

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0223.2
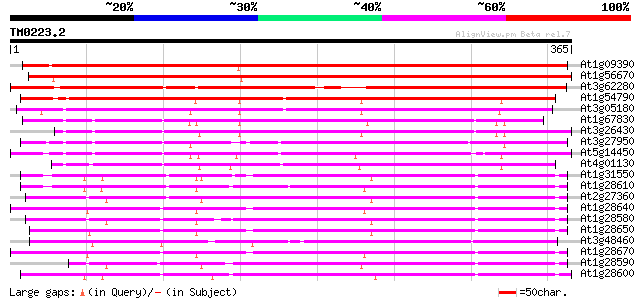
(365 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09390 putative lipase 479 e-135
At1g56670 lipase homolog (Lip-4) 459 e-129
At3g62280 putative protein 314 6e-86
At1g54790 298 3e-81
At3g05180 putative nodulin 278 3e-75
At1g67830 ENOD8-like protein 263 1e-70
At3g26430 nodulin, putative 254 7e-68
At3g27950 early nodule-specific protein, putative 251 4e-67
At5g14450 early nodule-specific protein - like 246 1e-65
At4g01130 putative acetyltransferase 224 7e-59
At1g31550 unknown protein 182 3e-46
At1g28610 lipase, putative 179 2e-45
At2g27360 putative lipase 177 1e-44
At1g28640 lipase, putative 174 7e-44
At1g28580 lipase like protein 170 1e-42
At1g28650 lipase, putative 170 1e-42
At3g48460 lipase - like protein 167 6e-42
At1g28670 lipase 166 2e-41
At1g28590 putative lipase (At1g28590) 165 4e-41
At1g28600 lipase like protein 164 9e-41
>At1g09390 putative lipase
Length = 370
Score = 479 bits (1233), Expect = e-135
Identities = 228/358 (63%), Positives = 279/358 (77%), Gaps = 4/358 (1%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
L +L F+ I + AV C VP V+F FGDSNSDTGGL +GLG+ I LPNGR+FF R
Sbjct: 12 LLVLLPFILILRQNLAVA-GGCQVPPVIFNFGDSNSDTGGLVAGLGYSIGLPNGRSFFQR 70
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNI 128
STGRLSDGRLVID LCQSLNT L PYLDSL GS F NGANFA+VGSSTLP+YVPF+LNI
Sbjct: 71 STGRLSDGRLVIDFLCQSLNTSLLNPYLDSLVGSKFQNGANFAIVGSSTLPRYVPFALNI 130
Query: 129 QVMQFLHFKARTLELVSAG---AKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVI 185
Q+MQFLHFK+R LEL S + +I + GFR ALY+IDIGQND+ADSF+K L+Y +V+
Sbjct: 131 QLMQFLHFKSRALELASISDPLKEMMIGESGFRNALYMIDIGQNDIADSFSKGLSYSRVV 190
Query: 186 KKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSA 245
K IP VI+EI++A+K LY+EG RKFWVHNTGPLGCLP+ L++ K D GCL++YN+A
Sbjct: 191 KLIPNVISEIKSAIKILYDEGGRKFWVHNTGPLGCLPQKLSMVHSKGFDKHGCLATYNAA 250
Query: 246 ARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPY 305
A+LFNE L H + LRT LK+A +VYVDIYAIK+DLIAN+ YGF PL CCG+GGPPY
Sbjct: 251 AKLFNEGLDHMCRDLRTELKEANIVYVDIYAIKYDLIANSNNYGFEKPLMACCGYGGPPY 310
Query: 306 NFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
N+++ +TCG G + CDEGSR+++WDG H+TE AN +A K+LS +STP TPF FFC
Sbjct: 311 NYNVNITCGNGGSKSCDEGSRFISWDGIHYTETANAIVAMKVLSMQHSTPPTPFHFFC 368
>At1g56670 lipase homolog (Lip-4)
Length = 373
Score = 459 bits (1180), Expect = e-129
Identities = 216/363 (59%), Positives = 274/363 (74%), Gaps = 10/363 (2%)
Query: 13 FLFLWITSSSAAVMM-------SKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTF 65
F F +IT S A+++ + C V+F FGDSNSDTGGL +GLG+PI PNGR F
Sbjct: 11 FSFFFITLVSLALLILRQPSRAASCTARPVIFNFGDSNSDTGGLVAGLGYPIGFPNGRLF 70
Query: 66 FHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFS 125
F RSTGRLSDGRL+ID LCQSLNT L PYLDSL + F NGANFA+ GS TLPK VPFS
Sbjct: 71 FRRSTGRLSDGRLLIDFLCQSLNTSLLRPYLDSLGRTRFQNGANFAIAGSPTLPKNVPFS 130
Query: 126 LNIQVMQFLHFKARTLELVSAGAK---NVINDEGFRAALYLIDIGQNDLADSFAKNLTYV 182
LNIQV QF HFK+R+LEL S+ I++ GF+ ALY+IDIGQND+A SFA+ +Y
Sbjct: 131 LNIQVKQFSHFKSRSLELASSSNSLKGMFISNNGFKNALYMIDIGQNDIARSFARGNSYS 190
Query: 183 QVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSY 242
Q +K IP +ITEI++++K LY+EG R+FW+HNTGPLGCLP+ L++ + KDLD GCL SY
Sbjct: 191 QTVKLIPQIITEIKSSIKRLYDEGGRRFWIHNTGPLGCLPQKLSMVKSKDLDQHGCLVSY 250
Query: 243 NSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGG 302
NSAA LFN+ L H ++LRT L+DAT++Y+DIYAIK+ LIAN+ +YGF +PL CCG+GG
Sbjct: 251 NSAATLFNQGLDHMCEELRTELRDATIIYIDIYAIKYSLIANSNQYGFKSPLMACCGYGG 310
Query: 303 PPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFF 362
PYN+++++TCG G VC+EGSR+++WDG H+TE AN +A K+LS YS P TPF FF
Sbjct: 311 TPYNYNVKITCGHKGSNVCEEGSRFISWDGIHYTETANAIVAMKVLSMHYSKPPTPFHFF 370
Query: 363 CHR 365
C R
Sbjct: 371 CRR 373
>At3g62280 putative protein
Length = 343
Score = 314 bits (804), Expect = 6e-86
Identities = 165/363 (45%), Positives = 229/363 (62%), Gaps = 26/363 (7%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLP 60
+SS F L L L ++S + +K +L FGDSNSDTGG+ +G+G PI LP
Sbjct: 5 VSSLQCFFLVLCLSLLVCSNSETSYKSNK---KPILINFGDSNSDTGGVLAGVGLPIGLP 61
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK 120
+G TFFHR TGRL DGRL++D C+ L +L+PYLDSLS +F G NFAV G++ LP
Sbjct: 62 HGITFFHRGTGRLGDGRLIVDFYCEHLKMTYLSPYLDSLS-PNFKRGVNFAVSGATALPI 120
Query: 121 YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADS-FAKNL 179
+ F L IQ+ QF+HFK R+ EL+S+G +++I+D GFR ALY+IDIGQNDL + + NL
Sbjct: 121 F-SFPLAIQIRQFVHFKNRSQELISSGRRDLIDDNGFRNALYMIDIGQNDLLLALYDSNL 179
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCL 239
TY V++KIP+++ EI+ A+ +G +HN DLD GC
Sbjct: 180 TYAPVVEKIPSMLLEIKKAI-----QGELAIHLHNDS---------------DLDPIGCF 219
Query: 240 SSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCG 299
+N A+ FN+ L +LR++ KDATLVYVDIY+IK+ L A+ YGF +PL CCG
Sbjct: 220 RVHNEVAKAFNKGLLSLCNELRSQFKDATLVYVDIYSIKYKLSADFKLYGFVDPLMACCG 279
Query: 300 FGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPF 359
+GG P N+D + TCGQPG +C + ++ + WDG H+TEAAN F+ +L+ YS P+
Sbjct: 280 YGGRPNNYDRKATCGQPGSTICRDVTKAIVWDGVHYTEAANRFVVDAVLTNRYSYPKNSL 339
Query: 360 EFF 362
+ F
Sbjct: 340 DRF 342
>At1g54790
Length = 383
Score = 298 bits (763), Expect = 3e-81
Identities = 160/366 (43%), Positives = 223/366 (60%), Gaps = 23/366 (6%)
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFH 67
S+ L L SS ++++ PA++ FGDSNSDTG L S +N P G+T+F+
Sbjct: 9 SIVLFILISLFLPSSFSIILK---YPAIIN-FGDSNSDTGNLISAGIENVNPPYGQTYFN 64
Query: 68 RSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----KYVP 123
+GR DGRL++D L ++ FL PYLDSL +F G NFA GS+ LP P
Sbjct: 65 LPSGRYCDGRLIVDFLLDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSP 124
Query: 124 FSLNIQVMQFLHFKARTLELVSAGA----KNVINDEGFRAALYLIDIGQNDLADSFAKNL 179
FS ++Q+ QF+ FK+R +EL+S K + + + LY+IDIGQND+A +F
Sbjct: 125 FSFDLQISQFIRFKSRAIELLSKTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSK- 183
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFG 237
T QV+ IP+++ E +K LY EG R W+HNTGPLGCL + +A LD FG
Sbjct: 184 TLDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFG 243
Query: 238 CLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVC 297
C+SS+N AA+LFN L+ S K + + DA + YVDI++IK +LIAN +++GF PL C
Sbjct: 244 CVSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMAC 303
Query: 298 CGFGGPPYNFDLRVTCGQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILS 349
CG GG P N+D R+TCGQ + C++ S Y+NWDG H+TEAAN F++S+IL+
Sbjct: 304 CGVGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYINWDGIHYTEAANEFVSSQILT 363
Query: 350 TDYSTP 355
YS P
Sbjct: 364 GKYSDP 369
>At3g05180 putative nodulin
Length = 379
Score = 278 bits (712), Expect = 3e-75
Identities = 157/370 (42%), Positives = 222/370 (59%), Gaps = 23/370 (6%)
Query: 5 LAFSLHLLFLFLWIT---SSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPN 61
L +L L LF+ I+ S A D PAV F FGDSNSDTG L+SGLGF
Sbjct: 4 LFHTLLRLLLFVAISHTLSPLAGSFRISNDFPAV-FNFGDSNSDTGELSSGLGFLPQPSY 62
Query: 62 GRTFFHRST-GRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---- 116
TFF T GR +GRL++D L ++++ +L PYLDS+S ++ G NFA S+
Sbjct: 63 EITFFRSPTSGRFCNGRLIVDFLMEAIDRPYLRPYLDSISRQTYRRGCNFAAAASTIQKA 122
Query: 117 TLPKYVPFSLNIQVMQFLHFKARTLELVSAGA---KNVINDEGFRAALYLIDIGQNDLAD 173
Y PF +QV QF+ FK++ L+L+ + + ++ F LY+ DIGQND+A
Sbjct: 123 NAASYSPFGFGVQVSQFITFKSKVLQLIQQDEELQRYLPSEYFFSNGLYMFDIGQNDIAG 182
Query: 174 SFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKK 231
+F T QV+ +P ++ ++ +K LY EGAR +W+HNTGPLGCL +++++ K
Sbjct: 183 AFYTK-TVDQVLALVPIILDIFQDGIKRLYAEGARNYWIHNTGPLGCLAQVVSIFGEDKS 241
Query: 232 DLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFS 291
LD FGC+S +N AA+LFN L+ +KL + ++ YVDI++IK DLI N +KYGF
Sbjct: 242 KLDEFGCVSDHNQAAKLFNLQLHGLFKKLPQQYPNSRFTYVDIFSIKSDLILNHSKYGFD 301
Query: 292 NPLTVCCGFGGPPYNFDLRVTCGQPG--------YQVCDEGSRYVNWDGTHHTEAANTFI 343
+ + VCCG GGPP N+D +V CG+ + C + S+YVNWDG H+TEAAN F+
Sbjct: 302 HSIMVCCGTGGPPLNYDDQVGCGKTARSNGTIITAKPCYDSSKYVNWDGIHYTEAANRFV 361
Query: 344 ASKILSTDYS 353
A IL+ YS
Sbjct: 362 ALHILTGKYS 371
>At1g67830 ENOD8-like protein
Length = 372
Score = 263 bits (672), Expect = 1e-70
Identities = 152/361 (42%), Positives = 215/361 (59%), Gaps = 26/361 (7%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
L LF ++S S + +C PA+ F FGDSNSDTGGL++ G P+G +FF
Sbjct: 5 LSSLFALSLLSSLSPSTHAHQCHFPAI-FNFGDSNSDTGGLSAAFG-QAGPPHGSSFFGS 62
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGS------STLPK-- 120
GR DGRLVID + +SL +L+ +LDS+ GS+F++GANFA GS STL +
Sbjct: 63 PAGRYCDGRLVIDFIAESLGLPYLSAFLDSV-GSNFSHGANFATAGSPIRALNSTLRQSG 121
Query: 121 YVPFSLNIQVMQFLHFKARTLELVSAGA--KNVIND-EGFRAALYLIDIGQNDLADSFAK 177
+ PFSL++Q +QF +F R+ + S G K ++ + + F ALY DIGQNDL +
Sbjct: 122 FSPFSLDVQFVQFYNFHNRSQTVRSRGGVYKTMLPESDSFSKALYTFDIGQNDLTAGYFA 181
Query: 178 NLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKK--DLDL 235
N T QV ++P +I++ NA+K++Y +G R FW+HNTGP+GCL ++ K D D
Sbjct: 182 NKTVEQVETEVPEIISQFMNAIKNIYGQGGRYFWIHNTGPIGCLAYVIERFPNKASDFDS 241
Query: 236 FGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLT 295
GC+S N A+ FN AL + +LR+ L +A + YVD+Y++KH+L +A +GF L
Sbjct: 242 HGCVSPLNHLAQQFNHALKQAVIELRSSLSEAAITYVDVYSLKHELFVHAQGHGFKGSLV 301
Query: 296 VCCGFGGPPYNFDLRVTCGQ----PGYQV-----CDEGSRYVNWDGTHHTEAANTFIASK 346
CCG GG YN++ + CG G +V CDE + V WDG H T+AAN FI K
Sbjct: 302 SCCGHGG-KYNYNKGIGCGMKKIVKGKEVYIGKPCDEPDKAVVWDGVHFTQAANKFIFDK 360
Query: 347 I 347
I
Sbjct: 361 I 361
>At3g26430 nodulin, putative
Length = 380
Score = 254 bits (648), Expect = 7e-68
Identities = 147/358 (41%), Positives = 200/358 (55%), Gaps = 26/358 (7%)
Query: 30 CDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNT 89
C+ PA+ F FGDSNSDTGGL++ G PNG+TFFH +GR SDGRL+ID + + L
Sbjct: 27 CNFPAI-FNFGDSNSDTGGLSASFG-QAPYPNGQTFFHSPSGRFSDGRLIIDFIAEELGL 84
Query: 90 RFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYV--------PFSLNIQVMQFLHFKARTL 141
+L +LDS+ GS+F++GANFA GS+ P P SL++Q++QF F R+
Sbjct: 85 PYLNAFLDSI-GSNFSHGANFATAGSTVRPPNATIAQSGVSPISLDVQLVQFSDFITRSQ 143
Query: 142 ELVSAGA---KNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENA 198
+ + G K + E F ALY DIGQNDL N+T Q+ IP V ++ N
Sbjct: 144 LIRNRGGVFKKLLPKKEYFSQALYTFDIGQNDLTAGLKLNMTSDQIKAYIPDVHDQLSNV 203
Query: 199 VKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGCLSSYNSAARLFNEALYHS 256
++ +Y++G R+FW+HNT PLGCLP +L +D GC N AR +N L
Sbjct: 204 IRKVYSKGGRRFWIHNTAPLGCLPYVLDRFPVPASQIDNHGCAIPRNEIARYYNSELKRR 263
Query: 257 SQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQ- 315
+LR L +A YVDIY+IK LI A K GF PL CCG GG YNF+ + CG
Sbjct: 264 VIELRKELSEAAFTYVDIYSIKLTLITQAKKLGFRYPLVACCGHGG-KYNFNKLIKCGAK 322
Query: 316 ---PGYQV-----CDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFCHR 365
G ++ C++ S V+WDG H TE N++I +I +S P P + C R
Sbjct: 323 VMIKGKEIVLAKSCNDVSFRVSWDGIHFTETTNSWIFQQINDGAFSDPPLPVKSACTR 380
>At3g27950 early nodule-specific protein, putative
Length = 361
Score = 251 bits (641), Expect = 4e-67
Identities = 153/363 (42%), Positives = 206/363 (56%), Gaps = 20/363 (5%)
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFH 67
+L ++ L L T +A + S C+ PAV F FGDSNSDTG +++ +G + PNG FF
Sbjct: 7 TLAIIVLLLGFTEKLSA-LSSSCNFPAV-FNFGDSNSDTGAISAAIG-EVPPPNGVAFFG 63
Query: 68 RSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---TLPKYVPF 124
RS GR SDGRL+ID + ++L +LTPYLDS+ G+++ +GANFA GS TL + PF
Sbjct: 64 RSAGRHSDGRLIIDFITENLTLPYLTPYLDSV-GANYRHGANFATGGSCIRPTLACFSPF 122
Query: 125 SLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQV 184
L QV QF+HFK RTL L N ND F ALY +DIGQNDLA F +N+T Q+
Sbjct: 123 HLGTQVSQFIHFKTRTLSLY-----NQTND--FSKALYTLDIGQNDLAIGF-QNMTEEQL 174
Query: 185 IKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNS 244
IP +I A+K LY EGAR F +HNTGP GCLP +L D +GCL N+
Sbjct: 175 KATIPLIIENFTIALKLLYKEGARFFSIHNTGPTGCLPYLLKAFPAIPRDPYGCLKPLNN 234
Query: 245 AARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPP 304
A FN+ L + +L+ L + YVD+Y+ K++LI A GF +P CC G
Sbjct: 235 VAIEFNKQLKNKITQLKKELPSSFFTYVDVYSAKYNLITKAKALGFIDPFDYCC-VGAIG 293
Query: 305 YNFDLRVTCGQPGYQV----CDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFE 360
T G ++ C +++WDG H+TE AN +A++IL S P P +
Sbjct: 294 RGMGCGKTIFLNGTELYSSSCQNRKNFISWDGIHYTETANMLVANRILDGSISDPPLPTQ 353
Query: 361 FFC 363
C
Sbjct: 354 KAC 356
>At5g14450 early nodule-specific protein - like
Length = 389
Score = 246 bits (629), Expect = 1e-65
Identities = 155/389 (39%), Positives = 215/389 (54%), Gaps = 35/389 (8%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLP 60
M S+ FS LL LF TS S + C PA+ + FGDSNSDTGG+++ PI P
Sbjct: 11 MVSSTVFSWLLLCLFAVTTSVS---VQPTCTFPAI-YNFGDSNSDTGGISAAFE-PIRDP 65
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---- 116
G+ FFHR TGR SDGRL ID + + L +L+ YL+SL GS+F +GANFA GS+
Sbjct: 66 YGQGFFHRPTGRDSDGRLTIDFIAERLGLPYLSAYLNSL-GSNFRHGANFATGGSTIRRQ 124
Query: 117 --TLPKY--VPFSLNIQVMQFLHFKARTLELVSA-----GAKNVINDEGFRAALYLIDIG 167
T+ +Y PFSL++Q+ QF FKAR+ L + + + E F ALY DIG
Sbjct: 125 NETIFQYGISPFSLDMQIAQFDQFKARSALLFTQIKSRYDREKLPRQEEFAKALYTFDIG 184
Query: 168 QNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPK---I 224
QNDL+ F + ++ Q+ IP ++ + +AV+++Y +G R FWVHNTGP GCLP
Sbjct: 185 QNDLSVGF-RTMSVDQLKATIPDIVNHLASAVRNIYQQGGRTFWVHNTGPFGCLPVNMFY 243
Query: 225 LALAQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIAN 284
+ LD GC+ + N A FN L + LR L A + YVD+Y K+++++N
Sbjct: 244 MGTPAPGYLDKSGCVKAQNEMAMEFNRKLKETVINLRKELTQAAITYVDVYTAKYEMMSN 303
Query: 285 ATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGY--------QVCDEGSRYVNWDGTHHT 336
K GF+NPL VCCG+ +D + CG G C V+WDG H+T
Sbjct: 304 PKKLGFANPLKVCCGY---HEKYD-HIWCGNKGKVNNTEIYGGSCPNPVMAVSWDGVHYT 359
Query: 337 EAANTFIASKILSTDYSTPRTPFEFFCHR 365
EAAN +A + L+ + P P C+R
Sbjct: 360 EAANKHVADRTLNGLLTDPPVPITRACYR 388
>At4g01130 putative acetyltransferase
Length = 382
Score = 224 bits (570), Expect = 7e-59
Identities = 141/350 (40%), Positives = 186/350 (52%), Gaps = 27/350 (7%)
Query: 28 SKCDVPAVLFVFGDSNSDTGGLTSGLGFPINL-PNGRTFFHRSTGRLSDGRLVIDLLCQS 86
SKCD A+ F FGDSNSDTGG + FP P G T+F + GR SDGRL+ID L +S
Sbjct: 28 SKCDFEAI-FNFGDSNSDTGGFWAA--FPAQSGPWGMTYFKKPAGRASDGRLIIDFLAKS 84
Query: 87 LNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYV--------PFSLNIQVMQFLHFKA 138
L FL+PYL S+ GS F +GANFA + S+ L PFSL IQ+ Q FK
Sbjct: 85 LGMPFLSPYLQSI-GSDFRHGANFATLASTVLLPNTSLFVSGISPFSLAIQLNQMKQFKV 143
Query: 139 RTLE---LVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEI 195
E L G K + + F +LY IGQND + A ++ +V +P VI +I
Sbjct: 144 NVDESHSLDRPGLKILPSKIVFGKSLYTFYIGQNDFTSNLA-SIGVERVKLYLPQVIGQI 202
Query: 196 ENAVKSLYNEGARKFWVHNTGPLGCLPKILA--LAQKKDLDLFGCLSSYNSAARLFNEAL 253
+K +Y G R F V N P+GC P IL DLD +GCL N A + +N L
Sbjct: 203 AGTIKEIYGIGGRTFLVLNLAPVGCYPAILTGYTHTDADLDKYGCLIPVNKAVKYYNTLL 262
Query: 254 YHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTC 313
+ + RT LK+AT++Y+D + I DL + YG + + CCG+GG PYNF+ ++ C
Sbjct: 263 NKTLSQTRTELKNATVIYLDTHKILLDLFQHPKSYGMKHGIKACCGYGGRPYNFNQKLFC 322
Query: 314 GQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTP 355
G + C + YV+WDG H TEAAN I+ IL S P
Sbjct: 323 GNTKVIGNFSTTAKACHDPHNYVSWDGIHATEAANHHISMAILDGSISYP 372
>At1g31550 unknown protein
Length = 394
Score = 182 bits (461), Expect = 3e-46
Identities = 131/376 (34%), Positives = 185/376 (48%), Gaps = 34/376 (9%)
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTG---GLTSGLGFPINL--PNG 62
SL L LF+ I SS S+C + FGDS +DTG GL+ P++ P G
Sbjct: 14 SLFLSTLFVSIVSSE-----SQCRNFESIISFGDSIADTGNLLGLSDHNNLPMSAFPPYG 68
Query: 63 RTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK-- 120
TFFH TGR SDGRL+ID + + L ++ PY S +G+ F G NFAV ++ L
Sbjct: 69 ETFFHHPTGRFSDGRLIIDFIAEFLGLPYVPPYFGSTNGN-FEKGVNFAVASATALESSF 127
Query: 121 ------YVP--FSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLA 172
+ P FSL +Q+ F L S +++I + A + + +IG ND
Sbjct: 128 LEEKGYHCPHNFSLGVQLKIFKQSLPNLCGLPS-DCRDMIGN----ALILMGEIGANDYN 182
Query: 173 DSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKD 232
F + +V + +P VI+ I +A+ L G R F V PLGC L L Q +
Sbjct: 183 FPFFQLRPLDEVKELVPLVISTISSAITELIGMGGRTFLVPGGFPLGCSVAFLTLHQTSN 242
Query: 233 LD----LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKY 288
++ L GCL N +E L +LR ++Y D Y L +KY
Sbjct: 243 MEEYDPLTGCLKWLNKFGEYHSEQLQEELNRLRKLNPHVNIIYADYYNASLRLGREPSKY 302
Query: 289 GFSNP-LTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKI 347
GF N L+ CCG GGP YNF+L +CG G + C + S+YV WDG H TEAA+ +A +
Sbjct: 303 GFINRHLSACCGVGGP-YNFNLSRSCGSVGVEACSDPSKYVAWDGLHMTEAAHKSMADGL 361
Query: 348 LSTDYSTPRTPFEFFC 363
+ Y+ P PF++ C
Sbjct: 362 VKGPYAIP--PFDWSC 375
>At1g28610 lipase, putative
Length = 383
Score = 179 bits (455), Expect = 2e-45
Identities = 132/374 (35%), Positives = 182/374 (48%), Gaps = 30/374 (8%)
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTG---GLTSGLGFPIN--LPNG 62
S L LF+ I SS ++C + FGDS +DTG GL+ P+ LP G
Sbjct: 9 SFFLSTLFVTIVSSQ-----TQCRNLESIISFGDSITDTGNLVGLSDRNHLPVTAFLPYG 63
Query: 63 RTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK-- 120
TFFH TGR +GR++ID + + L + P+ S +G+ F G NFAV G++ L
Sbjct: 64 ETFFHHPTGRSCNGRIIIDFIAEFLGLPHVPPFYGSKNGN-FEKGVNFAVAGATALETSI 122
Query: 121 ------YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADS 174
Y P S +Q FK L G+ D A + + +IG ND +
Sbjct: 123 LEKRGIYYPHSNISLGIQLKTFKESLPNL--CGSPTDCRDMIGNAFIIMGEIGGNDFNFA 180
Query: 175 FAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQ---KK 231
F N T +V + +P VIT+I +A+ L + G R F V PLGC L L Q K+
Sbjct: 181 FFVNKTS-EVKELVPLVITKISSAIVELVDMGGRTFLVPGNFPLGCSATYLTLYQTSNKE 239
Query: 232 DLD-LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGF 290
+ D L GCL+ N + +NE L +L ++Y D + L +K+GF
Sbjct: 240 EYDPLTGCLTWLNDFSEYYNEKLQAELNRLSKLYPHVNIIYGDYFNALLRLYQEPSKFGF 299
Query: 291 SN-PLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILS 349
+ PL CCG GGP YNF L CG G + C + S+YVNWDG H TEAA +IA +L
Sbjct: 300 MDRPLPACCGLGGP-YNFTLSKKCGSVGVKYCSDPSKYVNWDGVHMTEAAYKWIADGLLK 358
Query: 350 TDYSTPRTPFEFFC 363
Y+ P F + C
Sbjct: 359 GPYTIP--SFHWLC 370
>At2g27360 putative lipase
Length = 394
Score = 177 bits (448), Expect = 1e-44
Identities = 123/371 (33%), Positives = 176/371 (47%), Gaps = 23/371 (6%)
Query: 11 LLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPN------GRT 64
LL F+ + ++C + FGDS +DTG L GL P +LP G T
Sbjct: 9 LLSFFISTFLITVVTSQTRCRNFKSIISFGDSITDTGNLL-GLSSPNDLPESAFPPYGET 67
Query: 65 FFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVP- 123
FFH +GR SDGRL+ID + + L + P+ S +G+ F G NFAV G++ L V
Sbjct: 68 FFHHPSGRFSDGRLIIDFIAEFLGIPHVPPFYGSKNGN-FEKGVNFAVGGATALECSVLE 126
Query: 124 ------FSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAK 177
NI + L +L + + D A + + +IG ND
Sbjct: 127 EKGTHCSQSNISLGNQLKSFKESLPYLCGSSSPDCRDMIENAFILIGEIGGNDYNFPLFD 186
Query: 178 NLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLD--- 234
+V + +P VIT I +A+ L + GAR F V PLGC L L + + +
Sbjct: 187 RKNIEEVKELVPLVITTISSAISELVDMGARTFLVPGNFPLGCSVAYLTLYETPNKEEYN 246
Query: 235 -LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSN- 292
L GCL+ N + NE L ++LR ++Y D Y L+ +K+G +
Sbjct: 247 PLTGCLTWLNDFSVYHNEQLQAELKRLRNLYPHVNIIYGDYYNTLLRLMQEPSKFGLMDR 306
Query: 293 PLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDY 352
PL CCG GGP YNF + CG G + C + S+YVNWDG H TEAA +I+ +L+ Y
Sbjct: 307 PLPACCGLGGP-YNFTFSIKCGSKGVEYCSDPSKYVNWDGIHMTEAAYKWISEGVLTGPY 365
Query: 353 STPRTPFEFFC 363
+ P PF + C
Sbjct: 366 AIP--PFNWSC 374
>At1g28640 lipase, putative
Length = 390
Score = 174 bits (441), Expect = 7e-44
Identities = 123/384 (32%), Positives = 179/384 (46%), Gaps = 29/384 (7%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGL-----TSGLGF 55
M+S+L + L L+ T+ A S+C + FGDS +DTG + L
Sbjct: 1 MASSLEKLISSFLLVLYSTTIIVASSESRCRRFKSIISFGDSIADTGNYLHLSDVNHLPQ 60
Query: 56 PINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGS 115
LP G +FFH +GR SDGRL+ID + + L ++ Y S SF G NFAV G+
Sbjct: 61 SAFLPYGESFFHPPSGRYSDGRLIIDFIAEFLGLPYVPSYFGS-QNVSFDQGINFAVYGA 119
Query: 116 STLPK-----------YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLI 164
+ L + + SL++Q+ F S + ++ D + + +
Sbjct: 120 TALDRVFLVGKGIESDFTNVSLSVQLNIFKQILPNLCTSSSRDCREMLGD----SLILMG 175
Query: 165 DIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKI 224
+IG ND F + + ++ + +P VI I +A+ L + G + F V PLGC P
Sbjct: 176 EIGVNDYNYPFFEGKSINEIKQLVPLVIKAISSAIVDLIDLGGKTFLVPGNFPLGCYPAY 235
Query: 225 LALAQ---KKDLDLF-GCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHD 280
L L Q ++D D F GC+ N NE L ++L+ ++Y D Y
Sbjct: 236 LTLFQTAAEEDHDPFTGCIPRLNEFGEYHNEQLKTELKRLQELYDHVNIIYADYYNSLFR 295
Query: 281 LIANATKYGFSN-PLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAA 339
L KYGF N PL CCG GG YNF + CG G C S YVNWDG H TEA
Sbjct: 296 LYQEPVKYGFKNRPLAACCGVGGQ-YNFTIGKECGHRGVSCCQNPSEYVNWDGYHLTEAT 354
Query: 340 NTFIASKILSTDYSTPRTPFEFFC 363
+ +A IL+ Y++P F++ C
Sbjct: 355 HQKMAQVILNGTYASP--AFDWSC 376
>At1g28580 lipase like protein
Length = 390
Score = 170 bits (431), Expect = 1e-42
Identities = 122/375 (32%), Positives = 178/375 (46%), Gaps = 32/375 (8%)
Query: 11 LLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPN------GRT 64
L+F+FL + +KC + FGDS +DTG L GL P +LP+ G
Sbjct: 13 LVFIFLSTFVVTNVSSETKCREFKSIISFGDSIADTGNLL-GLSDPKDLPHMAFPPYGEN 71
Query: 65 FFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK---- 120
FFH TGR S+GRL+ID + + L + P+ S ++F G NFAV G++ L +
Sbjct: 72 FFHHPTGRFSNGRLIIDFIAEFLGLPLVPPFYGS-HNANFEKGVNFAVGGATALERSFLE 130
Query: 121 -------YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLAD 173
Y SL +Q+ F +L + G+ + D A + + +IG ND
Sbjct: 131 DRGIHFPYTNVSLGVQLNSF----KESLPSI-CGSPSDCRDMIENALILMGEIGGNDYNY 185
Query: 174 SFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDL 233
+F + ++ + +P VIT I +A+ L G R F V P+GC L Q ++
Sbjct: 186 AFFVDKGIEEIKELMPLVITTISSAITELIGMGGRTFLVPGEFPVGCSVLYLTSHQTSNM 245
Query: 234 D----LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYG 289
+ L GCL N E L +L+ ++Y D Y L K+G
Sbjct: 246 EEYDPLTGCLKWLNKFGENHGEQLRAELNRLQKLYPHVNIIYADYYNALFHLYQEPAKFG 305
Query: 290 FSN-PLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKIL 348
F N PL+ CCG GGP YN+ + CG + CD+ S+YV WDG H TEAA +A IL
Sbjct: 306 FMNRPLSACCGAGGP-YNYTVGRKCGTDIVESCDDPSKYVAWDGVHMTEAAYRLMAEGIL 364
Query: 349 STDYSTPRTPFEFFC 363
+ Y+ P PF++ C
Sbjct: 365 NGPYAIP--PFDWSC 377
>At1g28650 lipase, putative
Length = 385
Score = 170 bits (431), Expect = 1e-42
Identities = 115/370 (31%), Positives = 171/370 (46%), Gaps = 28/370 (7%)
Query: 14 LFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLT-----SGLGFPINLPNGRTFFHR 68
L L+ T+ A S+C + FGDS +DTG + L LP G +FFH
Sbjct: 16 LILYYTTIVVASSESRCRRYKSIISFGDSIADTGNYVHLSNVNNLPQAAFLPYGESFFHP 75
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK-------- 120
+GR SDGRLVID + + L ++ PY S SF G NFAV G++ L +
Sbjct: 76 PSGRYSDGRLVIDFIAEFLGLPYVPPYFGS-QNVSFNQGINFAVYGATALDRAFLVKQGI 134
Query: 121 ---YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAK 177
+ SL++Q+ F + + ++ D + + + +IG ND F +
Sbjct: 135 KSDFTNISLSVQLNTFKQILPNLCASSTRDCREMLGD----SLILMGEIGGNDYNYPFFE 190
Query: 178 NLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLD--- 234
+ ++ + +P +I I +A+ L + G + F V P+GC L L Q ++
Sbjct: 191 GKSINEIKELVPLIIKAISSAIVDLIDLGGKTFLVPGNFPIGCSTAYLTLFQTATVEHDP 250
Query: 235 LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSN-P 293
GC+ N NE L ++L+ ++Y D Y + L KYGF N P
Sbjct: 251 FTGCIPWLNKFGEHHNEQLKIELKQLQKLYPHVNIIYADYYNSLYGLFQEPAKYGFKNRP 310
Query: 294 LTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYS 353
L CCG GG YNF + CG+ G C S YVNWDG H TEA +A +L+ Y+
Sbjct: 311 LAACCGVGGQ-YNFTIGKECGENGVSYCQNPSEYVNWDGYHLTEATYQKMAQGLLNGRYT 369
Query: 354 TPRTPFEFFC 363
TP F++ C
Sbjct: 370 TP--AFDWSC 377
>At3g48460 lipase - like protein
Length = 381
Score = 167 bits (424), Expect = 6e-42
Identities = 116/361 (32%), Positives = 180/361 (49%), Gaps = 26/361 (7%)
Query: 14 LFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSG-----LGFPINLPNGRTFFHR 68
+ L+ T S+AA + + ++ FGDS +DTG SG G + P G TFF R
Sbjct: 17 ILLFSTISTAATIPNIHRPFNKIYAFGDSFTDTGNSRSGEGPAGFGHLSSPPYGMTFFRR 76
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLD----SLSGSSF-TNGANFAVVGSSTLPKYVP 123
T R SDGRL ID + +S+N FL PYL + +G++ T+G NFAV GS+ +
Sbjct: 77 PTNRYSDGRLTIDFVAESMNLPFLPPYLSLKTTNANGTATDTHGVNFAVSGSTVIKHAFF 136
Query: 124 FSLNIQVMQFLHFKARTLELVSAGAKNVINDEG-------FRAALYLI-DIGQNDLADSF 175
N+ L +++E A + + G F+ +L+ I +IG ND A +
Sbjct: 137 VKNNLS----LDMTPQSIETELAWFEKYLETLGTNQKVSLFKDSLFWIGEIGVNDYAYTL 192
Query: 176 AKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDL 235
++ I+++ I+ +++L N+G + V GCL ++LA + D D
Sbjct: 193 GSTVSS-DTIRELS--ISTFTRFLETLLNKGVKYMLVQGHPATGCLTLAMSLAAEDDRDS 249
Query: 236 FGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLT 295
GC+ S N+ + N AL ++LR + AT+VY D + +I + +KYG +
Sbjct: 250 LGCVQSANNQSYTHNLALQSKLKQLRIKYPSATIVYADYWNAYRAVIKHPSKYGITEKFK 309
Query: 296 VCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTP 355
CCG G PYNF + TCG VC + ++Y+NWDG H TEA +A L ++ P
Sbjct: 310 ACCGI-GEPYNFQVFQTCGTDAATVCKDPNQYINWDGVHLTEAMYKVMADMFLDGTFTRP 368
Query: 356 R 356
R
Sbjct: 369 R 369
>At1g28670 lipase
Length = 384
Score = 166 bits (419), Expect = 2e-41
Identities = 119/384 (30%), Positives = 176/384 (44%), Gaps = 29/384 (7%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGL-----TSGLGF 55
M+S+L + L L+ T+ A S+C + FGDS +DTG + L
Sbjct: 1 MASSLKKLISSFLLVLYSTTIIVASSESRCRRFKSIISFGDSIADTGNYLHLSDVNHLPQ 60
Query: 56 PINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGS 115
LP G +FFH +GR S+GRL+ID + + L ++ PY S SF G NFAV G+
Sbjct: 61 SAFLPYGESFFHPPSGRASNGRLIIDFIAEFLGLPYVPPYFGS-QNVSFEQGINFAVYGA 119
Query: 116 STLPK-----------YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLI 164
+ L + + SL++Q+ F + K ++ D + + +
Sbjct: 120 TALDRAFLLGKGIESDFTNVSLSVQLDTFKQILPNLCASSTRDCKEMLGD----SLILMG 175
Query: 165 DIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKI 224
+IG ND F + + ++ + +P ++ I +A+ L + G + F V P GC
Sbjct: 176 EIGGNDYNYPFFEGKSINEIKELVPLIVKAISSAIVDLIDLGGKTFLVPGGFPTGCSAAY 235
Query: 225 LALAQ---KKDLD-LFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHD 280
L L Q +KD D L GC N NE L ++L+ ++Y D + +
Sbjct: 236 LTLFQTVAEKDQDPLTGCYPLLNEFGEHHNEQLKTELKRLQKFYPHVNIIYADYHNSLYR 295
Query: 281 LIANATKYGFSN-PLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAA 339
KYGF N PL CCG GG YNF + CG G C S YVNWDG H TEAA
Sbjct: 296 FYQEPAKYGFKNKPLAACCGVGGK-YNFTIGKECGYEGVNYCQNPSEYVNWDGYHLTEAA 354
Query: 340 NTFIASKILSTDYSTPRTPFEFFC 363
+ IL+ Y+TP F++ C
Sbjct: 355 YQKMTEGILNGPYATP--AFDWSC 376
>At1g28590 putative lipase (At1g28590)
Length = 403
Score = 165 bits (417), Expect = 4e-41
Identities = 117/347 (33%), Positives = 165/347 (46%), Gaps = 32/347 (9%)
Query: 39 FGDSNSDTGGLTSGLGFPINLPN------GRTFFHRSTGRLSDGRLVIDLLCQSLNTRFL 92
FGDS +DTG L GL P +LP G TFFH TGR SDGRL+ID + + L +
Sbjct: 40 FGDSIADTGNLL-GLSDPNDLPASAFPPYGETFFHHPTGRYSDGRLIIDFIAEFLGFPLV 98
Query: 93 TPYLDSLSGSSFTNGANFAVVGSSTL-PKYVP----------FSLNIQVMQFLHFKARTL 141
P+ ++F G NFAV G++ L P ++ SL++Q+ F
Sbjct: 99 PPFY-GCQNANFKKGVNFAVAGATALEPSFLEERGIHSTITNVSLSVQLRSFTESLPNL- 156
Query: 142 ELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKS 201
G+ + D A + + +IG ND + + +V + +P VI I +A+
Sbjct: 157 ----CGSPSDCRDMIENALILMGEIGGNDYNFALFQRKPVKEVEELVPFVIATISSAITE 212
Query: 202 LYNEGARKFWVHNTGPLGCLPKILAL---AQKKDLD-LFGCLSSYNSAARLFNEALYHSS 257
L G R F V P+G L L + K++ D L GCL N + +N+ L
Sbjct: 213 LVCMGGRTFLVPGNFPIGYSASYLTLYKTSNKEEYDPLTGCLKWLNDFSEYYNKQLQEEL 272
Query: 258 QKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSN-PLTVCCGFGGPPYNFDLRVTCGQP 316
LR ++Y D Y L K+GF N PL CCG GG YNF+ CG
Sbjct: 273 NGLRKLYPHVNIIYADYYNALLRLFQEPAKFGFMNRPLPACCGVGGS-YNFNFSRRCGSV 331
Query: 317 GYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
G + CD+ S+YVN+DG H TEAA I+ +L Y+ P PF++ C
Sbjct: 332 GVEYCDDPSQYVNYDGIHMTEAAYRLISEGLLKGPYAIP--PFKWSC 376
>At1g28600 lipase like protein
Length = 393
Score = 164 bits (414), Expect = 9e-41
Identities = 120/376 (31%), Positives = 168/376 (43%), Gaps = 24/376 (6%)
Query: 8 SLHLLFLFLWITSSSAAVMM-SKCDVPAVLFVFGDSNSDTG---GLTSGLGFPINL--PN 61
SL L +FL+ T V + C + FGDS +DTG GL+ P+ P
Sbjct: 3 SLDSLVIFLFSTLFVTIVSSETPCPNFKSIISFGDSIADTGNLVGLSDRNQLPVTAFPPY 62
Query: 62 GRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKY 121
G TFFH TGR DGR+++D + + + ++ PY S +F G NFAV G++ L
Sbjct: 63 GETFFHHPTGRSCDGRIIMDFIAEFVGLPYVPPYFGS-KNRNFDKGVNFAVAGATALKSS 121
Query: 122 VPFSLNIQV-------MQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADS 174
IQ +Q FK L G+ + D A + + +IG ND
Sbjct: 122 FLKKRGIQPHTNVSLGVQLKSFKKSLPNL--CGSPSDCRDMIGNALILMGEIGGNDYNFP 179
Query: 175 FAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLD 234
F +V + +P VI I + + L G + F V P+GC L L + + D
Sbjct: 180 FFNRKPVKEVEELVPFVIASISSTITELIGMGGKTFLVPGEFPIGCSVVYLTLYKTSNKD 239
Query: 235 LF----GCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGF 290
+ GCL N +E L +LR ++Y D Y + K+GF
Sbjct: 240 EYDPSTGCLKWLNKFGEYHSEKLKVELNRLRKLYPHVNIIYADYYNSLLRIFKEPAKFGF 299
Query: 291 -SNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILS 349
P CCG GGP YNF+ CG G + C + S+YV WDG H TEAA +IA IL+
Sbjct: 300 MERPFPACCGIGGP-YNFNFTRKCGSVGVKSCKDPSKYVGWDGVHMTEAAYKWIADGILN 358
Query: 350 TDYSTPRTPFEFFCHR 365
Y+ P PF+ C R
Sbjct: 359 GPYANP--PFDRSCLR 372
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,222,319
Number of Sequences: 26719
Number of extensions: 354061
Number of successful extensions: 1240
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 754
Number of HSP's gapped (non-prelim): 129
length of query: 365
length of database: 11,318,596
effective HSP length: 101
effective length of query: 264
effective length of database: 8,619,977
effective search space: 2275673928
effective search space used: 2275673928
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0223.2


