

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.12
(414 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
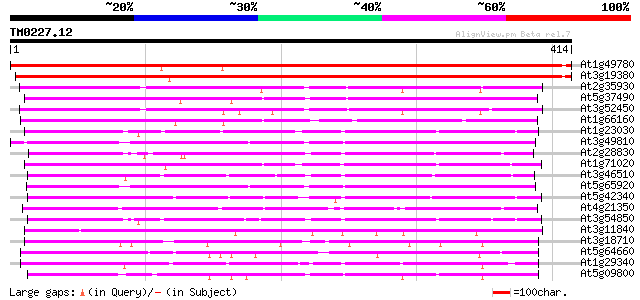
Score E
Sequences producing significant alignments: (bits) Value
At1g49780 unknown protein 527 e-150
At3g19380 unknown protein 521 e-148
At2g35930 unknown protein 204 6e-53
At5g37490 putative protein 199 3e-51
At3g52450 unknown protein 195 4e-50
At1g66160 unknown protein 163 1e-40
At1g23030 unknown protein 154 7e-38
At3g49810 unknown protein 149 2e-36
At2g28830 unknown protein 149 3e-36
At1g71020 unknown protein 149 4e-36
At3g46510 arm repeat containing protein homolog 148 6e-36
At5g65920 unknown protein (At5g65920) 147 1e-35
At5g42340 arm repeat containing protein 141 8e-34
At4g21350 putative protein 139 2e-33
At3g54850 putative protein 139 3e-33
At3g11840 unknown protein 135 4e-32
At3g18710 unknown protein 129 3e-30
At5g64660 unknown protein 128 5e-30
At1g29340 arm repeat-containing protein, putative 124 1e-28
At5g09800 putative protein 119 3e-27
>At1g49780 unknown protein
Length = 421
Score = 527 bits (1357), Expect = e-150
Identities = 270/423 (63%), Positives = 335/423 (78%), Gaps = 11/423 (2%)
Query: 1 MRGCLEPLDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRA 60
M G LEPLDLGIQIPYH+RCPISL+LM DPVT+ TGQTYDR+SI+SW+ GNTTCPVTR
Sbjct: 1 MPGNLEPLDLGIQIPYHFRCPISLDLMSDPVTISTGQTYDRTSIDSWIAMGNTTCPVTRV 60
Query: 61 PLTEFTLIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQ---ISSASAS 117
L++FTLIPNHTLRRLIQEWCVANR+ GVERIPTPKQPA+P VRSLL+Q I+ S
Sbjct: 61 ALSDFTLIPNHTLRRLIQEWCVANRSNGVERIPTPKQPADPISVRSLLSQASAITGTHVS 120
Query: 118 THHRLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIF-----DNRSSDELSHESLALLV 172
R ++RRLR LARDS+ NR LIA H+ R I++RI S EL ESLALLV
Sbjct: 121 VRSRAAAIRRLRGLARDSEKNRVLIAGHNAREILVRILFADIETTSLSSELVSESLALLV 180
Query: 173 MFPLAESDCAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSV 232
+ + E++C A+A+D ++G+++ LL S+++RVN+AALIEMV+ G S DL+ +S
Sbjct: 181 LLHMTETECEAVASDPSRVGFMTRLLFDSSIEIRVNAAALIEMVLTGAKSMDLKLIISGS 240
Query: 233 EGIHDGVVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRL-ADFEK 291
+ I +GV+++L++PIS RALK+G+KA+FALCLVKQ+R A++AGAP +LIDRL ADF++
Sbjct: 241 DSIFEGVLDLLKNPISSRRALKIGIKAIFALCLVKQTRHLAISAGAPGILIDRLAADFDR 300
Query: 292 CDAERALATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESE 351
CD ER LATVELLCR+P GCA F HALTVP++VK IL++SDRATEYAAGALL+LC+ E
Sbjct: 301 CDTERGLATVELLCRLPEGCAAFGEHALTVPLMVKTILRVSDRATEYAAGALLALCTAEE 360
Query: 352 RLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQDSIGNSDDFACSQLVV 411
R + EA AAG++TQLLLLVQSDCTERAKRKAQ+LLKLLRDSWP DS +SDDF S+ V
Sbjct: 361 RCRDEAAAAGLVTQLLLLVQSDCTERAKRKAQMLLKLLRDSWPDDSTVHSDDFNRSE--V 418
Query: 412 VPF 414
PF
Sbjct: 419 APF 421
>At3g19380 unknown protein
Length = 421
Score = 521 bits (1343), Expect = e-148
Identities = 273/418 (65%), Positives = 333/418 (79%), Gaps = 9/418 (2%)
Query: 5 LEPLDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGN-TTCPVTRAPLT 63
+EPLDLGIQIPYH+RCPISLELM+DPVTVCTGQTYDR+SIESWV+ GN TTCPVTRAPL+
Sbjct: 5 IEPLDLGIQIPYHFRCPISLELMQDPVTVCTGQTYDRASIESWVSIGNNTTCPVTRAPLS 64
Query: 64 EFTLIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASA---STHH 120
+FTLIPNHTLRRLIQEWCVANR+ GVERIPTPKQPA+P VR+LL+Q S+ + S
Sbjct: 65 DFTLIPNHTLRRLIQEWCVANRSNGVERIPTPKQPADPTSVRALLSQASAITGTHVSVRS 124
Query: 121 RLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRI-FDNRSSDELSHESLALLVMFPLAE- 178
R +LRRLR ARDSD NR LIA+H+ I+++I F +S EL ESLALLVM P+ E
Sbjct: 125 RAAALRRLRGFARDSDKNRVLIAAHNATEILIKILFSETTSSELVSESLALLVMLPITEP 184
Query: 179 SDCAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDG 238
+ +I++D ++ +L+ LL S++ RVN+AALIE+V GT SADL+ +S+ E + +G
Sbjct: 185 NQFVSISSDPGRVEFLTRLLFDSSIETRVNAAALIEIVSTGTKSADLKGSISNSESVFEG 244
Query: 239 VVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLA-DFEKCDAERA 297
V+++LR+PIS RALK+G+K LFALC VK +R A+ AGAP +LIDRLA DF++CD ERA
Sbjct: 245 VLDLLRNPISSRRALKIGIKTLFALCSVKSTRHIAITAGAPEILIDRLAADFDRCDTERA 304
Query: 298 LATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEA 357
LATVELLCR P GCA F HALTVP+LVK IL++SDRATEYAAGALL+LC+ ER + EA
Sbjct: 305 LATVELLCRTPEGCAAFGEHALTVPLLVKTILRVSDRATEYAAGALLALCTAEERWREEA 364
Query: 358 VAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSWPQ-DSIGNSDDFACSQLVVVPF 414
AGV+ QLLL+VQS+CTERAK+KAQ LLKLLRDSWP +S NSDDF CS VVPF
Sbjct: 365 AGAGVVVQLLLMVQSECTERAKKKAQKLLKLLRDSWPDYNSFANSDDFGCSS-QVVPF 421
>At2g35930 unknown protein
Length = 411
Score = 204 bits (520), Expect = 6e-53
Identities = 138/399 (34%), Positives = 220/399 (54%), Gaps = 22/399 (5%)
Query: 8 LDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGN-TTCPVTRAPLTEFT 66
+D I+IP + CPISLE+M+DPV V TG TYDR SIE W+ G +CPVT+ +T+
Sbjct: 6 MDEEIEIPPFFLCPISLEIMKDPVIVSTGITYDRDSIEKWLFAGKKNSCPVTKQDITDAD 65
Query: 67 LIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTH-HRLNSL 125
L PNHTLRRLIQ WC N ++GVERIPTP+ P + +S + ++ SAS+H +++ L
Sbjct: 66 LTPNHTLRRLIQSWCTLNASYGVERIPTPR----PPICKSEIEKLIRDSASSHENQVKCL 121
Query: 126 RRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSD-ELSHESLALLVMFPLAESDCAAI 184
+RLRQ+ ++ N+ + + V + I N S + L+ E+L LL +E+ +
Sbjct: 122 KRLRQIVSENATNKRCLEAAGVPEFLANIVSNDSENGSLTDEALNLLYHLETSETVLKNL 181
Query: 185 ---AADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVE 241
D + + L+ ++ + RV + L++ ++ AD ++ + VV+
Sbjct: 182 LNNKKDNNIVKSLTKIMQRGMYESRVYATLLLKNIL---EVADPMQSMTLKPEVFTEVVQ 238
Query: 242 ILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLAD----FEKCDAERA 297
IL IS +A K + L +C ++R +AV AG +V+I+ L D E+ E A
Sbjct: 239 ILDDRIS-QKATKAAMHILVNICPWGRNRHKAVEAGVISVIIELLMDESFTSERRGPEMA 297
Query: 298 LATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSL---CSESERLQ 354
+ ++LLC+ G AEF H + ++ K IL++S A++ A LLS+ C+ + L
Sbjct: 298 MVVLDLLCQCAEGRAEFLNHGAAIAVVCKKILRVSQTASDRAVRVLLSVGRFCA-TPALL 356
Query: 355 MEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLRDSW 393
E + GV+ +L L++Q C + K KA+ LLKL W
Sbjct: 357 HEMLQLGVVAKLCLVLQVSCGGKTKEKAKELLKLHARVW 395
>At5g37490 putative protein
Length = 435
Score = 199 bits (505), Expect = 3e-51
Identities = 127/388 (32%), Positives = 215/388 (54%), Gaps = 15/388 (3%)
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
I IP ++CPIS++LM+DPV + TG TYDR SIE+W+N+GN TCPVT LT F IPNH
Sbjct: 29 ITIPPEFQCPISIDLMKDPVIISTGITYDRVSIETWINSGNKTCPVTNTVLTTFDQIPNH 88
Query: 72 TLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNS--LRRLR 129
T+R++IQ WCV + ++RIPTP+ P P V + ++SSA+ + + +++
Sbjct: 89 TIRKMIQGWCVEKGSPLIQRIPTPRVPLMPCEVYEISRKLSSATRRGDYEKCGVIIEKIK 148
Query: 130 QLARDSDYNRSLIASHDVRRIVLRIFDNRSSDE----LSHESLALLV-MFPLAESDCAAI 184
+L +S+ NR + + V ++ FD S DE + +E L+LL MFP+ + +
Sbjct: 149 KLGDESEKNRKCVNENSVGWVLCDCFDKFSGDEKLTFMLNEILSLLTWMFPIGLEGISKL 208
Query: 185 AADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVE-GIHDGVVEIL 243
A+ ++ LL VR N+A +++ +++ + R +VE G+ + +V+++
Sbjct: 209 AS-ATSFRCVAGLLKSTDDSVRQNAAFIMKEILS---LDETRVHSFAVENGVAEALVKLI 264
Query: 244 RSPISYPRALKVGVKALFALCLVK-QSRQRAVAAGAPAVLIDRLADFEKCDAERALATVE 302
R +S + K + A++ + L K + + G ++ ++ + D E E+ALA ++
Sbjct: 265 RDSVS-SSSTKSSLIAIYQMVLQKPEIASEFLEIGLVSITVEMIVDAENSVCEKALAVLD 323
Query: 303 LLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQME-AVAAG 361
+C G E +AL +P+LVK I K+S+ AT + +L L + +E AV G
Sbjct: 324 AICETEHGREEVRKNALVMPLLVKKIAKVSELATRSSMSMILKLWKTGNTVAVEDAVRLG 383
Query: 362 VLTQLLLLVQSDCTERAKRKAQLLLKLL 389
++LL++Q E K KA LLK++
Sbjct: 384 AFQKVLLVLQVGYGEETKEKATELLKMM 411
>At3g52450 unknown protein
Length = 435
Score = 195 bits (495), Expect = 4e-50
Identities = 137/428 (32%), Positives = 224/428 (52%), Gaps = 51/428 (11%)
Query: 8 LDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGN-TTCPVTRAPLTEFT 66
+D I+IP + CPISL++M+DPV V TG TYDR SIE W+ +G +CPVT+ +TE
Sbjct: 1 MDQEIEIPSFFLCPISLDIMKDPVIVSTGITYDRESIEKWLFSGKKNSCPVTKQVITETD 60
Query: 67 LIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTH-HRLNSL 125
L PNHTLRRLIQ WC N ++G+ERIPTPK P + +S + ++ S+S+H +++ L
Sbjct: 61 LTPNHTLRRLIQSWCTLNASYGIERIPTPK----PPICKSEIEKLIKESSSSHLNQVKCL 116
Query: 126 RRLRQLARDSDYNRSLIASHDVRRIVLRIFD------NRSSDELSHESL----------- 168
+RLRQ+ ++ N+ + + +V + I N S LS +L
Sbjct: 117 KRLRQIVSENTTNKRCLEAAEVPEFLANIVSNSVDTYNSPSSSLSSSNLNDMCQSNMLEN 176
Query: 169 -----------ALLVMFPLAESDCAAIAADLDKIG-----YLSVLLSHQSLDVRVNSAAL 212
AL V++ L S+ A + +K G L+ ++ + R +A L
Sbjct: 177 RFDSSRSLMDEALSVLYHLDTSETALKSLLNNKKGTNLVKTLTKIMQRGIYESRAYAALL 236
Query: 213 IEMVVAGTHSADLRSQVSSVEGIHDGVVEILRSPISYPRALKVGVKALFALCLVKQSRQR 272
++ ++ AD + + V++IL IS+ +A + ++ L C ++R +
Sbjct: 237 LKKLL---EVADPMQIILLERELFGEVIQILHDQISH-KATRSAMQILVITCPWGRNRHK 292
Query: 273 AVAAGAPAVLIDRLAD----FEKCDAERALATVELLCRVPAGCAEFAGHALTVPMLVKII 328
AV G +++I+ L D E+ ++E A+ +++LC+ G AEF H + ++ K I
Sbjct: 293 AVEGGTISMIIELLMDDTFSSERRNSEMAMVVLDMLCQCAEGRAEFLNHGAAIAVVSKKI 352
Query: 329 LKISDRATEYAAGALLSL---CSESERLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLL 385
L++S +E A LLS+ C+ LQ E + GV+ +L L++Q C + K KA+ L
Sbjct: 353 LRVSQITSERAVRVLLSVGRFCATPSLLQ-EMLQLGVVAKLCLVLQVSCGNKTKEKAKEL 411
Query: 386 LKLLRDSW 393
LKL W
Sbjct: 412 LKLHARVW 419
>At1g66160 unknown protein
Length = 431
Score = 163 bits (413), Expect = 1e-40
Identities = 104/389 (26%), Positives = 194/389 (49%), Gaps = 20/389 (5%)
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
++ I IP ++CPIS ELM+DPV + +G TYDR +IE W +G TCPVT LT I
Sbjct: 28 EVEITIPSQFQCPISYELMKDPVIIASGITYDRENIEKWFESGYQTCPVTNTVLTSLEQI 87
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHR--LNSLR 126
PNHT+RR+IQ WC ++ G+ERIPTP+ P V + ++S+A+ + + +
Sbjct: 88 PNHTIRRMIQGWCGSSLGGGIERIPTPRVPVTSHQVSEICERLSAATRRGDYAACMEMVT 147
Query: 127 RLRQLARDSDYNRSLIASHDVRRIVLRIFD----NRSSDELSHESLALLV-MFPLAESDC 181
++ +L ++S+ NR + + ++ FD N ++ L E++++L M P+
Sbjct: 148 KMTRLGKESERNRKCVKENGAGLVLCVCFDAFSENANASLLLEETVSVLTWMLPIGLEGQ 207
Query: 182 AAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVE 241
+ + L LL + + L+E+ V H+ ++ + G+ + ++
Sbjct: 208 SKLTT-TSSFNRLVELLRNGDQNAAFLIKELLELNVTHVHA------LTKINGVQEAFMK 260
Query: 242 ILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATV 301
+ + +L + + +++ R + + ++ L D E E+AL +
Sbjct: 261 SINRDSTCVNSL---ISIHHMILTNQETVSRFLELDLVNITVEMLVDSENSVCEKALTVL 317
Query: 302 ELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQM-EAVAA 360
++C G + + L +P+LVK ILKIS++ + + +C + ++ EA+
Sbjct: 318 NVICETKEGREKVRRNKLVIPILVKKILKISEK--KDLVSVMWKVCKSGDGSEVEEALRL 375
Query: 361 GVLTQLLLLVQSDCTERAKRKAQLLLKLL 389
G +L++++Q C E K K LLK++
Sbjct: 376 GAFKKLVVMLQVGCGEGTKEKVTELLKMM 404
>At1g23030 unknown protein
Length = 612
Score = 154 bits (390), Expect = 7e-38
Identities = 119/385 (30%), Positives = 187/385 (47%), Gaps = 21/385 (5%)
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
+ IP + CP+SLELM+DPV V TGQTY+R+ I+ W++ GN TCP T+ L FTL PN+
Sbjct: 239 LTIPVDFLCPVSLELMKDPVIVATGQTYERAYIQRWIDCGNLTCPKTQQKLENFTLTPNY 298
Query: 72 TLRRLIQEWCVANRAFGVERIP-----TPKQPAEPALVRSLLNQISSASASTHHRLNSLR 126
LR LI WC + +E+ K + +++R+L+ ++S S ST R N++
Sbjct: 299 VLRSLISRWCAEH---NIEQPAGYINGRTKNSGDMSVIRALVQRLS--SRSTEDRRNAVS 353
Query: 127 RLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIAA 186
+R L++ S NR LIA +++ + S D + E+ V+ + +
Sbjct: 354 EIRSLSKRSTDNRILIAEAGAIPVLVNLL--TSEDVATQENAITCVLNLSIYENNKELIM 411
Query: 187 DLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEILRSP 246
+ + +L +++ R N+AA + AD + G +V++L +
Sbjct: 412 FAGAVTSIVQVLRAGTMEARENAAA----TLFSLSLADENKIIIGGSGAIPALVDLLEN- 466
Query: 247 ISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCD-AERALATVELLC 305
PR K ALF LC+ ++ RAV AG L+ L+D + + AL + +L
Sbjct: 467 -GTPRGKKDAATALFNLCIYHGNKGRAVRAGIVTALVKMLSDSTRHRMVDEALTILSVLA 525
Query: 306 RVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGVLTQ 365
+ A T+P L+ I+ R E AA LLSLC + G +
Sbjct: 526 NNQDAKSAIV-KANTLPALIGILQTDQTRNRENAAAILLSLCKRDTEKLITIGRLGAVVP 584
Query: 366 LLLLVQSDCTERAKRKAQLLLKLLR 390
L+ L ++ TER KRKA LL+LLR
Sbjct: 585 LMDLSKNG-TERGKRKAISLLELLR 608
>At3g49810 unknown protein
Length = 448
Score = 149 bits (377), Expect = 2e-36
Identities = 110/389 (28%), Positives = 188/389 (48%), Gaps = 14/389 (3%)
Query: 1 MRGCLEPLDLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRA 60
++ ++ LDL IP + CPISLE M+DPVT+CTGQTY+R +I W N G+ TCP T
Sbjct: 52 LKKMIKELDLQ-DIPSVFICPISLEPMQDPVTLCTGQTYERLNIHKWFNLGHLTCPTTMQ 110
Query: 61 PLTEFTLIPNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHH 120
L + T+ PN TL LI W ++ K+ +E R++ + A
Sbjct: 111 ELWDDTVTPNKTLHHLIYTWF-------SQKYVLMKKRSEDVQGRAIEILGTLKKAKGQA 163
Query: 121 RLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESD 180
R+++L L+Q+ R + ++ + +S + E +A+LV L +SD
Sbjct: 164 RVHALSELKQIVIAHLMARKTVVEEGGVSVISSLLGPFTSHAVGSEVVAILVSLDL-DSD 222
Query: 181 CAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVV 240
+ K+ + +L+ S + ++N A LI +V R+++ S + G++
Sbjct: 223 SKSGLMQPAKVSLIVDMLNDGSNETKINCARLIRGLV---EEKGFRAELVSSHSLLVGLM 279
Query: 241 EILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALAT 300
+++ + + ++ L + + KQ R V+ GA L+D L + E AL
Sbjct: 280 RLVKDK-RHRNGVSPALRLLKPISVHKQVRSLMVSIGAVPQLVDILPSLDPECLELALFV 338
Query: 301 VELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCS-ESERLQMEAVA 359
++ LC G A T+P V++++++S+ T YA L S+C E AV
Sbjct: 339 LDALCTDVEGRVAVKDSANTIPYTVRVLMRVSENCTNYALSILWSVCKLAPEECSPLAVE 398
Query: 360 AGVLTQLLLLVQSDCTERAKRKAQLLLKL 388
G+ +LLL++QS C K+++ LLKL
Sbjct: 399 VGLAAKLLLVIQSGCDAALKQRSAELLKL 427
>At2g28830 unknown protein
Length = 654
Score = 149 bits (376), Expect = 3e-36
Identities = 117/390 (30%), Positives = 191/390 (48%), Gaps = 35/390 (8%)
Query: 15 PYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTLR 74
P +RCPISLELM DPV V +GQTY+R I+ W+ G+ TCP T+ LT + PN+ LR
Sbjct: 257 PEEFRCPISLELMTDPVIVSSGQTYERECIKKWLEGGHLTCPKTQETLTSDIMTPNYVLR 316
Query: 75 RLIQEWCVANRAFGVERIPTPKQP--AEPALVRSLLNQISSASASTHHRLNSL------- 125
LI +WC +N G+E PK+P ++P+ S + SSA H+++ L
Sbjct: 317 SLIAQWCESN---GIE---PPKRPNISQPS---SKASSSSSAPDDEHNKIEELLLKLTSQ 367
Query: 126 ----RR-----LRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPL 176
RR +R LA+ +++NR IA+ +++ + + ++ ++ +
Sbjct: 368 QPEDRRSAAGEIRLLAKQNNHNRVAIAASGAIPLLVNLLTISNDSRTQEHAVTSILNLSI 427
Query: 177 AESDCAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIH 236
+ + I + + +L S++ R N+AA + + D G
Sbjct: 428 CQENKGKIVYSSGAVPGIVHVLQKGSMEARENAAATLFSLSV----IDENKVTIGAAGAI 483
Query: 237 DGVVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAER 296
+V +L R K ALF LC+ + ++ +AV AG VL+ L + E +
Sbjct: 484 PPLVTLLSE--GSQRGKKDAATALFNLCIFQGNKGKAVRAGLVPVLMRLLTEPESGMVDE 541
Query: 297 ALATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQME 356
+L+ + +L P G +E G A VP+LV I S R E +A L+ LCS +++ +E
Sbjct: 542 SLSILAILSSHPDGKSE-VGAADAVPVLVDFIRSGSPRNKENSAAVLVHLCSWNQQHLIE 600
Query: 357 AVAAGVLTQLLLLVQSDCTERAKRKAQLLL 386
A G++ LL+ + + T+R KRKA LL
Sbjct: 601 AQKLGIM-DLLIEMAENGTDRGKRKAAQLL 629
>At1g71020 unknown protein
Length = 628
Score = 149 bits (375), Expect = 4e-36
Identities = 116/390 (29%), Positives = 178/390 (44%), Gaps = 18/390 (4%)
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
+ IP + CPISLELM+DP V TGQTY+RS I+ W++ GN +CP T+ L FTL PN+
Sbjct: 241 LTIPEDFLCPISLELMKDPAIVSTGQTYERSFIQRWIDCGNLSCPKTQQKLENFTLTPNY 300
Query: 72 TLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISS--------ASASTHHRLN 123
LR LI +WC + + R L +S+ +S S R
Sbjct: 301 VLRSLISQWCTKHNIEQPGGYMNGRTKNSDGSFRDLSGDMSAIRALVCKLSSQSIEDRRT 360
Query: 124 SLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAA 183
++ +R L++ S NR LIA +++++ + E ++ ++ + E +
Sbjct: 361 AVSEIRSLSKRSTDNRILIAEAGAIPVLVKLLTSDGDTETQENAVTCILNLSIYEHNKEL 420
Query: 184 IAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEIL 243
I + + ++L S++ R N+AA + AD + G +V++L
Sbjct: 421 IML-AGAVTSIVLVLRAGSMEARENAAA----TLFSLSLADENKIIIGASGAIMALVDLL 475
Query: 244 RSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLAD-FEKCDAERALATVE 302
+ R K ALF LC+ + ++ RAV AG L+ L D + A+ AL +
Sbjct: 476 Q--YGSVRGKKDAATALFNLCIYQGNKGRAVRAGIVKPLVKMLTDSSSERMADEALTILS 533
Query: 303 LLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGV 362
+L A +P L+ + K R E AA LL LC + G
Sbjct: 534 VLASNQVAKTAIL-RANAIPPLIDCLQKDQPRNRENAAAILLCLCKRDTEKLISIGRLGA 592
Query: 363 LTQLLLLVQSDCTERAKRKAQLLLKLLRDS 392
+ L+ L D TERAKRKA LL+LLR S
Sbjct: 593 VVPLMEL-SRDGTERAKRKANSLLELLRKS 621
>At3g46510 arm repeat containing protein homolog
Length = 660
Score = 148 bits (373), Expect = 6e-36
Identities = 119/382 (31%), Positives = 185/382 (48%), Gaps = 20/382 (5%)
Query: 14 IPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTL 73
IP +RCPISLE+MRDPV V +GQTY+R+ IE W+ G++TCP T+ LT TL PN+ L
Sbjct: 256 IPDDFRCPISLEMMRDPVIVSSGQTYERTCIEKWIEGGHSTCPKTQQALTSTTLTPNYVL 315
Query: 74 RRLIQEWCVAN--------RAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSL 125
R LI +WC AN + ++ + PAE + L+ ++ A + + ++
Sbjct: 316 RSLIAQWCEANDIEPPKPPSSLRPRKVSSFSSPAEANKIEDLMWRL--AYGNPEDQRSAA 373
Query: 126 RRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIA 185
+R LA+ + NR IA +++ + S + S+ L+ + E++ AI
Sbjct: 374 GEIRLLAKRNADNRVAIAEAGAIPLLVGLLSTPDS-RIQEHSVTALLNLSICENNKGAIV 432
Query: 186 ADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEILRS 245
+ G + V L S++ R N+AA + + D G +V +L
Sbjct: 433 SAGAIPGIVQV-LKKGSMEARENAAATLFSLSV----IDENKVTIGALGAIPPLVVLLNE 487
Query: 246 PISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVELLC 305
R K ALF LC+ + ++ +A+ AG L L + + ALA + +L
Sbjct: 488 --GTQRGKKDAATALFNLCIYQGNKGKAIRAGVIPTLTRLLTEPGSGMVDEALAILAILS 545
Query: 306 RVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGVLTQ 365
P G A G + VP LV+ I S R E AA L+ LCS + +EA G++
Sbjct: 546 SHPEGKA-IIGSSDAVPSLVEFIRTGSPRNRENAAAVLVHLCSGDPQHLVEAQKLGLMGP 604
Query: 366 LLLLVQSDCTERAKRKAQLLLK 387
L+ L + T+R KRKA LL+
Sbjct: 605 LIDLA-GNGTDRGKRKAAQLLE 625
>At5g65920 unknown protein (At5g65920)
Length = 444
Score = 147 bits (370), Expect = 1e-35
Identities = 106/377 (28%), Positives = 184/377 (48%), Gaps = 13/377 (3%)
Query: 13 QIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHT 72
+IP + CPISLE M+DPVT+CTGQTY+RS+I W N G+ TCP T L + + PN T
Sbjct: 59 EIPSVFICPISLEPMQDPVTLCTGQTYERSNILKWFNIGHCTCPTTMQELWDDLVTPNKT 118
Query: 73 LRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQLA 132
L +LI W ++ K+ +E R++ + A ++++L L+Q+
Sbjct: 119 LHQLIYTWF-------SQKYVLMKKRSEDVQGRAIEILGTLRKAKGKAKVHALSELKQVV 171
Query: 133 RDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIAADLDKIG 192
+ + ++ + +S + E++A+LV L +SD A ++
Sbjct: 172 MAHAIAKKTVVDEGGVFVISSLLSPFTSHAVGSEAIAILVNLEL-DSDSKAGLMQPARVS 230
Query: 193 YLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEILRSPISYPRA 252
+ +L+ S++ ++N A LI +V R+++ S + G++ +++
Sbjct: 231 LMVDMLNDGSIETKINCARLIGRLV---EEKGFRAELVSSHSLLVGLMRLVKDR-RRRNG 286
Query: 253 LKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVELLCRVPAGCA 312
+ + L ++ + KQ R V GA L+D L + E AL ++ LC G
Sbjct: 287 VSPALTLLKSVSVHKQVRNLLVRIGAVPQLVDVLPCLDVECLESALFVLDSLCLESEGRI 346
Query: 313 EFAGHALTVPMLVKIILKISDRATEYAAGALLSLCS-ESERLQMEAVAAGVLTQLLLLVQ 371
T+P V++++K+S++ T YA L S+C SE AV G+ +LLL++Q
Sbjct: 347 ALKDSVNTIPHTVRLLMKVSEKCTNYAISILWSVCKLASEECSSLAVEVGLAAKLLLVIQ 406
Query: 372 SDCTERAKRKAQLLLKL 388
S C K+++ LLKL
Sbjct: 407 SGCDPALKQRSAELLKL 423
>At5g42340 arm repeat containing protein
Length = 656
Score = 141 bits (355), Expect = 8e-34
Identities = 113/382 (29%), Positives = 189/382 (48%), Gaps = 17/382 (4%)
Query: 14 IPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTL 73
+P+ + CPI+LE+M DPV + TGQTY++ SI+ W + G+ TCP TR L +L PN L
Sbjct: 286 LPHEFLCPITLEIMLDPVIIATGQTYEKESIQKWFDAGHKTCPKTRQELDHLSLAPNFAL 345
Query: 74 RRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQLAR 133
+ LI +WC N E+ +P E SLL + S+S R S++++R LAR
Sbjct: 346 KNLIMQWCEKNNFKIPEKEVSPDSQNEQKDEVSLLVEALSSSQLEEQR-RSVKQMRLLAR 404
Query: 134 DSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIAADLDKIGY 193
++ NR LIA+ +++++ S + ++ L+ + E + I+ + I
Sbjct: 405 ENPENRVLIANAGAIPLLVQLLSYPDSG-IQENAVTTLLNLSIDEVNKKLISNE-GAIPN 462
Query: 194 LSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGV---VEILRSPISYP 250
+ +L + + + R NSAA + S + + G+ +G+ V++L+
Sbjct: 463 IIEILENGNREARENSAAAL-------FSLSMLDENKVTIGLSNGIPPLVDLLQH--GTL 513
Query: 251 RALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVELLCRVPAG 310
R K + ALF L L ++ RA+ AG L++ L D + AL+ + LL P G
Sbjct: 514 RGKKDALTALFNLSLNSANKGRAIDAGIVQPLLNLLKDKNLGMIDEALSILLLLASHPEG 573
Query: 311 CAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGVLTQLLLLV 370
+ G + LV+ I + + + E A LL L S + + A+ GV L+ +
Sbjct: 574 -RQAIGQLSFIETLVEFIRQGTPKNKECATSVLLELGSNNSSFILAALQFGVYEYLVEIT 632
Query: 371 QSDCTERAKRKAQLLLKLLRDS 392
S T RA+RKA L++L+ S
Sbjct: 633 TSG-TNRAQRKANALIQLISKS 653
>At4g21350 putative protein
Length = 374
Score = 139 bits (351), Expect = 2e-33
Identities = 114/383 (29%), Positives = 195/383 (50%), Gaps = 19/383 (4%)
Query: 10 LGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFT-LI 68
+ +P +RCPISLE+M DPV + +G T+DR SI+ W+++GN TCP+T+ PL+E LI
Sbjct: 1 MAFDLPNDFRCPISLEIMSDPVILQSGHTFDRVSIQQWIDSGNRTCPITKLPLSETPYLI 60
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRL 128
PNH LR LI + A+ + P +Q + ++L++ + S S+S +L SL RL
Sbjct: 61 PNHALRSLILNF--AHVSLKESSRPRTQQEHSHSQSQALISTLVSQSSSNASKLESLTRL 118
Query: 129 RQLA-RDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAIAAD 187
+L RDS R + S VR + + + + L +SL+LL+ L + + + AD
Sbjct: 119 VRLTKRDSSIRRKVTESGAVRAALDCV--DSCNQVLQEKSLSLLLNLSLEDDNKVGLVAD 176
Query: 188 LDKIGYLSVLLSHQSLDVRVNSAALI-EMVVAGTHSADLRSQVSSVEGIHDGVVEILRSP 246
I + +L S D + +A L+ + V + A + S ++ + V +LR
Sbjct: 177 -GVIRRIVTVLRVGSPDCKAIAATLLTSLAVVEVNKATIGSYPDAISAL----VSLLR-- 229
Query: 247 ISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALATVELLCR 306
+ R K AL+ALC +R+R V G+ +L++ AD ERA+ + LL +
Sbjct: 230 VGNDRERKESATALYALCSFPDNRKRVVDCGSVPILVE-AAD---SGLERAVEVLGLLVK 285
Query: 307 VPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLCSESERLQMEAVAAGVLTQL 366
G E + + V +LV ++ + + +Y+ L LC S + E GV+ ++
Sbjct: 286 CRGGREEMSKVSGFVEVLVNVLRNGNLKGIQYSLFILNCLCCCSGEIVDEVKREGVV-EI 344
Query: 367 LLLVQSDCTERAKRKAQLLLKLL 389
+ + +E+ +R A +L+ L
Sbjct: 345 CFGFEDNESEKIRRNATILVHTL 367
>At3g54850 putative protein
Length = 632
Score = 139 bits (350), Expect = 3e-33
Identities = 124/393 (31%), Positives = 188/393 (47%), Gaps = 31/393 (7%)
Query: 14 IPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTL 73
IP ++RCPISLELM+DPV V TGQTY+RSSI+ W++ G+ TCP ++ L L PN+ L
Sbjct: 248 IPEYFRCPISLELMKDPVIVSTGQTYERSSIQKWLDAGHKTCPKSQETLLHAGLTPNYVL 307
Query: 74 RRLIQEWCVANRAFGVERIP-------------TPKQPAEPALVRSLLNQISSASASTHH 120
+ LI WC +N G+E +P + + V SLL ++ A+ +T
Sbjct: 308 KSLIALWCESN---GIE-LPQNQGSCRTTKIGGSSSSDCDRTFVLSLLEKL--ANGTTEQ 361
Query: 121 RLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESD 180
+ + LR LA+ + NR IA +++ + + H ALL + + E +
Sbjct: 362 QRAAAGELRLLAKRNVDNRVCIAEAGAIPLLVELLSSPDPRTQEHSVTALLNL-SINEGN 420
Query: 181 CAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVV 240
AI D I + +L + S++ R N+AA + + D G ++
Sbjct: 421 KGAI-VDAGAITDIVEVLKNGSMEARENAAATLFSLSV----IDENKVAIGAAGAIQALI 475
Query: 241 EILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDAERALAT 300
+L R K A+F LC+ + ++ RAV G L L D + ALA
Sbjct: 476 SLLEE--GTRRGKKDAATAIFNLCIYQGNKSRAVKGGIVDPLTRLLKDAGGGMVDEALAI 533
Query: 301 VELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLC-SESERLQMEAVA 359
+ +L G A A ++P+LV+II S R E AA L LC ERL + A
Sbjct: 534 LAILSTNQEGKTAIA-EAESIPVLVEIIRTGSPRNRENAAAILWYLCIGNIERLNV-ARE 591
Query: 360 AGVLTQLLLLVQSDCTERAKRKAQLLLKLLRDS 392
G L L ++ T+RAKRKA LL+L++ +
Sbjct: 592 VGADVALKELTENG-TDRAKRKAASLLELIQQT 623
>At3g11840 unknown protein
Length = 470
Score = 135 bits (340), Expect = 4e-32
Identities = 122/426 (28%), Positives = 185/426 (42%), Gaps = 45/426 (10%)
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLT-EFTLIPN 70
I+IP ++ CPISLE+M+DPVT +G TYDR +I W+ +CPVT+ PL + L PN
Sbjct: 22 IEIPNYFICPISLEIMKDPVTTVSGITYDRQNIVKWLEKV-PSCPVTKQPLPLDSDLTPN 80
Query: 71 HTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQ 130
H LRRLIQ WCV N GV RI TP+ P V + + R +L++L
Sbjct: 81 HMLRRLIQHWCVENETRGVVRISTPRVPPGKLNVVEEIKNLKKFGQEALGREETLQKLEV 140
Query: 131 LARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSH-------ESLALLVMFPLAESDCAA 183
LA D + R + + ++L + S DE ESL LL + + +D
Sbjct: 141 LAMDGNNRRLMCECGVHKSLILFVVKCTSEDEDGRRRIKGLDESLRLLHLIGIPSNDAKT 200
Query: 184 IAADLDKI-GYLSVLLSHQSLDVRVNSAALIEMVVAGTHS---------------ADLRS 227
I + D++ L+ +L + + + L+ + T S L+
Sbjct: 201 ILMENDRVMESLTWVLHQEDFLSKAYTIVLLRNLTEYTSSHIVERLNPEIFKGIIGFLKD 260
Query: 228 QVSSVEGIHDGVVEILR------------SPISYPRALKVGVKALFALCLVKQS----RQ 271
V+SV V E ++ S + + +K V A + L S R
Sbjct: 261 VVNSVNRTSPTVRETVQSSSRPSLGKTEPSKLDHSLVIKQAVTAALMILLETSSWSRNRS 320
Query: 272 RAVAAGAPAVLIDRLADF--EKCDAERALATVELLCRVPAGCAEFAGHALTVPMLVKIIL 329
V GA + LI+ + EK E L + LC G AE H + ++ K +L
Sbjct: 321 LLVDLGAVSELIELEISYTGEKRITELMLGVLSRLCCCANGRAEILAHRGGIAVVTKRLL 380
Query: 330 KISDRATEYAAGAL--LSLCSESERLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLK 387
++S A + A L +S S + E V G + +L ++ DC K KA+ +LK
Sbjct: 381 RVSPAADDRAISILTTVSKFSPENMVVEEMVNVGTVEKLCSVLGMDCGLNLKEKAKEILK 440
Query: 388 LLRDSW 393
D W
Sbjct: 441 DHFDEW 446
>At3g18710 unknown protein
Length = 415
Score = 129 bits (324), Expect = 3e-30
Identities = 111/402 (27%), Positives = 195/402 (47%), Gaps = 38/402 (9%)
Query: 12 IQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNH 71
I +P ++CPISL++MR PV++CTG TYDR+SI+ W++ GN TCP T L +PN
Sbjct: 10 ITVPSFFKCPISLDVMRSPVSLCTGVTYDRASIQRWLDGGNNTCPATMQLLKTKDFVPNL 69
Query: 72 TLRRLIQEW--CVANRAFG---VERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLR 126
TL+RLI W + R G V P+ ++ V LL ++ S L +L
Sbjct: 70 TLQRLINIWSDSIGRRHNGDSPVLNPPSGREVPTKEEVNVLLERLMS--------LENLM 121
Query: 127 RLRQLARDSDYNRSLIAS--HDVRRIVLRIFDNRSSDELSHESLALLVMFPLAESDCAAI 184
++ + +DSD NR ++ V +V I ++ EL ++ +L + + +
Sbjct: 122 KIVRFVKDSDSNREFLSKKMEFVPMLVDIIRTKKTKIELVIMAIRILDSIKVDRERLSNL 181
Query: 185 AADLDKIGYLSVLL---SHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVE 241
D L+ +L +L+ ++ S +++ + S D +S++ E DGV+
Sbjct: 182 MLANDGGDCLTAILLAIQRGNLESKIESVRVLDWI-----SFDAKSKLMIAE--RDGVLT 234
Query: 242 ILRSPISY-----PRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCD--- 293
+ IS P ++ + L + K+ R + +AA A + D L +
Sbjct: 235 EMMKSISITESSDPSLIEASLSFLITISKSKRVRSKLIAAKAITKIKDILLTETLTNVAV 294
Query: 294 AERALATVELLCRVPAGCAEFAG--HALTVPMLVKIILKISDRATEYAAGALLSLC---S 348
E++L +E L G E G + V +VK +LK+S ATE+A L LC
Sbjct: 295 TEKSLKLLETLSSKREGRLEICGDDNGRCVEGVVKKLLKVSTTATEHAVTILWCLCYVFR 354
Query: 349 ESERLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLR 390
E + ++ + +T+LL+++QS+C+ ++ A+ L+K+L+
Sbjct: 355 EDKTVEETVERSNGVTKLLVVIQSNCSAMVRQMAKDLIKVLK 396
>At5g64660 unknown protein
Length = 420
Score = 128 bits (322), Expect = 5e-30
Identities = 109/406 (26%), Positives = 187/406 (45%), Gaps = 34/406 (8%)
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
DL I +P +RCPISL++M+ PV++CTG TYDR+SI+ W++ GN TCP T L I
Sbjct: 5 DLCITVPTFFRCPISLDVMKSPVSLCTGVTYDRASIQRWLDGGNNTCPATMQILQNKDFI 64
Query: 69 PNHTLRRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRL 128
PN TL+RLI+ W + R P + ++++ R L ++
Sbjct: 65 PNRTLQRLIEIWSDSVRRRTCVESAELAAPTRDE-IADAIDRVKIEKEERDDR-EVLSKI 122
Query: 129 RQLARDSDYNRSLIASHD--VRRIVLRI----FDNRSSDE--LSHESLALLVMFPLAESD 180
+ R+SD NR +A D V+ +V I F+ S+ + + E++ +L SD
Sbjct: 123 VRFGRESDDNRGFLAGKDDFVKLLVDLINQVDFETTSAAKSLVVQEAVKILSTIRSKVSD 182
Query: 181 -----CAAIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGI 235
+ D++ + L +++++++ A L+E + S L ++
Sbjct: 183 RRRFSNLILTNGRDRLSVIVYLFKTGNVELKIDCAGLLEFIAVDAESKLLIAE------- 235
Query: 236 HDGVVEILRSPISYPRALKVGVKALFALCLVKQSR-------QRAVAAGAPAVLIDRLAD 288
DG++ L IS L + +L L + + + + +L D +
Sbjct: 236 RDGLITELMKSISKDSDLSLIESSLSCLIAISSPKRVKLNLLREKLIGDVTKLLSDSTSS 295
Query: 289 FEKCDAERALATVELLCRVPAGCAEF-AGHALTVPMLVKIILKISDRATEYAAGALLS-- 345
E+ L +E+L G +E G + +VK ++K+S ATE+A L S
Sbjct: 296 LSVSVTEKCLKLLEILASTKEGRSEICGGDGECLKTVVKKLMKVSTAATEHAVTVLWSVS 355
Query: 346 -LCSESERLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLR 390
L E + L+ GV T++LLL+QS+C+ +R LLK+ +
Sbjct: 356 YLFKEDKALEAVTSVNGV-TKILLLLQSNCSPAVRRMLTDLLKVFK 400
>At1g29340 arm repeat-containing protein, putative
Length = 729
Score = 124 bits (311), Expect = 1e-28
Identities = 108/401 (26%), Positives = 186/401 (45%), Gaps = 33/401 (8%)
Query: 9 DLGIQIPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLI 68
D I +P + CPISL+LM DPV + TGQTYDR+SI W+ G+ TCP T L + ++
Sbjct: 300 DTFITVPKDFVCPISLDLMTDPVIISTGQTYDRNSIARWIEEGHCTCPKTGQMLMDSRIV 359
Query: 69 PNHTLRRLIQEWCVA-------------NRAFGVERIPTPKQPAEPALVRSLLNQISSAS 115
PN L+ LI +WC A N +F A A V L+ ++ S
Sbjct: 360 PNRALKNLIVQWCTASGISYESEFTDSPNESFASALPTKAAVEANKATVSILIKYLADGS 419
Query: 116 ASTHHRLNSLRRLRQLARDSDYNRSLIASHDVRRIVLRIFDNRSSDELSHESLALLVMFP 175
+ + + R +R LA+ NR+ IA + R+ + ++ S+ ++
Sbjct: 420 QAA--QTVAAREIRLLAKTGKENRAYIAEAGAIPHLCRLLTSENA-IAQENSVTAMLNLS 476
Query: 176 LAESDCAAIAADLDKI-GYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEG 234
+ E + + I + D + +SVL+S +++ + N+AA + ++ H + + +++ V+
Sbjct: 477 IYEKNKSRIMEEGDCLESIVSVLVSGLTVEAQENAAATL-FSLSAVH--EYKKRIAIVDQ 533
Query: 235 IHDGVVEILRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLADFEKCDA 294
+ + +L++ PR K V AL+ L + R + G + L+ L + + A
Sbjct: 534 CVEALALLLQN--GTPRGKKDAVTALYNLSTHPDNCSRMIEGGGVSSLVGALKN--EGVA 589
Query: 295 ERALATVELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLC-----SE 349
E A + LL R G V L+ ++ + R E A ALL LC +
Sbjct: 590 EEAAGALALLVRQSLGAEAIGKEDSAVAGLMGMMRCGTPRGKENAVAALLELCRSGGAAV 649
Query: 350 SERLQMEAVAAGVLTQLLLLVQSDCTERAKRKAQLLLKLLR 390
+E++ AG+L LL T+RA+RKA L ++ +
Sbjct: 650 AEKVLRAPAIAGLLQTLLF----TGTKRARRKAASLARVFQ 686
>At5g09800 putative protein
Length = 409
Score = 119 bits (298), Expect = 3e-27
Identities = 98/392 (25%), Positives = 185/392 (47%), Gaps = 28/392 (7%)
Query: 14 IPYHYRCPISLELMRDPVTVCTGQTYDRSSIESWVNTGNTTCPVTRAPLTEFTLIPNHTL 73
+P ++CPISL++M+ PV++ TG TYDR SI+ W++ GN TCP T L +PN TL
Sbjct: 11 VPCFFKCPISLDVMKSPVSLSTGVTYDRVSIQRWLDDGNNTCPATMQILQNKEFVPNLTL 70
Query: 74 RRLIQEWCVANRAFGVERIPTPKQPAEPALVRSLLNQISSASASTHHRLNSLRRLRQLAR 133
RLI W + + R + P R ++I++A ++ ++ + AR
Sbjct: 71 HRLIDHW-----SDSINRRADSESPESDTPTR---DEINAAIERFRIENDARSKILRFAR 122
Query: 134 DSDYNRSLIASHD--VRRIVLRIFDNRSSDE----LSHESLALLVM-----FPLAESDCA 182
+SD NR +A D V +V I D+R+ + L E++ +L M F
Sbjct: 123 ESDENREFLAGKDDFVAMLVDLISDSRNFSDSQLLLVGEAVKILSMIRRKIFDRRRLSNL 182
Query: 183 AIAADLDKIGYLSVLLSHQSLDVRVNSAALIEMVVAGTHSADLRSQVSSVEGIHDGVVEI 242
+ D + +L+ + ++++ +A++E + A+ + ++ EG+ ++++
Sbjct: 183 ILTNGGDCLTSFFLLIKRGNPKLKIDCSAVLEFIAV---DAESKLIIAKGEGLVTEIIKL 239
Query: 243 LRSPISYPRALKVGVKALFALCLVKQSRQRAVAAGAPAVLIDRLAD--FEKCDAERALAT 300
+ S S ++ + L A+ K+ + + L L D E+ L
Sbjct: 240 ISSD-SDSSLIEANLSLLIAIASSKRVKLALIREKLVTKLTSLLTDPTTSVSVTEKCLKL 298
Query: 301 VELLCRVPAGCAEFAGHALTVPMLVKIILKISDRATEYAAGALLSLC--SESERLQMEAV 358
+E + G +E + V +V ++K+S ATE+A L S+C + ++ Q +
Sbjct: 299 LEAISSCKEGRSEIC-DGVCVETVVNKLMKVSTAATEHAVTVLWSVCYLFKEKKAQDAVI 357
Query: 359 AAGVLTQLLLLVQSDCTERAKRKAQLLLKLLR 390
+T++LLL+QS+C+ + LLK+ +
Sbjct: 358 RINGVTKILLLLQSNCSLTVRHMLTDLLKVFK 389
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,075,127
Number of Sequences: 26719
Number of extensions: 297601
Number of successful extensions: 1033
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 75
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 827
Number of HSP's gapped (non-prelim): 138
length of query: 414
length of database: 11,318,596
effective HSP length: 102
effective length of query: 312
effective length of database: 8,593,258
effective search space: 2681096496
effective search space used: 2681096496
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0227.12


