

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.1
(556 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
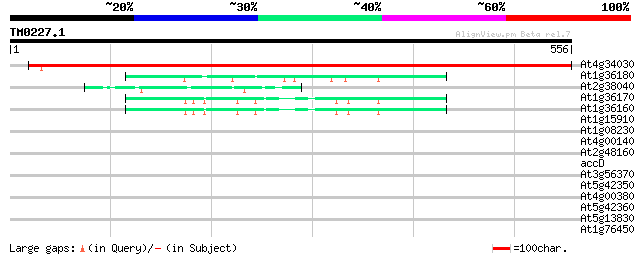
Score E
Sequences producing significant alignments: (bits) Value
At4g34030 3-methylcrotonyl-CoA carboxylase non-biotinylated subu... 910 0.0
At1g36180 acetyl-CoA carboxylase, putative 45 1e-04
At2g38040 carboxyltransferase alpha subunit (CAC3) 45 1e-04
At1g36170 acetyl-CoA carboxylase, putative, 5' partial 44 2e-04
At1g36160 hypothetical protein, 3' partial 44 2e-04
At1g15910 32 1.2
At1g08230 unknown protein 32 1.2
At4g00140 hypothetical protein 30 2.7
At2g48160 unknown protein 30 2.7
accD -chloroplast genome- carboxytransferase beta subunit 30 3.5
At3g56370 putative protein 30 3.5
At5g42350 putative protein 30 4.6
At4g00380 30 4.6
At5g42360 putative protein 29 7.8
At5g13830 FtsJ (dbj|BAA83750.1) 29 7.8
At1g76450 unknown protein 29 7.8
>At4g34030 3-methylcrotonyl-CoA carboxylase non-biotinylated subunit
(MCCB)
Length = 587
Score = 910 bits (2352), Expect = 0.0
Identities = 437/546 (80%), Positives = 495/546 (90%), Gaps = 8/546 (1%)
Query: 19 RRGFCLGILPHS--------NGGATAMEDLLSQLQSHVHKALAGGGAEAVKRNTSRNKLL 70
++GFC+GILP + + AME +LS+L+SH+ K LAGGG EAVKRN SRNKLL
Sbjct: 42 QKGFCVGILPDGVDRNSEAFSSNSIAMEGILSELRSHIKKVLAGGGEEAVKRNRSRNKLL 101
Query: 71 PRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGVVTGIGPVHGRLCMFVANDPTVKGG 130
PRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGG++TGIGP+HGR+CMF+ANDPTVKGG
Sbjct: 102 PRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGIITGIGPIHGRICMFMANDPTVKGG 161
Query: 131 TYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIFYNQALMSA 190
TYYPIT+KKHLRAQEIA++C+LPC+YLVDSGGA+LPKQA+VFPD+ENFGR+FYN+++MS+
Sbjct: 162 TYYPITIKKHLRAQEIAARCRLPCIYLVDSGGAYLPKQAEVFPDKENFGRVFYNESVMSS 221
Query: 191 EGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVKAATGEEVSAEDLGGA 250
+GIPQIA+VLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVKAATGEEVSAEDLGGA
Sbjct: 222 DGIPQIAIVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGPPLVKAATGEEVSAEDLGGA 281
Query: 251 TVHCKTSGVSDYFAQDEFHALALGRNIIKNLHMAGKDVLANGLQNLSYEYKEPLYDVNEL 310
TVHC SGVSDYFAQDE H LA+GRNI+KNLHMA K + + + YKEPLYD+NEL
Sbjct: 282 TVHCTVSGVSDYFAQDELHGLAIGRNIVKNLHMAAKQGMEGTFGSKNLVYKEPLYDINEL 341
Query: 311 RSIAPTDLKKQFDVRSVIARIVDGSEFDEFKKLYGTTLVTGFARIYGQPVGIIGNNGILF 370
RSIAP D K+QFDVRS+IARIVDGSEFDEFKK YGTTLVTGFARIYGQ VGIIGNNGILF
Sbjct: 342 RSIAPVDHKQQFDVRSIIARIVDGSEFDEFKKQYGTTLVTGFARIYGQTVGIIGNNGILF 401
Query: 371 NESALKGAHFIEICTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCAKVPKI 430
NESALKGAHFIE+C+QR IPLVFLQNITGFMVGSR+EANGIAK+GAKMVMAVSCAKVPKI
Sbjct: 402 NESALKGAHFIELCSQRKIPLVFLQNITGFMVGSRAEANGIAKAGAKMVMAVSCAKVPKI 461
Query: 431 TIIVGGSFGAGNYAMCGRAYSPNFMFLWPNARISVMGGAQAAGVLSQIEKTNKKKQGIQW 490
TII G SFGAGNYAMCGRAYSP+FMF+WPNARI +MGGAQAAGVL+QIE+ KK+QGI+W
Sbjct: 462 TIITGASFGAGNYAMCGRAYSPDFMFIWPNARIGIMGGAQAAGVLTQIERATKKRQGIKW 521
Query: 491 SKEEEEKFKTKVVEAYEVEGSPYYSTARLWDDGIIDPADTRKVIGLSISASLNRAIEDTK 550
++EEEE FK K V+AYE E +PYYSTARLWDDG+IDP DTRKV+GL +SA+LNR +EDT+
Sbjct: 522 TEEEEEAFKKKTVDAYEREANPYYSTARLWDDGVIDPCDTRKVLGLCLSAALNRPLEDTR 581
Query: 551 FGVFRM 556
FGVFRM
Sbjct: 582 FGVFRM 587
>At1g36180 acetyl-CoA carboxylase, putative
Length = 2359
Score = 45.1 bits (105), Expect = 1e-04
Identities = 89/410 (21%), Positives = 140/410 (33%), Gaps = 96/410 (23%)
Query: 115 GRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADV--- 171
GR + VAND T K G++ P L E+A KLP +YL + GA L +V
Sbjct: 1697 GRKLLIVANDVTFKAGSFGPREDAFFLAVTELACTKKLPLIYLAANSGARLGVAEEVKAC 1756
Query: 172 ----FPDRENFGRIFYNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVM------VK 221
+ D + G F L S + A + S A +P+ V+
Sbjct: 1757 FKVGWSDEVSPGNDFQYIYLSSED----YARIGSSVIAHEVKLPSGETRWVIDTIVGKED 1812
Query: 222 GNGTIFLAGPPLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHAL---------- 271
G G L G + A + E V ++ G+ Y A+ +
Sbjct: 1813 GLGVENLTGSGAIAGAYSRAYN-ETFTLTFVSGRSVGIGAYLARLGMRCIQRLDQPIILT 1871
Query: 272 -------ALGRNIIKN---------------LHMAGKDVLANGLQNLSYEYKEPLYDVNE 309
LGR + + +H+ D L L++ P Y
Sbjct: 1872 GFSTLNKLLGREVYSSHMQLGGPKIMGTNGVVHLTVSDDLEGVSAILNWLSYIPAYVGGP 1931
Query: 310 LRSIAPTD---------LKKQFDVRSVIARI-----------VDGSEFDEFKKLYGTTLV 349
L +AP D + D R+ IA I D + F E + + T+V
Sbjct: 1932 LPVLAPLDPPERTVEYIPENSCDPRAAIAGINDNTGKWLGGIFDKNSFVETLEGWARTVV 1991
Query: 350 TGFARIYGQPVGIIG--------------------------NNGILFNESALKGAHFIEI 383
TG A++ G P+G++ + F +SA K A +
Sbjct: 1992 TGRAKLGGIPIGVVAVETQTVMHVIPADPGQLDSHERVVPQAGQVWFPDSAAKTAQALMD 2051
Query: 384 CTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCAKVPKITII 433
+ +PL + N GF G R GI ++G+ +V + + P I
Sbjct: 2052 FNREQLPLFIIANWRGFSGGQRDLFEGILQAGSAIVENLRTYRQPVFVYI 2101
>At2g38040 carboxyltransferase alpha subunit (CAC3)
Length = 769
Score = 44.7 bits (104), Expect = 1e-04
Identities = 60/253 (23%), Positives = 101/253 (39%), Gaps = 60/253 (23%)
Query: 75 IDRLLDPGSSFLEL-SQLAGHELYEEPLPSGGVVTGIGPVHGRLCMFVANDPTVKG---- 129
+D + + F+EL AG Y++P +VTGIG + G+ MF+ + KG
Sbjct: 165 LDHIHNITDKFMELHGDRAG---YDDP----AIVTGIGTIDGKRYMFIGHQ---KGRNTK 214
Query: 130 -------GTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVFPDRENFGRIF 182
G P +K LR A P V +D+ GA+ +++ E I
Sbjct: 215 ENIMRNFGMPTPHGYRKALRMMYYADHHGFPIVTFIDTPGAYADLKSEELGQGE---AIA 271
Query: 183 YNQALMSAEGIPQIALVLGSCTAGGAYIPAMADESVMVKGNGTIFLAGP----------- 231
N M +P +++V+G +GGA A++ +M++ N ++A P
Sbjct: 272 NNLRTMFGLKVPILSIVIGEGGSGGALAIGCANKMLMLE-NAVFYVASPEACAAILWKTS 330
Query: 232 ---------------PLVKAATGEEVSAEDLGGATVHCKTSGVSDYFAQDEFHALALGRN 276
LVK + + E LGGA H S S +A+ N
Sbjct: 331 KAAPEAAEKLRITSKELVKLNVADGIIPEPLGGA--HADPSWTSQQI------KIAINEN 382
Query: 277 IIKNLHMAGKDVL 289
+ + M+G+++L
Sbjct: 383 MNEFGKMSGEELL 395
Score = 30.4 bits (67), Expect = 2.7
Identities = 52/235 (22%), Positives = 86/235 (36%), Gaps = 52/235 (22%)
Query: 338 DEFKKLYGT-------TLVTGFARIYGQPVGIIG-------------NNGILFNESALKG 377
D+F +L+G +VTG I G+ IG N G+ K
Sbjct: 173 DKFMELHGDRAGYDDPAIVTGIGTIDGKRYMFIGHQKGRNTKENIMRNFGMPTPHGYRKA 232
Query: 378 AHFIEICTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCAKVPKITIIVGGS 437
+ P+V + G +SE G ++ A + + KVP ++I++G
Sbjct: 233 LRMMYYADHHGFPIVTFIDTPGAYADLKSEELGQGEAIANNLRTMFGLKVPILSIVIGEG 292
Query: 438 FGAGNYAMCGRAYSPNFMFLWPNARISVMGGAQAAGVLSQIEKTNKKKQGIQWSKEEEEK 497
G+G G A N M + NA V A +L + K + E EK
Sbjct: 293 -GSGGALAIGCA---NKMLMLENAVFYVASPEACAAILWKTSKA---------APEAAEK 339
Query: 498 FKTKVVEAYEVEGSPYYSTARLWDDGII---------DPADTRKVIGLSISASLN 543
+ E ++ + DGII DP+ T + I ++I+ ++N
Sbjct: 340 LRITSKELVKLNVA----------DGIIPEPLGGAHADPSWTSQQIKIAINENMN 384
>At1g36170 acetyl-CoA carboxylase, putative, 5' partial
Length = 1865
Score = 44.3 bits (103), Expect = 2e-04
Identities = 95/428 (22%), Positives = 140/428 (32%), Gaps = 132/428 (30%)
Query: 115 GRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVF-- 172
GR + +AND T K G++ P L E+A KLP +YL + GA L +V
Sbjct: 1203 GRKLLVIANDVTFKAGSFGPREDAFFLAVTELACAKKLPLIYLAANSGARLGVAEEVKAC 1262
Query: 173 ----------------------PDRENFGR--IFYNQALMSAE-------------GIPQ 195
D E G I + L S E GI
Sbjct: 1263 FKVGWSDEISPENGFQYIYLSPEDHERIGSSVIAHEVKLSSGETRWVIDTIVGKEDGI-G 1321
Query: 196 IALVLGSCTAGGAYIPAMADESVMVKGNG----------------------TIFLAGPPL 233
+ + GS GAY A + + +G I L G
Sbjct: 1322 VENLTGSGAIAGAYSKAYNETFTLTFVSGRTVGIGAYLARLGMRCIQRLDQPIILTGFST 1381
Query: 234 VKAATGEEV--SAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLHMAGKDVLAN 291
+ G EV S LGG + T+GV D+ + G + N
Sbjct: 1382 LNKLLGREVYSSHMQLGGPKI-MGTNGVVHLTVSDD---------------LEGVSAILN 1425
Query: 292 GLQNLSYEYKEPLYDVNELRSIAPTDLKKQF---------DVRSVIARIVDG-------- 334
L + P Y L +AP D ++ D R+ IA + D
Sbjct: 1426 WLSYI------PAYVGGPLPVLAPLDPPERIVEYVPENSCDPRAAIAGVKDNTGKWLGGI 1479
Query: 335 ---SEFDEFKKLYGTTLVTGFARIYGQPVGIIG--------------------------N 365
+ F E + + T+VTG A++ G PVG++
Sbjct: 1480 FDKNSFIETLEGWARTVVTGRAKLGGIPVGVVAVETQTVMQIIPADPGQLDSHERVVPQA 1539
Query: 366 NGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCA 425
+ F +SA K A + + +PL L N GF G R GI ++G+ +V +
Sbjct: 1540 GQVWFPDSAAKTAQALMDFNREELPLFILANWRGFSGGQRDLFEGILQAGSTIVENLRTY 1599
Query: 426 KVPKITII 433
+ P I
Sbjct: 1600 RQPVFVYI 1607
>At1g36160 hypothetical protein, 3' partial
Length = 2252
Score = 44.3 bits (103), Expect = 2e-04
Identities = 95/428 (22%), Positives = 140/428 (32%), Gaps = 132/428 (30%)
Query: 115 GRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGAFLPKQADVF-- 172
GR + +AND T K G++ P L E+A KLP +YL + GA L +V
Sbjct: 1626 GRKLLVIANDVTFKAGSFGPREDAFFLAVTELACAKKLPLIYLAANSGARLGVAEEVKAC 1685
Query: 173 ----------------------PDRENFGR--IFYNQALMSAE-------------GIPQ 195
D E G I + L S E GI
Sbjct: 1686 FKVGWSDEISPENGFQYIYLSPEDHERIGSSVIAHEVKLSSGETRWVIDTIVGKEDGI-G 1744
Query: 196 IALVLGSCTAGGAYIPAMADESVMVKGNG----------------------TIFLAGPPL 233
+ + GS GAY A + + +G I L G
Sbjct: 1745 VENLTGSGAIAGAYSKAYNETFTLTFVSGRTVGIGAYLARLGMRCIQRLDQPIILTGFST 1804
Query: 234 VKAATGEEV--SAEDLGGATVHCKTSGVSDYFAQDEFHALALGRNIIKNLHMAGKDVLAN 291
+ G EV S LGG + T+GV D+ + G + N
Sbjct: 1805 LNKLLGREVYSSHMQLGGPKI-MGTNGVVHLTVSDD---------------LEGVSAILN 1848
Query: 292 GLQNLSYEYKEPLYDVNELRSIAPTDLKKQF---------DVRSVIARIVDG-------- 334
L + P Y L +AP D ++ D R+ IA + D
Sbjct: 1849 WLSYI------PAYVGGPLPVLAPLDPPERIVEYVPENSCDPRAAIAGVKDNTGKWLGGI 1902
Query: 335 ---SEFDEFKKLYGTTLVTGFARIYGQPVGIIG--------------------------N 365
+ F E + + T+VTG A++ G PVG++
Sbjct: 1903 FDKNSFIETLEGWARTVVTGRAKLGGIPVGVVAVETQTVMQIIPADPGQLDSHERVVPQA 1962
Query: 366 NGILFNESALKGAHFIEICTQRNIPLVFLQNITGFMVGSRSEANGIAKSGAKMVMAVSCA 425
+ F +SA K A + + +PL L N GF G R GI ++G+ +V +
Sbjct: 1963 GQVWFPDSAAKTAQALMDFNREELPLFILANWRGFSGGQRDLFEGILQAGSTIVENLRTY 2022
Query: 426 KVPKITII 433
+ P I
Sbjct: 2023 RQPVFVYI 2030
>At1g15910
Length = 634
Score = 31.6 bits (70), Expect = 1.2
Identities = 15/38 (39%), Positives = 19/38 (49%)
Query: 484 KKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWD 521
KK +W KE KT +VE E S Y+T LW+
Sbjct: 569 KKLKREWGKEVHNAVKTALVEMNEYNASGRYTTPELWN 606
>At1g08230 unknown protein
Length = 422
Score = 31.6 bits (70), Expect = 1.2
Identities = 21/77 (27%), Positives = 37/77 (47%), Gaps = 7/77 (9%)
Query: 293 LQNLSYEYKEPLYDVNELRSIAPTDLKKQFDVRSVIARIVDGSEFDEFKKLYGTTLVTGF 352
L ++ Y +P+ D+ L S+ KK+F +R+VI R+V S F + T+V
Sbjct: 291 LSAVAVVYLQPINDI--LESVISDPTKKEFSIRNVIPRLVVRSLF-----VVMATIVAAM 343
Query: 353 ARIYGQPVGIIGNNGIL 369
+G ++G G +
Sbjct: 344 LPFFGDVNSLLGAFGFI 360
>At4g00140 hypothetical protein
Length = 257
Score = 30.4 bits (67), Expect = 2.7
Identities = 15/35 (42%), Positives = 21/35 (59%)
Query: 174 DRENFGRIFYNQALMSAEGIPQIALVLGSCTAGGA 208
+++ RI N+A + A GIP IAL L AGG+
Sbjct: 37 EKQRLSRIAENKARLDALGIPAIALSLQGSVAGGS 71
>At2g48160 unknown protein
Length = 1366
Score = 30.4 bits (67), Expect = 2.7
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 4/73 (5%)
Query: 481 TNKKKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWDDGIIDPADTRKVIGLSISA 540
T +KK +EEEK +T V + +V ST ++D + T++ IG +
Sbjct: 805 TGEKKNDCDAIVKEEEKIETGVCQGQKVVSCDVQSTRESYEDALCSLVRTKESIGRATCL 864
Query: 541 SLNRAIEDTKFGV 553
A++ KFGV
Sbjct: 865 ----AMDLMKFGV 873
>accD -chloroplast genome- carboxytransferase beta subunit
Length = 488
Score = 30.0 bits (66), Expect = 3.5
Identities = 36/149 (24%), Positives = 58/149 (38%), Gaps = 27/149 (18%)
Query: 68 KLLPRERIDRLLDPGS------SFLELSQLAGH---ELYEEPLPSG--------GVVTGI 110
K+ ERI+ +DPG+ + + H E Y+ + S V TG
Sbjct: 255 KMSSSERIELSIDPGTWNPMDEDMVSADPIKFHSKEEPYKNRIDSAQKTTGLTDAVQTGT 314
Query: 111 GPVHGRLCMFVANDPTVKGGTYYPITVKKHLRAQEIASQCKLPCVYLVDSGGA------- 163
G ++G D GG+ + +K R E A+ LP + + SGGA
Sbjct: 315 GQLNGIPVALGVMDFRFMGGSMGSVVGEKITRLIEYATNQCLPLILVCSSGGARMQEGSL 374
Query: 164 ---FLPKQADVFPDRENFGRIFYNQALMS 189
+ K + V D ++ ++FY L S
Sbjct: 375 SLMQMAKISSVLCDYQSSKKLFYISILTS 403
>At3g56370 putative protein
Length = 964
Score = 30.0 bits (66), Expect = 3.5
Identities = 34/115 (29%), Positives = 54/115 (46%), Gaps = 32/115 (27%)
Query: 25 GILPHSNGGATAMEDLLSQLQSHVHKALAGGGAEAVKRNT-------SRNKLL---PRE- 73
G++P GGA ++E+L +L++++ L G ++K + S NKLL P E
Sbjct: 438 GMIPRETGGAVSLEEL--RLENNL---LEGNIPSSIKNCSSLRSLILSHNKLLGSIPPEL 492
Query: 74 -RIDRLLDPGSSFLELS-----QLA----------GHELYEEPLPSGGVVTGIGP 112
++ RL + SF EL+ QLA H LP+GG+ G+ P
Sbjct: 493 AKLTRLEEVDLSFNELAGTLPKQLANLGYLHTFNISHNHLFGELPAGGIFNGLSP 547
>At5g42350 putative protein
Length = 563
Score = 29.6 bits (65), Expect = 4.6
Identities = 15/45 (33%), Positives = 26/45 (57%)
Query: 62 RNTSRNKLLPRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGV 106
+N +++ L R+++DRL S L + G L++EPL SG +
Sbjct: 330 QNGTKSHRLIRQKLDRLNRNSSKRFVLIAIGGTGLFDEPLDSGEI 374
>At4g00380
Length = 662
Score = 29.6 bits (65), Expect = 4.6
Identities = 14/38 (36%), Positives = 18/38 (46%)
Query: 484 KKQGIQWSKEEEEKFKTKVVEAYEVEGSPYYSTARLWD 521
KK +W KE K +VE E S Y T+ LW+
Sbjct: 597 KKLKREWGKEVHNAVKAALVEMNEYNASGRYPTSELWN 634
>At5g42360 putative protein
Length = 563
Score = 28.9 bits (63), Expect = 7.8
Identities = 14/45 (31%), Positives = 26/45 (57%)
Query: 62 RNTSRNKLLPRERIDRLLDPGSSFLELSQLAGHELYEEPLPSGGV 106
+N +++ L R+++DRL S L + G +++EPL SG +
Sbjct: 330 QNGTKSHRLIRQKLDRLNRNSSKRFVLIAIGGTGIFDEPLDSGEI 374
>At5g13830 FtsJ (dbj|BAA83750.1)
Length = 224
Score = 28.9 bits (63), Expect = 7.8
Identities = 19/48 (39%), Positives = 26/48 (53%), Gaps = 5/48 (10%)
Query: 68 KLLPRERIDRLLDPGSSFLELSQLAGHELYEE-----PLPSGGVVTGI 110
KLL ++ +L+ PGSS L+L G L PL SGG+V G+
Sbjct: 26 KLLQIQKQYKLIKPGSSVLDLGCAPGAWLQVACQSLGPLRSGGIVVGM 73
>At1g76450 unknown protein
Length = 247
Score = 28.9 bits (63), Expect = 7.8
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query: 333 DGSEFDEFKKL--YGTTLVTGFARIYGQPVGIIGNNGILFNESALKGAHFIEICTQ 386
D + + F K+ + TLV+G R + +PVG+ L + A KG ++IE Q
Sbjct: 142 DFTRMESFGKVEAFAETLVSGLDRSWQKPVGVTAK---LIDSRASKGFYYIEYTLQ 194
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,657,208
Number of Sequences: 26719
Number of extensions: 561098
Number of successful extensions: 1378
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1357
Number of HSP's gapped (non-prelim): 23
length of query: 556
length of database: 11,318,596
effective HSP length: 104
effective length of query: 452
effective length of database: 8,539,820
effective search space: 3859998640
effective search space used: 3859998640
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0227.1


