredoxin m4 121 2e-28
At4g03520 putative M-type thioredoxin 102 1e-22
At1g03680 putative thioredoxin-m 94 4e-20
At1g76760 thioredoxin-like protein 89 1e-18
At1g43560 unknown protein 77 4e-15
At1g50320 thioredoxin like protein 66 9e-12
At4g12170 unknown protein (At4g12170) 62 2e-10
At3g51030 thioredoxin h 62 2e-10
At1g19730 thioredoxin 62 2e-10
At1g52990 hypothetical protein 61 3e-10
At3g06730 thioredoxin, putative 56 1e-08
At5g42980 thioredoxin (clone GIF1) (pir||S58118) 55 2e-08
At4g04950 putative thioredoxin 54 4e-08
At5g39950 thioredoxin 54 5e-08
At3g08710 thioredoxin-like protein 54 6e-08
At4g37200 thiol-disulfide interchange like protein 51 3e-07
At2g32920 putative protein disulfide isomerase 51 3e-07
At1g59730 unknown protein 51 4e-07
At4g32580 unknown protein (At4g32580) 50 5e-07
>At2g15570 putative thioredoxin M
Length = 173
Score = 174 bits (440), Expect = 3e-44
Identities = 89/170 (52%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>At3g15360 thioredoxin m4
Length = 193
Score = 121 bits (304), Expect = 2e-28
Identities = 68/194 (35%), Positives = 109/194 (56%), Gaps = 15/194 (7%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MAS+ S+++T S ++S SSS S SR ++P+ F + + L Q+
Sbjct: 1 MASLLDSVTVTRVFSLPIAASVSSSSAAP---SVSRRRISPARFLEFRGLKSSRSLVTQS 57
Query: 61 PP-------HLPRSHKIRSLRQETRATPI-----TKDLWENSILKSDIPVLVEFYASWCG 108
+ R +I Q+T A + + W+ +L+SD+PVLVEF+A WCG
Sbjct: 58 ASLGANRRTRIARGGRIACEAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCG 117
Query: 109 PCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGT 168
PCRM+H ++D +AK++AG+ + +NTD A+ Y I++VP V++FK G+K D++IG
Sbjct: 118 PCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTANRYGIRSVPTVIIFKGGEKKDSIIGA 177
Query: 169 MPKEFYVNAIERVL 182
+P+E IER L
Sbjct: 178 VPRETLEKTIERFL 191
>At4g03520 putative M-type thioredoxin
Length = 186
Score = 102 bits (254), Expect = 1e-22
Identities = 53/169 (31%), Positives = 92/169 (54%), Gaps = 5/169 (2%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSL 73
SSS S+SS SS + SS + SLS P L H+P L R+ +
Sbjct: 22 SSSPSASSLSSRRMFAVLPESSGLRIRLSLS-PASLTSIHQP----RVSRLRRAVVCEAQ 76
Query: 74 RQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLV 133
T + W++ +LK+ PV+V+F+A WCGPC+M+ +++D+A+ Y G++ + +
Sbjct: 77 ETTTDIQVVNDSTWDSLVLKATGPVVVDFWAPWCGPCKMIDPLVNDLAQHYTGKIKFYKL 136
Query: 134 NTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
NTD Y ++++P +M+F G+K D +IG +PK +++++ L
Sbjct: 137 NTDESPNTPGQYGVRSIPTIMIFVGGEKKDTIIGAVPKTTLTSSLDKFL 185
>At1g03680 putative thioredoxin-m
Length = 179
Score = 94.0 bits (232), Expect = 4e-20
Identities = 36/103 (34%), Positives = 68/103 (65%), Gaps = 1/103 (0%)
Query: 81 PITKD-LWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM 139
P+ D W++ +LK+D PV V+F+A WCGPC+M+ +++++A++YAG+ + +NTD
Sbjct: 77 PVVNDSTWDSLVLKADEPVFVDFWAPWCGPCKMIDPIVNELAQKYAGQFKFYKLNTDESP 136
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Y ++++P +M+F +G+K D +IG + K+ +I + L
Sbjct: 137 ATPGQYGVRSIPTIMIFVNGEKKDTIIGAVSKDTLATSINKFL 179
>At1g76760 thioredoxin-like protein
Length = 172
Score = 89.0 bits (219), Expect = 1e-18
Identities = 61/190 (32%), Positives = 95/190 (49%), Gaps = 28/190 (14%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MASIS S S S +S SS S+ +SRS FP R+ L
Sbjct: 1 MASISLSSSTVPSLNSKESSGVSAF--------ASRSISAVKFQFPVR-RVRTGDL---- 47
Query: 61 PPHLPRSHKIRSLRQETRATPI-------TKDLWENSILKSDIPVLVEFYASWCGPCRMV 113
K SL TR TP T D +E+ ++ SD PVLV++YA+WCGPC+ +
Sbjct: 48 --------KFPSLSSTTRCTPRRIEAKKQTFDSFEDLLVNSDKPVLVDYYATWCGPCQFM 99
Query: 114 HRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEF 173
+++++++ ++ ++T+ IA+ Y+I+A+P +LFKDG+ CD G + +
Sbjct: 100 VPILNEVSETLKDKIQVVKIDTEKYPSIANKYKIEALPTFILFKDGEPCDRFEGALTAKQ 159
Query: 174 YVNAIERVLK 183
+ IE LK
Sbjct: 160 LIQRIEDSLK 169
>At1g43560 unknown protein
Length = 167
Score = 77.4 bits (189), Expect = 4e-15
Identities = 39/131 (29%), Positives = 73/131 (54%), Gaps = 8/131 (6%)
Query: 61 PPHLPRSHKIRSLRQETRATPITK--------DLWENSILKSDIPVLVEFYASWCGPCRM 112
P + R ++S TR P+T + +++ + SD PVLV+FYA+WCGPC++
Sbjct: 34 PSQIRRFGSVQSPSSSTRFAPLTVRAAKKQTFNSFDDLLQNSDKPVLVDFYATWCGPCQL 93
Query: 113 VHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKE 172
+ +++++++ + ++T+ +A+ Y+I+A+P +LFKDG+ D G +P
Sbjct: 94 MVPILNEVSETLKDIIAVVKIDTEKYPSLANKYQIEALPTFILFKDGKLWDRFEGALPAN 153
Query: 173 FYVNAIERVLK 183
V IE L+
Sbjct: 154 QLVERIENSLQ 164
>At1g50320 thioredoxin like protein
Length = 182
Score = 66.2 bits (160), Expect = 9e-12
Identities = 41/152 (26%), Positives = 79/152 (51%), Gaps = 19/152 (12%)
Query: 29 DPFLSSS----RSHLNPSL-SFPFHLRITHKPLHLQNPPHLPRSHKIRSLRQETRAT--- 80
D +SSS RS+L P + S ++ KPL + + + SL R T
Sbjct: 2 DSIVSSSTILMRSYLTPPVRSCSPATSVSVKPLSSVQVTSVAANRHLLSLSSGARRTRKS 61
Query: 81 -----------PITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLN 129
I + + +++L+S PVLVEF A+WCGPC++++ ++ +++EY +L
Sbjct: 62 SSSVIRCGGIKEIGESEFSSTVLESAQPVLVEFVATWCGPCKLIYPAMEALSQEYGDKLT 121
Query: 130 CFLVNTDTDMQIADDYEIKAVPVVMLFKDGQK 161
++ D + ++ ++++ +P +LFKDG++
Sbjct: 122 IVKIDHDANPKLIAEFKVYGLPHFILFKDGKE 153
>At4g12170 unknown protein (At4g12170)
Length = 128
Score = 61.6 bits (148), Expect = 2e-10
Identities = 26/85 (30%), Positives = 51/85 (59%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W + +++S +PV+V F A C C + ++ + EY L + V+TD ++++A DY
Sbjct: 34 WNSLVIQSKVPVIVVFIAKDCAECGSLMPELEFLDSEYEYMLKFYTVDTDEELELAKDYR 93
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPK 171
I+ P+ ++FK G++ + V+G P+
Sbjct: 94 IEYHPITIVFKGGEEKERVLGYYPQ 118
>At3g51030 thioredoxin h
Length = 114
Score = 61.6 bits (148), Expect = 2e-10
Identities = 36/94 (38%), Positives = 51/94 (53%), Gaps = 6/94 (6%)
Query: 83 TKDLWENSILK---SDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL-VNTDTD 138
T + W + K S V+V+F ASWCGPCR + D+AK+ L FL V+TD
Sbjct: 13 TVETWNEQLQKANESKTLVVVDFTASWCGPCRFIAPFFADLAKKLPNVL--FLKVDTDEL 70
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKE 172
+A D+ I+A+P M K+G+ D V+G E
Sbjct: 71 KSVASDWAIQAMPTFMFLKEGKILDKVVGAKKDE 104
>At1g19730 thioredoxin
Length = 119
Score = 61.6 bits (148), Expect = 2e-10
Identities = 27/100 (27%), Positives = 55/100 (55%), Gaps = 3/100 (3%)
Query: 83 TKDLWENSILK---SDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM 139
T D+W + K S+ ++++F ASWC PCRM+ + +D+AK++ F V+ D
Sbjct: 13 TNDVWTVQLDKAKESNKLIVIDFTASWCPPCRMIAPIFNDLAKKFMSSAIFFKVDVDELQ 72
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIE 179
+A ++ ++A+P + K G+ D ++G ++ ++
Sbjct: 73 SVAKEFGVEAMPTFVFIKAGEVVDKLVGANKEDLQAKIVK 112
>At1g52990 hypothetical protein
Length = 313
Score = 61.2 bits (147), Expect = 3e-10
Identities = 28/82 (34%), Positives = 47/82 (57%), Gaps = 1/82 (1%)
Query: 92 LKSDIP-VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAV 150
L S P V+V F A WCGPCR + +++ + EY + VN DT+++ + ++I +
Sbjct: 223 LNSQTPHVMVMFTARWCGPCRDMIPILNKMDSEYKNEFKFYTVNFDTEIRFTERFDISYL 282
Query: 151 PVVMLFKDGQKCDAVIGTMPKE 172
P ++FK G++ V G PK+
Sbjct: 283 PTTLVFKGGEQMAKVTGADPKK 304
>At3g06730 thioredoxin, putative
Length = 183
Score = 55.8 bits (133), Expect = 1e-08
Identities = 23/77 (29%), Positives = 45/77 (57%), Gaps = 2/77 (2%)
Query: 91 ILKSD--IPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
++K D +P++V+FYA+WCGPC ++ + ++ +A EY V+TD + + A D +++
Sbjct: 88 LVKGDRKVPLIVDFYATWCGPCILMAQELEMLAVEYESNAIIVKVDTDDEYEFARDMQVR 147
Query: 149 AVPVVMLFKDGQKCDAV 165
+P + DA+
Sbjct: 148 GLPTLFFISPDPSKDAI 164
>At5g42980 thioredoxin (clone GIF1) (pir||S58118)
Length = 118
Score = 55.1 bits (131), Expect = 2e-08
Identities = 23/79 (29%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Query: 98 VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFK 157
++++F A+WC PCR + V D+AK++ + F V+ D +A++++++A+P + K
Sbjct: 30 IVIDFTATWCPPCRFIAPVFADLAKKHLD-VVFFKVDVDELNTVAEEFKVQAMPTFIFMK 88
Query: 158 DGQKCDAVIGTMPKEFYVN 176
+G+ + V+G +E N
Sbjct: 89 EGEIKETVVGAAKEEIIAN 107
>At4g04950 putative thioredoxin
Length = 488
Score = 54.3 bits (129), Expect = 4e-08
Identities = 25/82 (30%), Positives = 47/82 (56%), Gaps = 1/82 (1%)
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
+++ +S PV++ F+ASWC + + +V +A ++ R + F V + +I++ Y +
Sbjct: 15 DNLRQSGAPVVLHFWASWCDASKQMDQVFSHLATDFP-RAHFFRVEAEEHPEISEAYSVA 73
Query: 149 AVPVVMLFKDGQKCDAVIGTMP 170
AVP + FKDG+ D + G P
Sbjct: 74 AVPYFVFFKDGKTVDTLEGADP 95
>At5g39950 thioredoxin
Length = 133
Score = 53.9 bits (128), Expect = 5e-08
Identities = 26/84 (30%), Positives = 49/84 (57%), Gaps = 1/84 (1%)
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
N I +S+ ++V+F ASWCGPCRM+ I +A ++ ++ ++ D +A ++ +
Sbjct: 41 NEIKESNKLLVVDFSASWCGPCRMIEPAIHAMADKF-NDVDFVKLDVDELPDVAKEFNVT 99
Query: 149 AVPVVMLFKDGQKCDAVIGTMPKE 172
A+P +L K G++ + +IG E
Sbjct: 100 AMPTFVLVKRGKEIERIIGAKKDE 123
>At3g08710 thioredoxin-like protein
Length = 140
Score = 53.5 bits (127), Expect = 6e-08
Identities = 26/93 (27%), Positives = 51/93 (53%), Gaps = 4/93 (4%)
Query: 83 TKDLWENSILKSDIP---VLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM 139
TK+ W++ + ++D V+ F A+WCGPC++V +++++++ L LV+ D
Sbjct: 30 TKESWDDKLAEADRDGKIVVANFSATWCGPCKIVAPFFIELSEKHSS-LMFLLVDVDELS 88
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKE 172
+ ++IKA P K+GQ+ ++G E
Sbjct: 89 DFSSSWDIKATPTFFFLKNGQQIGKLVGANKPE 121
>At4g37200 thiol-disulfide interchange like protein
Length = 261
Score = 51.2 bits (121), Expect = 3e-07
Identities = 25/90 (27%), Positives = 49/90 (53%), Gaps = 3/90 (3%)
Query: 92 LKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM--QIADDYEIKA 149
L + P +VEFYA WC CR + + I ++Y ++N ++N D Q D++ ++
Sbjct: 135 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDKVNFVMLNVDNTKWEQELDEFGVEG 194
Query: 150 VP-VVMLFKDGQKCDAVIGTMPKEFYVNAI 178
+P L ++G + V+G +P+++ V +
Sbjct: 195 IPHFAFLDREGNEEGNVVGRLPRQYLVENV 224
>At2g32920 putative protein disulfide isomerase
Length = 440
Score = 51.2 bits (121), Expect = 3e-07
Identities = 24/79 (30%), Positives = 40/79 (50%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T +++ +L S+ VLVEF+A WCG C+ + + +A G ++ D
Sbjct: 35 LTASNFKSKVLNSNGVVLVEFFAPWCGHCKALTPTWEKVANILKGVATVAAIDADAHQSA 94
Query: 142 ADDYEIKAVPVVMLFKDGQ 160
A DY IK P + +F G+
Sbjct: 95 AQDYGIKGFPTIKVFVPGK 113
Score = 43.9 bits (102), Expect = 5e-05
Identities = 18/70 (25%), Positives = 39/70 (55%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
+++ +++S+ +VEF+A WCG C+ + AK G++ VN D + I ++
Sbjct: 172 FDDLVIESNELWIVEFFAPWCGHCKKLAPEWKRAAKNLQGKVKLGHVNCDVEQSIMSRFK 231
Query: 147 IKAVPVVMLF 156
++ P +++F
Sbjct: 232 VQGFPTILVF 241
>At1g59730 unknown protein
Length = 129
Score = 50.8 bits (120), Expect = 4e-07
Identities = 28/92 (30%), Positives = 50/92 (53%), Gaps = 2/92 (2%)
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
+S+ S+ ++++F A WCGPC+ + + +IA +Y+ + V+ D M +A Y
Sbjct: 37 DSMKGSNKLLVIDFTAVWCGPCKAMEPRVREIASKYSEAVFA-RVDVDRLMDVAGTYRAI 95
Query: 149 AVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
+P + K G++ D V+G P E V IE+
Sbjct: 96 TLPAFVFVKRGEEIDRVVGAKPDEL-VKKIEQ 126
>At4g32580 unknown protein (At4g32580)
Length = 160
Score = 50.4 bits (119), Expect = 5e-07
Identities = 23/82 (28%), Positives = 45/82 (54%), Gaps = 1/82 (1%)
Query: 89 NSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIK 148
+++ S P+++ F+ASWC + + +V +A ++ R + F V + +I++ Y +
Sbjct: 15 DNLRHSGAPLVLHFWASWCDASKQMDQVFSHLATDFP-RAHFFRVEAEEHPEISEAYSVA 73
Query: 149 AVPVVMLFKDGQKCDAVIGTMP 170
VP + FKDG+ D + G P
Sbjct: 74 LVPYFVFFKDGKTVDTLEGADP 95
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,281,741
Number of Sequences: 26719
Number of extensions: 190861
Number of successful extensions: 3667
Number of sequences better than 10.0: 228
Number of HSP's better than 10.0 without gapping: 162
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 2448
Number of HSP's gapped (non-prelim): 651
length of query: 184
length of database: 11,318,596
effective HSP length: 93
effective length of query: 91
effective length of database: 8,833,729
effective search space: 803869339
effective search space used: 803869339
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0227.2


