

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0221.11
(142 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
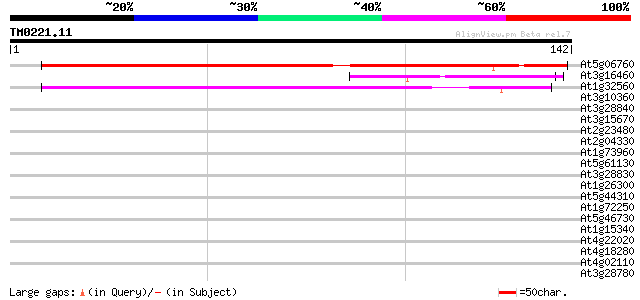
Score E
Sequences producing significant alignments: (bits) Value
At5g06760 late embryogenesis abundant protein LEA like 152 4e-38
At3g16460 putative jasmonate inducible protein (MDC8.9/AT3g16460) 47 3e-06
At1g32560 late-embryogenesis abundant protein, putative 45 1e-05
At3g10360 putative RNA binding protein 37 0.003
At3g28840 unknown protein 37 0.005
At3g15670 LEA76 homologue type2 35 0.010
At2g23480 hypothetical protein 35 0.018
At2g04330 hypothetical protein 34 0.030
At1g73960 unknown protein 33 0.039
At5g61130 predicted GPI-anchored protein 33 0.051
At3g28830 hypothetical protein 33 0.051
At1g26300 unknown protein 33 0.051
At5g44310 late embryogenesis abundant protein-like 33 0.067
At1g72250 kinesin, putative 33 0.067
At5g46730 unknown protein 32 0.087
At1g15340 unknown protein 32 0.087
At4g22020 glycine-rich protein 32 0.11
At4g18280 unknown protein 32 0.11
At4g02110 predicted protein of unknown function 32 0.11
At3g28780 histone-H4-like protein 32 0.11
>At5g06760 late embryogenesis abundant protein LEA like
Length = 158
Score = 152 bits (385), Expect = 4e-38
Identities = 86/160 (53%), Positives = 101/160 (62%), Gaps = 32/160 (20%)
Query: 9 ETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAELDKLA 68
++MKETA+NI ASAKSG +KTKAT++EKAEK+ DPVQK+MATQ KE K++QAE+ K
Sbjct: 2 QSMKETASNIAASAKSGMDKTKATLEEKAEKMKTRDPVQKQMATQVKEDKINQAEMQKRE 61
Query: 69 AREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTG------- 121
R+HNAA K++A A GTG G GTA +STTG G G HQ SALPGHGTG
Sbjct: 62 TRQHNAAMKEAAGA----GTGLGLGTATHSTTGQVGHGTGTHQMSALPGHGTGQLTDRVV 117
Query: 122 --------------------HNTRVGGNPNATGYGTGGTY 141
HNT VGG ATGYGTGG Y
Sbjct: 118 EGTAVTDPIGRNTGTGRTTAHNTHVGGG-GATGYGTGGGY 156
>At3g16460 putative jasmonate inducible protein (MDC8.9/AT3g16460)
Length = 705
Score = 47.4 bits (111), Expect = 3e-06
Identities = 25/52 (48%), Positives = 26/52 (49%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
GTGTGTGT + TG G G T G GTG T GG TG GTG
Sbjct: 198 GTGTGTGTGTGTGTGGTGTGTGTGGTGTGTGTGTGTGTGTGGTGTGTGTGTG 249
Score = 42.7 bits (99), Expect = 6e-05
Identities = 25/54 (46%), Positives = 26/54 (47%), Gaps = 10/54 (18%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGGT 140
GTGTGTGT + TG T G GTG T GG TG GTGGT
Sbjct: 137 GTGTGTGTGTGTGTG----------TGTGTGTGTGTGTGTGGTGTGTGTGTGGT 180
Score = 42.7 bits (99), Expect = 6e-05
Identities = 27/55 (49%), Positives = 27/55 (49%), Gaps = 2/55 (3%)
Query: 87 GTGTGTGTAAYST-TGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGGT 140
GTGTGTGT T TG G G T G GTG T G TG GTGGT
Sbjct: 170 GTGTGTGTGGTGTGTGTGGTGTGT-GTGTGTGTGTGTGTGTGTGGTGTGTGTGGT 223
Score = 40.0 bits (92), Expect = 4e-04
Identities = 23/52 (44%), Positives = 25/52 (47%), Gaps = 4/52 (7%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
GTGTGTGT + TG G G + GTG T GG TG GTG
Sbjct: 151 GTGTGTGTGTGTGTGTGGTGTGTGTGTG----GTGTGTGTGGTGTGTGTGTG 198
Score = 37.4 bits (85), Expect = 0.003
Identities = 21/52 (40%), Positives = 22/52 (41%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
GTGTGTG T G G + GTG T GG TG GTG
Sbjct: 181 GTGTGTGGTGTGTGTGTGTGTGTGTGTGTGTGGTGTGTGTGGTGTGTGTGTG 232
Score = 37.0 bits (84), Expect = 0.004
Identities = 26/63 (41%), Positives = 27/63 (42%), Gaps = 12/63 (19%)
Query: 87 GTGTGT---------GTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGT 137
GTGTGT GT + TG G G T G GTG T G TG GT
Sbjct: 155 GTGTGTGTGTGTGGTGTGTGTGTGGTGTGTGTGGTGTGTGTGTGTGT---GTGTGTGTGT 211
Query: 138 GGT 140
GGT
Sbjct: 212 GGT 214
Score = 35.8 bits (81), Expect = 0.008
Identities = 22/52 (42%), Positives = 24/52 (45%), Gaps = 5/52 (9%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
GTGTGTGT + TG G + G GTG T G TG GTG
Sbjct: 188 GTGTGTGTGTGTGTGT-----GTGTGTGTGGTGTGTGTGGTGTGTGTGTGTG 234
Score = 33.1 bits (74), Expect = 0.051
Identities = 23/52 (44%), Positives = 24/52 (45%), Gaps = 5/52 (9%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
GTGTGTGT + TG G G T G GTG G TG GTG
Sbjct: 190 GTGTGTGTGTGTGTGT-GTGTGTGGT----GTGTGTGGTGTGTGTGTGTGTG 236
Score = 31.6 bits (70), Expect = 0.15
Identities = 21/53 (39%), Positives = 22/53 (40%), Gaps = 1/53 (1%)
Query: 89 GTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNP-NATGYGTGGT 140
GTGTGT T G G + G GTG T G A G TGGT
Sbjct: 213 GTGTGTGTGGTGTGTGTGTGTGTGTGGTGTGTGTGTGSGAQKLEAQGNSTGGT 265
Score = 28.5 bits (62), Expect = 1.3
Identities = 16/35 (45%), Positives = 18/35 (50%), Gaps = 1/35 (2%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTG 121
GTGTGTGT T G GA + A G+ TG
Sbjct: 230 GTGTGTGTGGTGTGTGTGTGSGAQKLEA-QGNSTG 263
>At1g32560 late-embryogenesis abundant protein, putative
Length = 134
Score = 45.1 bits (105), Expect = 1e-05
Identities = 36/130 (27%), Positives = 60/130 (45%), Gaps = 10/130 (7%)
Query: 9 ETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAELDKLA 68
++ K+ +++ ++AK +A EKAE+ A +KE+A Q+++AK ++A +D
Sbjct: 2 QSAKQKISDMASTAKEKMVICQAKADEKAERAMARTKEEKEIAHQRRKAKEAEANMDMHM 61
Query: 69 AREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHN-TRVG 127
A+ +A K A + + T G + A M GHG GHN T V
Sbjct: 62 AKAAHAEDKLMAKQSHYHVTDHGPHVPQQAPVPAPAPVM---------GHGYGHNPTGVT 112
Query: 128 GNPNATGYGT 137
P T + T
Sbjct: 113 SVPPQTYHPT 122
>At3g10360 putative RNA binding protein
Length = 1003
Score = 37.4 bits (85), Expect = 0.003
Identities = 39/151 (25%), Positives = 60/151 (38%), Gaps = 35/151 (23%)
Query: 26 KEKTKATVQEKAEKVTAGDPVQ-------KEMATQKKEAKVSQAELD------------- 65
K+ T T + E TA D EM+ K+ SQ +++
Sbjct: 331 KKNTFGTAIQNCESYTAADVADTLSRLNMSEMSQVKENHMQSQLQVELENQSDVMRYIPN 390
Query: 66 --KLAAREHNAATKQSAAAAAHMGTGTGTGTA-AYSTTGAHGQPMGAHQTSALPG-HGTG 121
K A R+ N A + +A+ G +G G + ST G+HGQ +TS+ + T
Sbjct: 391 GHKKALRQQNTAETKDHLFSANYGGMSGYGASLGASTVGSHGQVNIPKRTSSSASLYSTS 450
Query: 122 HNTRVG-----------GNPNATGYGTGGTY 141
++R+G GN N T + T G Y
Sbjct: 451 DHSRLGSVGLSDVNIRNGNINGTDFSTAGGY 481
>At3g28840 unknown protein
Length = 391
Score = 36.6 bits (83), Expect = 0.005
Identities = 34/142 (23%), Positives = 52/142 (35%), Gaps = 8/142 (5%)
Query: 4 AKKATETMKETAANIGASAKSGKE-----KTKATVQEKAEKV-TAGDPVQKEMATQKKEA 57
+ K +T+ +G +SG + K + A+ T G V A +
Sbjct: 118 SSKVMDTLLSMGKTLGTQQQSGSSVMSLGQRKEMIMSMAKWAQTIGQFVVSAAAKSGNKI 177
Query: 58 KVSQAELDKLAAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPG 117
+S +D + A A S A+ TGTG+ A + A G A T+A G
Sbjct: 178 DISSLGIDGIDASAAGAG--DSTASGGVSATGTGSYGAGAGGSSASGSDTAAGGTTATGG 235
Query: 118 HGTGHNTRVGGNPNATGYGTGG 139
+ G A+G GT G
Sbjct: 236 TTAAGGSTAAGGTTASGAGTAG 257
Score = 31.2 bits (69), Expect = 0.19
Identities = 22/67 (32%), Positives = 25/67 (36%)
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGY 135
T S A A T +G GTAA TT A +A G G T GG + G
Sbjct: 277 TTASDAGTAAGTTASGAGTAAGGTTAAGAGAAAGAGAAAGAGAAAGGTTAAGGVSGSYGS 336
Query: 136 GTGGTYN 142
G N
Sbjct: 337 SGGSASN 343
Score = 28.5 bits (62), Expect = 1.3
Identities = 20/65 (30%), Positives = 25/65 (37%), Gaps = 1/65 (1%)
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGY 135
T + AA T +G GTA Y T GA T+A + T G + G
Sbjct: 237 TAAGGSTAAGGTTASGAGTAGYGATAGGATASGA-GTAAGGTTASDAGTAAGTTASGAGT 295
Query: 136 GTGGT 140
GGT
Sbjct: 296 AAGGT 300
Score = 28.1 bits (61), Expect = 1.6
Identities = 22/64 (34%), Positives = 28/64 (43%), Gaps = 5/64 (7%)
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGY 135
T A AAA G G G AA TT A G G++ +S G+ NT G + T
Sbjct: 301 TAAGAGAAAGAGAAAGAGAAAGGTTAA-GGVSGSYGSSG----GSASNTNGGTSSCGTSA 355
Query: 136 GTGG 139
+ G
Sbjct: 356 CSNG 359
>At3g15670 LEA76 homologue type2
Length = 225
Score = 35.4 bits (80), Expect = 0.010
Identities = 22/83 (26%), Positives = 36/83 (42%)
Query: 6 KATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAELD 65
KA E + + A+ G++KT T Q +K K+ +Q +A +A
Sbjct: 16 KAQEKTGQAMGTMRDKAEEGRDKTSQTAQTAQQKAHETAQSAKDKTSQTAQAAQQKAHET 75
Query: 66 KLAAREHNAATKQSAAAAAHMGT 88
+A+E + T Q+A AH T
Sbjct: 76 AQSAKEKTSQTAQTAQQKAHETT 98
Score = 30.4 bits (67), Expect = 0.33
Identities = 29/105 (27%), Positives = 42/105 (39%), Gaps = 11/105 (10%)
Query: 5 KKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMA---TQKKEAKVSQ 61
+KA ET + +A++ ++K T Q EK + ++ A TQ + K SQ
Sbjct: 48 QKAHETAQSAKDKTSQTAQAAQQKAHETAQSAKEKTSQTAQTAQQKAHETTQAAKEKTSQ 107
Query: 62 A------ELDKLAA--REHNAATKQSAAAAAHMGTGTGTGTAAYS 98
A DK + E A K A AA T G A Y+
Sbjct: 108 AGDKAREAKDKAGSYLSETGEAIKNKAQDAAQYTKETAQGAAQYT 152
>At2g23480 hypothetical protein
Length = 705
Score = 34.7 bits (78), Expect = 0.018
Identities = 33/97 (34%), Positives = 42/97 (43%), Gaps = 9/97 (9%)
Query: 18 IGASAKSGKEKTKATVQEKAEKVTAGD-----PVQKE--MATQKKEAKVSQAELDKLAAR 70
I A A + EK KA ++ EKV A P +KE A KKE + A+ K+ A
Sbjct: 16 IEAPAPAKTEKVKAPAEKVKEKVPAKKAKVQAPAKKEKVQAPAKKEKVQAPAKKAKVQAT 75
Query: 71 EHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPM 107
AT Q+ A A M T T A +T PM
Sbjct: 76 AKTTATAQTTATA--MATTAAPTTTAPTTAPTTESPM 110
>At2g04330 hypothetical protein
Length = 564
Score = 33.9 bits (76), Expect = 0.030
Identities = 33/91 (36%), Positives = 40/91 (43%), Gaps = 8/91 (8%)
Query: 18 IGASAKSGKEKTKATVQEKAEKVTAGDPVQKE--MATQKKEAKVSQAELDKLAAREHNAA 75
I A A + EK KA AEKV P +K A KKE + A+ K+ A A
Sbjct: 16 IEAPAPTKTEKVKAP----AEKVKEKVPAKKAKVQAPAKKEKVQAPAKKAKVQAPAKTTA 71
Query: 76 TKQSAAAAAHMGTGTGTGTAAYSTTGAHGQP 106
T Q+ A A M T TAA +TT P
Sbjct: 72 TAQTIATA--MATNAAPTTAALTTTAPTTAP 100
>At1g73960 unknown protein
Length = 1273
Score = 33.5 bits (75), Expect = 0.039
Identities = 23/60 (38%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Query: 25 GKEKTKATVQEKAEKVTAG-DPVQKEMATQKKEAKVSQAELDKLAAREHNAATKQSAAAA 83
GKEK K +EK EK DPV E KKE K + E+ KL + + A K+ + A
Sbjct: 1169 GKEKEKKKDKEKKEKKRKREDPVYLEKKRLKKEKKRKEKEMAKLVSSTTDPAKKKIESVA 1228
>At5g61130 predicted GPI-anchored protein
Length = 201
Score = 33.1 bits (74), Expect = 0.051
Identities = 27/91 (29%), Positives = 35/91 (37%), Gaps = 9/91 (9%)
Query: 53 QKKEAKVSQAELDKLAAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQT 112
QKK D A ++ + A +G+ T TT G P T
Sbjct: 75 QKKGQSPGSCNFDGTATPTNSDPSYTGCAFPTSASGSSGSTTVTPGTTNPKGSPT----T 130
Query: 113 SALPGHGTGHNTRVGGNPNATGYG---TGGT 140
+ LPG GT N+ GNP +G TGGT
Sbjct: 131 TTLPGSGT--NSPYSGNPTNGVFGGNSTGGT 159
>At3g28830 hypothetical protein
Length = 536
Score = 33.1 bits (74), Expect = 0.051
Identities = 22/85 (25%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Query: 15 AANIGASAKSGKEKTKATVQEKA-EKVTAGDPVQKEMATQKKEAKVSQAELDKLAAREHN 73
A++ +S KSG + +V K+ E ++G +AT+ KE+ A + +
Sbjct: 197 ASSESSSTKSGSVSSSGSVSTKSKESSSSGSSASGSVATKSKESSGGSAATKSKESSGGS 256
Query: 74 AATKQSAAAAAHMGTGTGTGTAAYS 98
AATK ++ TG +G+ + S
Sbjct: 257 AATKSKESSGGSATTGKTSGSPSGS 281
>At1g26300 unknown protein
Length = 213
Score = 33.1 bits (74), Expect = 0.051
Identities = 29/102 (28%), Positives = 49/102 (47%), Gaps = 12/102 (11%)
Query: 3 GAKKATETMKETAANIGASAKSGKEKTKATVQ--EKAEKVTAGDPVQKEMATQKKEAKVS 60
G K T T +A + ++ K+ + ++ +KA++ A D V A K++K
Sbjct: 16 GKKSQTITQSSSATFVNLVTETAKKSKELALEASKKADQFNAADFV----AETAKKSKEF 71
Query: 61 QAELDKLAAREHNAATKQS------AAAAAHMGTGTGTGTAA 96
AE K A + AA KQ+ + A +GTGTG+G+ +
Sbjct: 72 AAEASKKADQFKVAALKQADQIQNIKSIADIIGTGTGSGSGS 113
>At5g44310 late embryogenesis abundant protein-like
Length = 331
Score = 32.7 bits (73), Expect = 0.067
Identities = 22/65 (33%), Positives = 28/65 (42%)
Query: 2 QGAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQ 61
+GA +A + ET A KEKTK +E EKV G + A KE +
Sbjct: 157 EGASRAADKAYETKEKAKDKAYDVKEKTKDFAEETKEKVNEGASRAADKAYDVKEKTKNY 216
Query: 62 AELDK 66
AE K
Sbjct: 217 AEQTK 221
Score = 32.3 bits (72), Expect = 0.087
Identities = 26/87 (29%), Positives = 36/87 (40%), Gaps = 4/87 (4%)
Query: 2 QGAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQ 61
+GA +A + ET A KEKTK +E +KV G + A + KE +
Sbjct: 117 EGASRAADKAYETKEKAKDKAYDVKEKTKDYAEEAKDKVNEGASRAADKAYETKEKAKDK 176
Query: 62 A----ELDKLAAREHNAATKQSAAAAA 84
A E K A E + A+ AA
Sbjct: 177 AYDVKEKTKDFAEETKEKVNEGASRAA 203
Score = 30.4 bits (67), Expect = 0.33
Identities = 21/77 (27%), Positives = 33/77 (42%)
Query: 4 AKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAE 63
A++ E + E A+ A KEKTK ++ +KV G + A + K+ AE
Sbjct: 188 AEETKEKVNEGASRAADKAYDVKEKTKNYAEQTKDKVNEGASRAADKAEETKDKAKDYAE 247
Query: 64 LDKLAAREHNAATKQSA 80
K A + K+ A
Sbjct: 248 DSKEKAEDMAHGFKEKA 264
>At1g72250 kinesin, putative
Length = 1195
Score = 32.7 bits (73), Expect = 0.067
Identities = 18/77 (23%), Positives = 41/77 (52%), Gaps = 1/77 (1%)
Query: 1 MQGAKKATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVS 60
M+G + M+ET + A K K K T+Q+K +++ + V++++A Q + K++
Sbjct: 845 MKGKDEQIRKMEETMYGLEAKIKERDTKNK-TLQDKVKELESQLLVERKLARQHVDTKIA 903
Query: 61 QAELDKLAAREHNAATK 77
+ + + E+N + +
Sbjct: 904 EQQTKQQTEDENNTSKR 920
>At5g46730 unknown protein
Length = 268
Score = 32.3 bits (72), Expect = 0.087
Identities = 22/67 (32%), Positives = 26/67 (37%)
Query: 73 NAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNA 132
N A + A A+ G G G + G G GAH S G G G + A
Sbjct: 147 NGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGA 206
Query: 133 TGYGTGG 139
GYG GG
Sbjct: 207 GGYGGGG 213
Score = 32.0 bits (71), Expect = 0.11
Identities = 26/73 (35%), Positives = 30/73 (40%), Gaps = 6/73 (8%)
Query: 74 AATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGT-----GHNTRVGG 128
AA+ A+ A G G G G AA G G G SA G G+ GG
Sbjct: 94 AASSGGYASGAGEGGGGGYGGAAGGHAGGGGGGSGGGGGSAYGAGGEHASGYGNGAGEGG 153
Query: 129 NPNATGYGTGGTY 141
A+GYG GG Y
Sbjct: 154 GAGASGYG-GGAY 165
Score = 29.6 bits (65), Expect = 0.56
Identities = 20/68 (29%), Positives = 28/68 (40%), Gaps = 8/68 (11%)
Query: 79 SAAAAAHMGTGTGTGTAAYSTTGA-------HGQPMGAHQTSALPGHGTGHNTRVG-GNP 130
S + H G G G ++ +GA +G G H G G G + G G
Sbjct: 81 SGGGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAGGGGGGSGGGGGSAYGAGGE 140
Query: 131 NATGYGTG 138
+A+GYG G
Sbjct: 141 HASGYGNG 148
Score = 27.7 bits (60), Expect = 2.1
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 9/59 (15%)
Query: 81 AAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGG 139
A+ G G G G AA S G + + A G G G+ GG+ G G+GG
Sbjct: 80 ASGGGSGHGGGGGGAASS---------GGYASGAGEGGGGGYGGAAGGHAGGGGGGSGG 129
Score = 26.6 bits (57), Expect = 4.8
Identities = 21/60 (35%), Positives = 25/60 (41%), Gaps = 14/60 (23%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGA-------HQTSALPGHGTGHNTRVGGNPNATGYGTGG 139
G G G AY GAHG G+ + A G+G GH GG G+G GG
Sbjct: 213 GGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGHG---GGG----GHGGGG 265
>At1g15340 unknown protein
Length = 384
Score = 32.3 bits (72), Expect = 0.087
Identities = 27/106 (25%), Positives = 43/106 (40%), Gaps = 3/106 (2%)
Query: 2 QGAKKATETMKETAANIGASAKSGKE--KTKAT-VQEKAEKVTAGDPVQKEMATQKKEAK 58
+ A+ E KE I + K E KT+A V ++ EK AG Q E+A +KE +
Sbjct: 135 ENAEAEKEKEKEGVTEIAEAEKENNEGEKTEAEKVNKEGEKTEAGKEGQTEIAEAEKEKE 194
Query: 59 VSQAELDKLAAREHNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHG 104
+AE + A + + + G+G A + G
Sbjct: 195 GEKAEAENKEAEVVRDKKESMEVDTSELEKKAGSGEGAEEPSKVEG 240
>At4g22020 glycine-rich protein
Length = 396
Score = 32.0 bits (71), Expect = 0.11
Identities = 22/68 (32%), Positives = 26/68 (37%), Gaps = 5/68 (7%)
Query: 79 SAAAAAHMGTGTGTGTAAYSTTGAHGQPMG-----AHQTSALPGHGTGHNTRVGGNPNAT 133
S AA G G G G+ G +G G S G G G GG N +
Sbjct: 245 SGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGGGGGGGGGGNGS 304
Query: 134 GYGTGGTY 141
GYG+G Y
Sbjct: 305 GYGSGSGY 312
Score = 30.4 bits (67), Expect = 0.33
Identities = 21/60 (35%), Positives = 25/60 (41%), Gaps = 1/60 (1%)
Query: 80 AAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGG 139
+ A A G G +G A G G+ GA+ S G G G VGG G G GG
Sbjct: 162 SGAGAGAGVGGSSGGAGGGGGGGGGEGGGANGGSG-HGSGAGAGAGVGGAAGGVGGGGGG 220
Score = 30.0 bits (66), Expect = 0.43
Identities = 17/54 (31%), Positives = 22/54 (40%), Gaps = 5/54 (9%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTGGT 140
G G G G + Y G G G + G+GH G+ + G G GGT
Sbjct: 84 GYGAGAGMSGYGGVGGGGGRGGGEGGGSSANGGSGH-----GSGSGAGAGVGGT 132
Score = 30.0 bits (66), Expect = 0.43
Identities = 23/72 (31%), Positives = 29/72 (39%), Gaps = 11/72 (15%)
Query: 79 SAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQ-------TSALPGHGTGHNT----RVG 127
SA + G+G+G G TTG G G +S GHG+G VG
Sbjct: 112 SANGGSGHGSGSGAGAGVGGTTGGVGGGGGGGGGGGEGGGSSGGSGHGSGSGAGAGAGVG 171
Query: 128 GNPNATGYGTGG 139
G+ G G GG
Sbjct: 172 GSSGGAGGGGGG 183
Score = 28.1 bits (61), Expect = 1.6
Identities = 20/57 (35%), Positives = 24/57 (42%), Gaps = 4/57 (7%)
Query: 87 GTGTG---TGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVG-GNPNATGYGTGG 139
G+G G +G A G G G + GHG+G VG GN G G GG
Sbjct: 237 GSGAGGGVSGAAGGGGGGGGGGGSGGSKVGGGYGHGSGFGGGVGFGNSGGGGGGGGG 293
Score = 27.3 bits (59), Expect = 2.8
Identities = 17/52 (32%), Positives = 24/52 (45%), Gaps = 6/52 (11%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGTG 138
G+G G+G+ A + G G GA G G G GG +G+G+G
Sbjct: 155 GSGHGSGSGAGAGAGVGGSSGGA------GGGGGGGGGEGGGANGGSGHGSG 200
Score = 26.9 bits (58), Expect = 3.7
Identities = 16/61 (26%), Positives = 29/61 (47%), Gaps = 6/61 (9%)
Query: 78 QSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPNATGYGT 137
+ ++A+ G+G G+G+ A + G +G G G G GG+ +G+G+
Sbjct: 107 EGGGSSANGGSGHGSGSGAGAGVGGTTGGVGG------GGGGGGGGGEGGGSSGGSGHGS 160
Query: 138 G 138
G
Sbjct: 161 G 161
Score = 26.6 bits (57), Expect = 4.8
Identities = 18/68 (26%), Positives = 22/68 (31%), Gaps = 6/68 (8%)
Query: 72 HNAATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNTRVGGNPN 131
H A + + G G G G G+ H G G+G VGG
Sbjct: 81 HGEGYGAGAGMSGYGGVGGGGGRGGGEGGGSSANGGSGH------GSGSGAGAGVGGTTG 134
Query: 132 ATGYGTGG 139
G G GG
Sbjct: 135 GVGGGGGG 142
>At4g18280 unknown protein
Length = 148
Score = 32.0 bits (71), Expect = 0.11
Identities = 20/55 (36%), Positives = 25/55 (45%), Gaps = 2/55 (3%)
Query: 87 GTGTGTGTAAYSTTGAHGQPMGAHQTSALPGH--GTGHNTRVGGNPNATGYGTGG 139
G G G G+ + S TG G T GH GTGH GG+ + G G+ G
Sbjct: 86 GAGFGFGSGSGSGTGFGSGSGGGGATDGGSGHGSGTGHAGEGGGSGSGNGEGSPG 140
Score = 27.7 bits (60), Expect = 2.1
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 5/66 (7%)
Query: 74 AATKQSAAAAAHMGTGTGTGTAAYSTTGAHGQPMG-AHQTSALPGHGTGHNTRVGGNPNA 132
+ T +AAA T +G S+ AHG G + + PG G G+ + G +P+
Sbjct: 27 SGTPSNAAATPTSPTEGSSG----SSGSAHGPNWGYSWGWGSAPGGGYGYGSGSGSSPDG 82
Query: 133 TGYGTG 138
G G G
Sbjct: 83 GGKGAG 88
>At4g02110 predicted protein of unknown function
Length = 1293
Score = 32.0 bits (71), Expect = 0.11
Identities = 24/84 (28%), Positives = 36/84 (42%), Gaps = 8/84 (9%)
Query: 7 ATETMKETAANIGASAKSGKEKTKATVQE--------KAEKVTAGDPVQKEMATQKKEAK 58
AT+ AA + K GK + +ATV+E K KV+ + K T KK+
Sbjct: 823 ATKDPSYAAAQLEVDTKKGKRRKQATVEENRLQTPSVKKAKVSKKEDGAKANNTVKKDIW 882
Query: 59 VSQAELDKLAAREHNAATKQSAAA 82
+ AE+ + A + N S A
Sbjct: 883 IHSAEVKENVAVDENCGDVSSDGA 906
>At3g28780 histone-H4-like protein
Length = 614
Score = 32.0 bits (71), Expect = 0.11
Identities = 34/138 (24%), Positives = 51/138 (36%), Gaps = 11/138 (7%)
Query: 7 ATETMKETAANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAELDK 66
+ ET E+A+ A SG E T G+ +A+ S +
Sbjct: 406 SAETSDESASG---GAASGGESASGGAASGGSAETGGESTSSGVASGGSTGSESAS---- 458
Query: 67 LAAREHNAATKQSAAAAAHMGT--GTGTGTAAYSTTGAHGQPMGAHQTSALPGHGTGHNT 124
A +T+ + AAA T G+GT T S G G ++S+ G +
Sbjct: 459 -AGAASGGSTEANGGAAAGGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGTS 517
Query: 125 RVGGNPNAT-GYGTGGTY 141
+AT G +GGTY
Sbjct: 518 ESASGGSATAGGASGGTY 535
Score = 29.3 bits (64), Expect = 0.74
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 6/67 (8%)
Query: 78 QSAAAAAHMGTGTGTGTAAYSTTGAHGQPM--GAHQT---SALPGHGTGHNTRVGGNPNA 132
+SA+ A G G +G +A T G G+ G+ +T SA G +G + GG +
Sbjct: 280 ESASGGAASGAGAASGASA-KTGGESGEAASGGSAETGGESASAGAASGGSAETGGESGS 338
Query: 133 TGYGTGG 139
G +GG
Sbjct: 339 GGAASGG 345
Score = 27.7 bits (60), Expect = 2.1
Identities = 29/140 (20%), Positives = 55/140 (38%), Gaps = 6/140 (4%)
Query: 7 ATETMKETA---ANIGASAKSGKEKTKATVQEKAEKVTAGDPVQKEMATQKKEAKVSQAE 63
+ ET E+A A G SA++G E E + G T ++
Sbjct: 312 SAETGGESASAGAASGGSAETGGESGSGGAASGGESASGGATSGGSPETGGSAETGGESA 371
Query: 64 LDKLAAREHNAATKQSAAAAAHMG---TGTGTGTAAYSTTGAHGQPMGAHQTSALPGHGT 120
A+ +A+ +++ + G TG +G +A ++ + + SA G +
Sbjct: 372 SGGAASGGESASGGAASSGSVESGGESTGATSGGSAETSDESASGGAASGGESASGGAAS 431
Query: 121 GHNTRVGGNPNATGYGTGGT 140
G + GG ++G +GG+
Sbjct: 432 GGSAETGGESTSSGVASGGS 451
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.301 0.116 0.312
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,915,116
Number of Sequences: 26719
Number of extensions: 115758
Number of successful extensions: 825
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 512
Number of HSP's gapped (non-prelim): 321
length of query: 142
length of database: 11,318,596
effective HSP length: 89
effective length of query: 53
effective length of database: 8,940,605
effective search space: 473852065
effective search space used: 473852065
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0221.11


