

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0223.17
(88 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
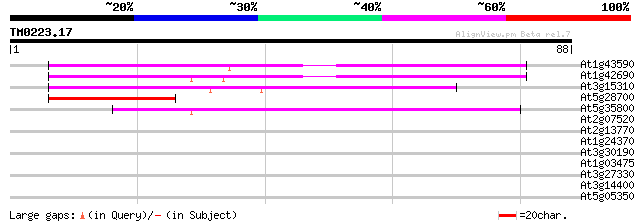
Score E
Sequences producing significant alignments: (bits) Value
At1g43590 hypothetical protein 58 7e-10
At1g42690 unknown protein 55 6e-09
At3g15310 unknown protein 44 2e-05
At5g28700 putative protein 40 2e-04
At5g35800 putative protein 39 5e-04
At2g07520 pseudogene 35 0.005
At2g13770 hypothetical protein 33 0.025
At1g24370 hypothetical protein 33 0.032
At3g30190 hypothetical protein 31 0.094
At1g03475 coproporphyrinogen III oxidase (LIN2) 30 0.27
At3g27330 hypothetical protein 27 1.8
At3g14400 unknown protein 27 1.8
At5g05350 putative protein 25 5.2
>At1g43590 hypothetical protein
Length = 168
Score = 58.2 bits (139), Expect = 7e-10
Identities = 37/85 (43%), Positives = 43/85 (50%), Gaps = 15/85 (17%)
Query: 7 ILYACIILHNMIVEDEREGYNGNFVYD----------QVDNDIITAEVSNGPIPSFATFL 56
I+ ACIILHNMIVEDER+GY V D QVD T SN +
Sbjct: 82 IMRACIILHNMIVEDERDGYTQYDVSDFAHPESASSSQVDFTYSTDMPSN-----LGNMM 136
Query: 57 ERRGHMSQREIHRQLQADLVEHIWE 81
R + R H L+ADLVEHIW+
Sbjct: 137 ATRTRLRDRTKHEHLKADLVEHIWQ 161
>At1g42690 unknown protein
Length = 333
Score = 55.1 bits (131), Expect = 6e-09
Identities = 34/85 (40%), Positives = 44/85 (51%), Gaps = 15/85 (17%)
Query: 7 ILYACIILHNMIVEDEREGYN----GNFVY------DQVDNDIITAEVSNGPIPSFATFL 56
I+ ACIILHNMIVED+R+GY FV+ QVD T SN +
Sbjct: 247 IMRACIILHNMIVEDKRDGYTQFDVSEFVHPESASSSQVDFTYATVMPSN-----LGNMM 301
Query: 57 ERRGHMSQREIHRQLQADLVEHIWE 81
+ R H +L+ADLVEH+W+
Sbjct: 302 ATGARVRDRIKHEELKADLVEHVWQ 326
>At3g15310 unknown protein
Length = 415
Score = 43.5 bits (101), Expect = 2e-05
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query: 7 ILYACIILHNMIVEDEREGYNGNF---VYDQVDND---IITAEVSNGPIPSFATFLERRG 60
I+ ACIILHNMIVEDER+GYN F + QV+ + + S G + + R
Sbjct: 346 IMKACIILHNMIVEDERDGYNIQFDVSEFLQVEGNQTPQVDLSYSTGMPLNIENMMGMRN 405
Query: 61 HMSQREIHRQ 70
+ + +H+Q
Sbjct: 406 QLRDQNMHQQ 415
>At5g28700 putative protein
Length = 292
Score = 40.0 bits (92), Expect = 2e-04
Identities = 17/20 (85%), Positives = 19/20 (95%)
Query: 7 ILYACIILHNMIVEDEREGY 26
I+ ACIILHNMIVEDER+GY
Sbjct: 234 IMRACIILHNMIVEDERDGY 253
>At5g35800 putative protein
Length = 73
Score = 38.9 bits (89), Expect = 5e-04
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Query: 17 MIVEDEREGYN---GNFVYDQVDNDIITAEVSNGPIPSFATFLERRGHMSQREIHRQLQA 73
MIV DERE Y ++ + S +P FA+ + R + +H +LQA
Sbjct: 1 MIVNDERETYAQHWSDYNQSEASGSSTPQPFSIEVLPEFASHVRARSELRDSNLHHELQA 60
Query: 74 DLVEHIW 80
LV++IW
Sbjct: 61 GLVKYIW 67
>At2g07520 pseudogene
Length = 222
Score = 35.4 bits (80), Expect = 0.005
Identities = 17/29 (58%), Positives = 23/29 (78%), Gaps = 1/29 (3%)
Query: 7 ILYACIILHNMIVEDEREGYNGNFV-YDQ 34
I+ +CIILHNMIVE+ER+ Y ++ YDQ
Sbjct: 157 IMRSCIILHNMIVENERDTYAQHWTDYDQ 185
>At2g13770 hypothetical protein
Length = 244
Score = 33.1 bits (74), Expect = 0.025
Identities = 19/52 (36%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Query: 7 ILYACIILHNMIVEDEREGYNGNFVYDQVDNDIITAEVSNGPIPSFATFLER 58
I+ +CII+HNMI+EDER+ + ++V+ I E++ F FL R
Sbjct: 194 IMTSCIIMHNMIIEDERD--IDAPIEERVEVPIEEVEMTGDDDTRFQEFLAR 243
>At1g24370 hypothetical protein
Length = 413
Score = 32.7 bits (73), Expect = 0.032
Identities = 12/18 (66%), Positives = 17/18 (93%)
Query: 7 ILYACIILHNMIVEDERE 24
I+ +CII+HNMI+EDER+
Sbjct: 229 IMTSCIIMHNMIIEDERD 246
>At3g30190 hypothetical protein
Length = 263
Score = 31.2 bits (69), Expect = 0.094
Identities = 11/15 (73%), Positives = 15/15 (99%)
Query: 10 ACIILHNMIVEDERE 24
+CII+HNMI+EDER+
Sbjct: 225 SCIIMHNMIIEDERD 239
>At1g03475 coproporphyrinogen III oxidase (LIN2)
Length = 386
Score = 29.6 bits (65), Expect = 0.27
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Query: 21 DEREGYNGNFVYDQVDND-----IITAEVSNGPIPSFATFLERRGHMSQREIHRQLQ 72
DER G G F D D D E +N +P++ +E+R M E H+ Q
Sbjct: 263 DERRGLGGIFFDDLNDYDQEMLLSFATECANSVVPAYIPIVEKRKDMEFTEQHKAWQ 319
>At3g27330 hypothetical protein
Length = 913
Score = 26.9 bits (58), Expect = 1.8
Identities = 18/62 (29%), Positives = 25/62 (40%), Gaps = 9/62 (14%)
Query: 4 CMQILYACIILHNMIVEDEREGYNGNFVYDQVDNDIITAEVSNGPIPSFATFLERRGHMS 63
C A +L ++ G F+YD +D I AE+ N LERRG+
Sbjct: 283 CTMTRNAAAVLREWVMYHAGIGVQRWFIYDNNSDDDIIAEIEN---------LERRGYNI 333
Query: 64 QR 65
R
Sbjct: 334 SR 335
>At3g14400 unknown protein
Length = 661
Score = 26.9 bits (58), Expect = 1.8
Identities = 10/21 (47%), Positives = 16/21 (75%)
Query: 66 EIHRQLQADLVEHIWELSESR 86
+I +L+ADLV IWE+S+ +
Sbjct: 639 QIPEELKADLVNRIWEISKKK 659
>At5g05350 putative protein
Length = 410
Score = 25.4 bits (54), Expect = 5.2
Identities = 17/56 (30%), Positives = 24/56 (42%), Gaps = 1/56 (1%)
Query: 32 YD-QVDNDIITAEVSNGPIPSFATFLERRGHMSQREIHRQLQADLVEHIWELSESR 86
YD Q D + V GP+ + LERR + ++ + V IWE E R
Sbjct: 266 YDPQGDEENQVQAVGEGPVSNRKLSLERRYSFASNDVSNPEWREGVLDIWEAQEVR 321
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,887,341
Number of Sequences: 26719
Number of extensions: 65581
Number of successful extensions: 236
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 223
Number of HSP's gapped (non-prelim): 13
length of query: 88
length of database: 11,318,596
effective HSP length: 64
effective length of query: 24
effective length of database: 9,608,580
effective search space: 230605920
effective search space used: 230605920
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0223.17


