

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
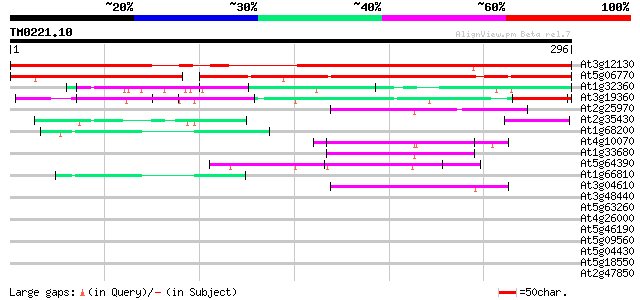
Query= TM0221.10
(296 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g12130 unknown protein 294 3e-80
At5g06770 unknown protein 201 5e-52
At1g32360 RING-H2 zinc finger like protein ATL6 66 3e-11
At3g19360 unknown protein 63 2e-10
At2g25970 pseudogene 62 5e-10
At2g35430 hypothetical protein 59 2e-09
At1g68200 putative zinc finger protein 51 7e-07
At4g10070 putative DNA-directed RNA polymerase 45 5e-05
At1g33680 single-strand nucleic acid-binding protein, putative 44 8e-05
At5g64390 HEN4 44 1e-04
At1g66810 hypothetical protein 42 3e-04
At3g04610 putative RNA-binding protein 41 7e-04
At3g48440 unknown protein 40 0.002
At5g63260 unknown protein 39 0.004
At4g26000 nucleic acid binding protein like 38 0.006
At5g46190 unknown protein 38 0.007
At5g09560 putative protein 38 0.007
At5g04430 putative RNA-binding protein 37 0.013
At5g18550 zinc finger -like protein 37 0.017
At2g47850 unknown protein 37 0.017
>At3g12130 unknown protein
Length = 248
Score = 294 bits (753), Expect = 3e-80
Identities = 164/301 (54%), Positives = 190/301 (62%), Gaps = 62/301 (20%)
Query: 1 MDFRKRGRPEPG-FNSNGG-IKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFL 58
MD RKRGRPE G FNSNGG KKSKQE+ES S+G+GSKSKPCTKFFST+GCPFGE+CHFL
Sbjct: 1 MDTRKRGRPEAGSFNSNGGGYKKSKQEMESYSTGLGSKSKPCTKFFSTSGCPFGENCHFL 60
Query: 59 HYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGD 118
HYVPGGYNAV+ M N+ PP+P S N + G +F
Sbjct: 61 HYVPGGYNAVSQMTNMG--------------PPIPQVSR--------NMQGSGNGGRF-- 96
Query: 119 KCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEAS 178
GR E PG + FG +ATA+ SV+AS
Sbjct: 97 ---------------------------------SGRGESGPGHVSNFGDSATARFSVDAS 123
Query: 179 LAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLT 238
LAGAIIGKGGV+SKQICRQTG KLSI++HE DPNL+NI LEG+ +QI EAS MVKDL+
Sbjct: 124 LAGAIIGKGGVSSKQICRQTGVKLSIQDHERDPNLKNIVLEGTLEQISEASAMVKDLIGR 183
Query: 239 LQMSA--PPKSNQGGAGGPGGHGH-HGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELR 295
L +A PP GG GG G G H +NFKTK+CE F+KG CTFG+RCHFAHG AELR
Sbjct: 184 LNSAAKKPPGGGLGGGGGMGSEGKPHPGSNFKTKICERFSKGNCTFGDRCHFAHGEAELR 243
Query: 296 K 296
K
Sbjct: 244 K 244
>At5g06770 unknown protein
Length = 240
Score = 201 bits (510), Expect = 5e-52
Identities = 107/206 (51%), Positives = 138/206 (66%), Gaps = 17/206 (8%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPI----------GHAPAGR 150
K++ C KF + GC FGD CHF H G + A + RP P GR
Sbjct: 38 KSKPCTKFFSTSGCPFGDNCHFLHYV-PGGYNAAAQMTNLRPPVSQVSRNMQGSGGPGGR 96
Query: 151 IGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESD 210
GR +P GP + FGA+ T+KISV+ASLAGAIIGKGG++SKQICR+TGAKLSI++HE D
Sbjct: 97 FSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKDHERD 155
Query: 211 PNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKL 270
PNL+ IELEG+F+QI AS MV++L+ L P+ G GGP G H GS N+KTK+
Sbjct: 156 PNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQ----GIGGPEGKPHPGS-NYKTKI 210
Query: 271 CENFAKGTCTFGERCHFAHGPAELRK 296
C+ ++KG CT+G+RCHFAHG +ELR+
Sbjct: 211 CDRYSKGNCTYGDRCHFAHGESELRR 236
Score = 133 bits (334), Expect = 1e-31
Identities = 62/93 (66%), Positives = 71/93 (75%), Gaps = 2/93 (2%)
Query: 1 MDFRKRGRPEPG--FNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFL 58
MD RKRGRPE NSNGG K+SKQE+ES+S+G+GSKSKPCTKFFST+GCPFG++CHFL
Sbjct: 1 MDARKRGRPEAAASHNSNGGFKRSKQEMESISTGLGSKSKPCTKFFSTSGCPFGDNCHFL 60
Query: 59 HYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPP 91
HYVPGGYNA A M NL P RN+ P
Sbjct: 61 HYVPGGYNAAAQMTNLRPPVSQVSRNMQGSGGP 93
Score = 28.5 bits (62), Expect = 4.5
Identities = 11/28 (39%), Positives = 16/28 (56%), Gaps = 1/28 (3%)
Query: 32 GVGSKSKPCTKFFSTAGCPFGESCHFLH 59
G K+K C ++ S C +G+ CHF H
Sbjct: 203 GSNYKTKICDRY-SKGNCTYGDRCHFAH 229
>At1g32360 RING-H2 zinc finger like protein ATL6
Length = 384
Score = 65.9 bits (159), Expect = 3e-11
Identities = 57/196 (29%), Positives = 77/196 (39%), Gaps = 33/196 (16%)
Query: 31 SGVGSKSKPCTKFFSTAGCPFGESCHFLH-------------YVPGGYNAVAHMMNLAPS 77
SG K + C KF++ GCP+GESC FLH PGGY + + S
Sbjct: 169 SGRSFKGRHCKKFYTEEGCPYGESCTFLHDEASRNRESVAISLGPGGYGSGGGGGSGGGS 228
Query: 78 AQA--PPRNVAAPPPPVPNGSTPAV--------KTRICNKFNTAEGCKFGDKCHFAHGEW 127
NV +GS + KTRICNK+ C FG KCHFAHG
Sbjct: 229 VGGGGSSSNVVVLGGGGGSGSGSGIQILKPSNWKTRICNKWEITGYCPFGAKCHFAHGAA 288
Query: 128 ELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPG---------PATGFGANATAKISVEAS 178
EL + ++ G +P ++ P G P A+A+ V
Sbjct: 289 ELHRFGGGLVEEEGKDGVSPNPDTKQTVQNPKGLSDTTTLLSPGVPHNADASYHTGVALQ 348
Query: 179 LA-GAIIGKGGVNSKQ 193
A A+ K G+ + Q
Sbjct: 349 RASSAVTQKPGIRTHQ 364
Score = 46.6 bits (109), Expect = 2e-05
Identities = 58/220 (26%), Positives = 85/220 (38%), Gaps = 44/220 (20%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPG 160
KT++C KF A C + C+FAH EL + P++ + I A G M P
Sbjct: 93 KTKLCCKFR-AGTCPYITNCNFAHTVEEL-RRPPPNWQE---IVAAHEEERSGGMGTPTV 147
Query: 161 PATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEG 220
S+ +S A + G + K+ + G ES L +
Sbjct: 148 SVVEIPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPYG----ESCTFLHD----- 198
Query: 221 SFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGP--------------GGHGHHGS--- 263
EAS + + ++L GG+GG GG G GS
Sbjct: 199 ------EASRNRESVAISLGPGGYGSGGGGGSGGGSVGGGGSSSNVVVLGGGGGSGSGSG 252
Query: 264 ------NNFKTKLCENFA-KGTCTFGERCHFAHGPAELRK 296
+N+KT++C + G C FG +CHFAHG AEL +
Sbjct: 253 IQILKPSNWKTRICNKWEITGYCPFGAKCHFAHGAAELHR 292
Score = 43.1 bits (100), Expect = 2e-04
Identities = 33/108 (30%), Positives = 46/108 (42%), Gaps = 18/108 (16%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYV------PGGYNAV--AHMMNLAPSAQAPPRNVAA 87
K+K C KF CP+ +C+F H V P + + AH + P +V
Sbjct: 93 KTKLCCKF-RAGTCPYITNCNFAHTVEELRRPPPNWQEIVAAHEEERSGGMGTPTVSVVE 151
Query: 88 PPPP---VPN------GSTPAVKTRICNKFNTAEGCKFGDKCHFAHGE 126
P +P+ S + K R C KF T EGC +G+ C F H E
Sbjct: 152 IPREEFQIPSLVSSTAESGRSFKGRHCKKFYTEEGCPYGESCTFLHDE 199
Score = 39.7 bits (91), Expect = 0.002
Identities = 18/35 (51%), Positives = 21/35 (59%)
Query: 262 GSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
G FKTKLC F GTC + C+FAH ELR+
Sbjct: 88 GKMFFKTKLCCKFRAGTCPYITNCNFAHTVEELRR 122
>At3g19360 unknown protein
Length = 386
Score = 62.8 bits (151), Expect = 2e-10
Identities = 38/115 (33%), Positives = 55/115 (47%), Gaps = 21/115 (18%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHY------VPGGYNAVAHMMNLAPSAQAPPRNVAAP- 88
+ K C KF CP+G+ C+F+H G + ++++ +A P + A+
Sbjct: 185 RMKLCRKFCFGEECPYGDRCNFIHEDLSKFREDSGKLRESSVISVGATAADQPSDTASNL 244
Query: 89 -----------PPPVPNGS---TPAVKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
P P+ NG T KTR+C KF+ C FGDKCHFAHG+ EL
Sbjct: 245 IEVNRQGSIPVPAPMNNGGVVKTVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 60.8 bits (146), Expect = 8e-10
Identities = 65/233 (27%), Positives = 92/233 (38%), Gaps = 27/233 (11%)
Query: 76 PSAQAPPRNVAAPPPPVPNGSTPAV-KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIA 134
PS+ P + PPPV G+ KTR+C KF A C+ G+ C+FAHG +L +
Sbjct: 80 PSSSNPWMVPSLNPPPVNKGTANIFYKTRMCAKFR-AGTCRNGELCNFAHGIEDLRQ--- 135
Query: 135 PSFDDHRPIGHAPAG--------RIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGK 186
P + +G PAG R R P P KI + L
Sbjct: 136 PPSNWQEIVGPPPAGQDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFG 195
Query: 187 GGVNSKQICRQTGAKLSIREHESDPNLRNIELEG----SFDQIKEASNMVKDLLLTLQMS 242
C LS + E LR + + DQ + ++ + ++ +
Sbjct: 196 EECPYGDRCNFIHEDLS-KFREDSGKLRESSVISVGATAADQPSDTASNLIEVNRQGSIP 254
Query: 243 APPKSNQGGAGGPGGHGHHGSNNFKTKLCENF-AKGTCTFGERCHFAHGPAEL 294
P N GG + +KT+LC F G C FG++CHFAHG AEL
Sbjct: 255 VPAPMNNGGVVK--------TVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/86 (32%), Positives = 39/86 (44%), Gaps = 10/86 (11%)
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
R+ P P +NGG+ K+ V K++ C KF T CPFG+ CHF H
Sbjct: 249 RQGSIPVPAPMNNGGVVKT----------VYWKTRLCMKFDITGQCPFGDKCHFAHGQAE 298
Query: 64 GYNAVAHMMNLAPSAQAPPRNVAAPP 89
+N+V + A +A A A P
Sbjct: 299 LHNSVGRVEGEAMNAVASVNKQAVVP 324
Score = 41.6 bits (96), Expect = 5e-04
Identities = 16/31 (51%), Positives = 22/31 (70%)
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
+KT++C F GTC GE C+FAHG +LR+
Sbjct: 105 YKTRMCAKFRAGTCRNGELCNFAHGIEDLRQ 135
Score = 37.4 bits (85), Expect = 0.010
Identities = 29/118 (24%), Positives = 43/118 (35%), Gaps = 44/118 (37%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRN---VAAPPP-- 90
K++ C KF C GE C+F H + + PP N + PPP
Sbjct: 106 KTRMCAKF-RAGTCRNGELCNFAHGIE--------------DLRQPPSNWQEIVGPPPAG 150
Query: 91 -------------------PVPNGSTP-----AVKTRICNKFNTAEGCKFGDKCHFAH 124
PV N + ++ ++C KF E C +GD+C+F H
Sbjct: 151 QDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFGEECPYGDRCNFIH 208
>At2g25970 pseudogene
Length = 632
Score = 61.6 bits (148), Expect = 5e-10
Identities = 38/107 (35%), Positives = 58/107 (53%), Gaps = 5/107 (4%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQIK 226
T KI + G IIGKGG K + Q+GAK+ + R+ ++DPN R ++L G+ DQI
Sbjct: 135 TKKIDIPNMRVGVIIGKGGETIKYLQLQSGAKIQVTRDMDADPNCATRTVDLTGTPDQIS 194
Query: 227 EASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCEN 273
+A ++ D+L + A + GG GG G G++ F K+ N
Sbjct: 195 KAEQLITDVL--QEAEAGNTAGSGGGGGRRMGGQAGADQFVMKIPNN 239
Score = 35.0 bits (79), Expect = 0.048
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 4/60 (6%)
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDPN----LRNIELEGSFDQIKEASNMVKDLL 236
G IIGKGG K + +TGA++ + P R ++++G +QI+ A +V +++
Sbjct: 242 GLIIGKGGETIKSMQAKTGARIQVIPLHLPPGDPTPERTLQIDGITEQIEHAKQLVNEII 301
>At2g35430 hypothetical protein
Length = 180
Score = 59.3 bits (142), Expect = 2e-09
Identities = 48/142 (33%), Positives = 57/142 (39%), Gaps = 50/142 (35%)
Query: 14 NSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFG-ESCHFLHYVPGGYNAVA 69
NS+G SK+ S SS +G K+K C KF CP+ SCHF H
Sbjct: 48 NSDGAESPSKKTRSSSSSEIGKSFFKTKLCFKF-RAGTCPYSASSCHFAHS--------- 97
Query: 70 HMMNLAPSAQAPPRNVAAPPPPV--------------------PNGS------TPAVKTR 103
A + PP PPPP P G+ +P KTR
Sbjct: 98 -----AEELRLPP-----PPPPNWQETVTEASRNRESFAVSLGPRGNVAQTLKSPNWKTR 147
Query: 104 ICNKFNTAEGCKFGDKCHFAHG 125
ICNK+ T C FG CHFAHG
Sbjct: 148 ICNKWQTTGYCPFGSHCHFAHG 169
Score = 41.2 bits (95), Expect = 7e-04
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 262 GSNNFKTKLCENFAKGTCTF-GERCHFAHGPAELR 295
G + FKTKLC F GTC + CHFAH ELR
Sbjct: 68 GKSFFKTKLCFKFRAGTCPYSASSCHFAHSAEELR 102
Score = 40.4 bits (93), Expect = 0.001
Identities = 16/31 (51%), Positives = 21/31 (67%), Gaps = 1/31 (3%)
Query: 263 SNNFKTKLCENF-AKGTCTFGERCHFAHGPA 292
S N+KT++C + G C FG CHFAHGP+
Sbjct: 141 SPNWKTRICNKWQTTGYCPFGSHCHFAHGPS 171
Score = 27.7 bits (60), Expect = 7.7
Identities = 12/29 (41%), Positives = 14/29 (47%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT++C KF CHFAH EL
Sbjct: 73 KTKLCFKFRAGTCPYSASSCHFAHSAEEL 101
>At1g68200 putative zinc finger protein
Length = 308
Score = 51.2 bits (121), Expect = 7e-07
Identities = 33/125 (26%), Positives = 49/125 (38%), Gaps = 32/125 (25%)
Query: 17 GGIKKSKQE----LESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMM 72
GG KK QE +E + G+ +K++ C K+ T CP+G+ C F H + + H
Sbjct: 202 GGGKKEDQEEEIEVEVYNQGM-TKTELCNKWQETGTCPYGDHCQFAHGIKELRPVIRH-- 258
Query: 73 NLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKH 132
P KT +C + C +G +CHF H E K
Sbjct: 259 -------------------------PRYKTEVCRMVLAGDNCPYGHRCHFRHSLSEQEKL 293
Query: 133 IAPSF 137
+A F
Sbjct: 294 VAAGF 298
Score = 37.7 bits (86), Expect = 0.007
Identities = 16/30 (53%), Positives = 21/30 (69%), Gaps = 1/30 (3%)
Query: 267 KTKLCENFAK-GTCTFGERCHFAHGPAELR 295
KT+LC + + GTC +G+ C FAHG ELR
Sbjct: 224 KTELCNKWQETGTCPYGDHCQFAHGIKELR 253
Score = 37.7 bits (86), Expect = 0.007
Identities = 15/29 (51%), Positives = 18/29 (61%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT +CNK+ C +GD C FAHG EL
Sbjct: 224 KTELCNKWQETGTCPYGDHCQFAHGIKEL 252
Score = 32.0 bits (71), Expect = 0.41
Identities = 13/32 (40%), Positives = 18/32 (55%), Gaps = 1/32 (3%)
Query: 266 FKTKLCENFAKG-TCTFGERCHFAHGPAELRK 296
+KT++C G C +G RCHF H +E K
Sbjct: 261 YKTEVCRMVLAGDNCPYGHRCHFRHSLSEQEK 292
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 45.1 bits (105), Expect = 5e-05
Identities = 26/81 (32%), Positives = 45/81 (55%), Gaps = 3/81 (3%)
Query: 168 NATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQ 224
+ T +I V +S G +IGKGG + + +GAK+ I R+ E+DP+ LR +E+ GS
Sbjct: 198 STTRRIDVPSSKVGVLIGKGGETIRYLQFNSGAKIQILRDSEADPSSALRPVEIIGSVAC 257
Query: 225 IKEASNMVKDLLLTLQMSAPP 245
I+ A ++ ++ + P
Sbjct: 258 IESAEKLISAVIAEAEAGGSP 278
Score = 42.7 bits (99), Expect = 2e-04
Identities = 36/114 (31%), Positives = 52/114 (45%), Gaps = 14/114 (12%)
Query: 161 PAT-GFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLS-IREHESDPNL--RNI 216
P+T G +I V G IIG+GG K + ++GA+ I +H L R +
Sbjct: 286 PSTHAIGIPEQIEIKVPNDKVGLIIGRGGETIKNMQTRSGARTQLIPQHAEGDGLKERTV 345
Query: 217 ELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAG-------GPGGHGHHGS 263
+ G QI A++M+KD+ + +A P S GG GPGG GS
Sbjct: 346 RISGDKMQIDIATDMIKDV---MNQNARPSSYSGGYNQPAYRPQGPGGPPQWGS 396
>At1g33680 single-strand nucleic acid-binding protein, putative
Length = 407
Score = 44.3 bits (103), Expect = 8e-05
Identities = 25/81 (30%), Positives = 47/81 (57%), Gaps = 3/81 (3%)
Query: 168 NATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQ 224
+ T +I V +S G +IGKGG + + +GAK+ I R+ E+DP+ LR +E+ G+
Sbjct: 231 STTRRIDVPSSKVGTLIGKGGEMVRYLQVNSGAKIQIRRDAEADPSSALRPVEIIGTVSC 290
Query: 225 IKEASNMVKDLLLTLQMSAPP 245
I++A ++ ++ ++ P
Sbjct: 291 IEKAEKLINAVIAEVEAGGVP 311
>At5g64390 HEN4
Length = 857
Score = 43.9 bits (102), Expect = 1e-04
Identities = 43/154 (27%), Positives = 66/154 (41%), Gaps = 11/154 (7%)
Query: 106 NKFNTAEGCK--FGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAG---RIGGRMEPPPG 160
++FN + G FG + H + APS P AP G GG G
Sbjct: 702 DRFNPSAGYSHNFGRRFTMDHSDNSHHLTEAPSRLWASPPPAAPRGLSDASGGLSSARAG 761
Query: 161 PATGFG------ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLR 214
G G N T +I V A+ + G+ G N +Q+ + +GA++ I E + R
Sbjct: 762 HVLGSGHKSAIVTNTTVEIRVPANAMSFVYGEQGYNLEQLRQISGARVIIHEPPLGTSDR 821
Query: 215 NIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSN 248
I + G+ DQ + A N++ +LT + S K N
Sbjct: 822 IIVISGTPDQTQAAQNLLHAFILTGETSLSKKYN 855
Score = 40.8 bits (94), Expect = 9e-04
Identities = 21/67 (31%), Positives = 39/67 (57%), Gaps = 5/67 (7%)
Query: 167 ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP-----NLRNIELEGS 221
++ TA++ V S G ++GKGGV ++ + TGA + I + E +P N + +++ G
Sbjct: 540 SSITARLVVPTSQIGCVLGKGGVIVSEMRKTTGAAIQILKVEQNPKCISENDQVVQITGE 599
Query: 222 FDQIKEA 228
F ++EA
Sbjct: 600 FPNVREA 606
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/61 (37%), Positives = 37/61 (59%), Gaps = 4/61 (6%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHE----SDPNLRNIELEGSFDQIKE 227
++ E+S AGA+IGKGG I ++TG K+SIR +D + +E+EG+ +K+
Sbjct: 153 RLLTESSHAGAVIGKGGQMVGSIRKETGCKISIRIENLPICADTDDEMVEVEGNAIAVKK 212
Query: 228 A 228
A
Sbjct: 213 A 213
Score = 32.7 bits (73), Expect = 0.24
Identities = 22/83 (26%), Positives = 37/83 (44%)
Query: 155 MEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLR 214
M+PPP + KI AG +IG GG + + +TGA +++ D R
Sbjct: 438 MKPPPSEVEVGNQDVVFKILCSTENAGGVIGTGGKVVRMLHSETGAFINVGNTLDDCEER 497
Query: 215 NIELEGSFDQIKEASNMVKDLLL 237
I + S + ++S K ++L
Sbjct: 498 LIAVTASENPECQSSPAQKAIML 520
Score = 31.2 bits (69), Expect = 0.70
Identities = 16/42 (38%), Positives = 24/42 (57%)
Query: 168 NATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHES 209
+A ++ S GA+IGK G KQ+ + TGAK+ + E S
Sbjct: 46 HAAFRLLCPLSHVGAVIGKSGNVIKQLQQSTGAKIRVEEPPS 87
>At1g66810 hypothetical protein
Length = 310
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/100 (25%), Positives = 37/100 (37%), Gaps = 28/100 (28%)
Query: 25 ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRN 84
ELE G+ K++ C K+ T C +G++C F H + + H
Sbjct: 224 ELEVYRQGM-MKTELCNKWQETGACCYGDNCQFAHGIDELRPVIRH-------------- 268
Query: 85 VAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAH 124
P KT +C T C +G +CHF H
Sbjct: 269 -------------PRYKTEVCRMMVTGAMCPYGHRCHFRH 295
Score = 38.1 bits (87), Expect = 0.006
Identities = 15/30 (50%), Positives = 19/30 (63%)
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
+KT +CNK+ C +GD C FAHG EL
Sbjct: 233 MKTELCNKWQETGACCYGDNCQFAHGIDEL 262
Score = 35.4 bits (80), Expect = 0.037
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Query: 267 KTKLCENFAK-GTCTFGERCHFAHGPAELR 295
KT+LC + + G C +G+ C FAHG ELR
Sbjct: 234 KTELCNKWQETGACCYGDNCQFAHGIDELR 263
Score = 30.4 bits (67), Expect = 1.2
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 1/25 (4%)
Query: 266 FKTKLCENFAKGT-CTFGERCHFAH 289
+KT++C G C +G RCHF H
Sbjct: 271 YKTEVCRMMVTGAMCPYGHRCHFRH 295
>At3g04610 putative RNA-binding protein
Length = 577
Score = 41.2 bits (95), Expect = 7e-04
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEAS 229
T ++ + S A A+IG G N R +GA ++I+E P +E+ G+ Q++ A
Sbjct: 459 TQQMQIPLSYADAVIGTSGSNISYTRRLSGATVTIQETRGVPGEMTVEVSGTGSQVQTAV 518
Query: 230 NMVKDLLLTLQMSAP--PKSNQGGAGGPGGHGHHGS 263
++++ + AP P++ G + HGS
Sbjct: 519 QLIQNFMAEAGAPAPAQPQTVAPEQQGYNPYATHGS 554
>At3g48440 unknown protein
Length = 448
Score = 39.7 bits (91), Expect = 0.002
Identities = 29/99 (29%), Positives = 40/99 (40%), Gaps = 20/99 (20%)
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C +F T GC +GE+C F H +P A A +N P P + NGS
Sbjct: 164 CKYYFRTGGCKYGETCRFNHTIPKSGLASAPELNFLGLPLRP--GEVECPYYMRNGS--- 218
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAHGE-WELGKHIAPSF 137
CK+G +C F H + +G +PSF
Sbjct: 219 --------------CKYGAECKFNHPDPTTIGGTDSPSF 243
>At5g63260 unknown protein
Length = 435
Score = 38.5 bits (88), Expect = 0.004
Identities = 27/89 (30%), Positives = 31/89 (34%), Gaps = 19/89 (21%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K C +F T GC +GESC F +HM A P N P
Sbjct: 150 KLMECKYYFRTGGCKYGESCRF-----------SHMKEHNSPASVPELNFLGLP------ 192
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAH 124
P K C + CKFG C F H
Sbjct: 193 IRPGEKE--CPFYMRNGSCKFGSDCKFNH 219
Score = 28.9 bits (63), Expect = 3.5
Identities = 24/90 (26%), Positives = 29/90 (31%), Gaps = 5/90 (5%)
Query: 22 SKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAP 81
S EL L + K C + C FG C F H P V +P +
Sbjct: 182 SVPELNFLGLPIRPGEKECPFYMRNGSCKFGSDCKFNHPDPTAIGGVD-----SPLYRGN 236
Query: 82 PRNVAAPPPPVPNGSTPAVKTRICNKFNTA 111
+P P ST TR N TA
Sbjct: 237 NGGSFSPKAPSQASSTSWSSTRHMNGTGTA 266
>At4g26000 nucleic acid binding protein like
Length = 495
Score = 38.1 bits (87), Expect = 0.006
Identities = 23/90 (25%), Positives = 46/90 (50%), Gaps = 4/90 (4%)
Query: 162 ATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGS 221
++ F + + + S A IIG G N I R++GA ++I+E P+ +E++G+
Sbjct: 334 SSAFVTQVSQTMQIPFSYAEDIIGVEGANIAYIRRRSGATITIKE-SPHPDQITVEIKGT 392
Query: 222 FDQIKEASNMVKDLLLTLQMSAPPKSNQGG 251
Q++ A ++++ ++ P S GG
Sbjct: 393 TSQVQTAEQLIQEFIIN---HKEPVSVSGG 419
Score = 28.1 bits (61), Expect = 5.9
Identities = 12/41 (29%), Positives = 24/41 (58%)
Query: 164 GFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
G+ + ++ V + GAIIG+ G K++C +T A++ +
Sbjct: 69 GWPGDCVFRMIVPVTKVGAIIGRKGDFIKKMCEETRARIKV 109
>At5g46190 unknown protein
Length = 644
Score = 37.7 bits (86), Expect = 0.007
Identities = 18/75 (24%), Positives = 40/75 (53%)
Query: 164 GFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFD 223
G + T ++ + A+ G ++G+GG N I R +GA + I + ++ R + G+ +
Sbjct: 569 GLPQSTTMEVRIPANAVGKVMGRGGGNLDNIRRISGAMIEISDSKNSHGGRVALISGTSE 628
Query: 224 QIKEASNMVKDLLLT 238
Q + A N+ + +++
Sbjct: 629 QKRTAENLFQAFIMS 643
Score = 30.8 bits (68), Expect = 0.91
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 5/62 (8%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHE-----SDPNLRNIELEGSFDQIK 226
++ V + + G IIGK G +I ++T A + I + +DPN +E+ G ++
Sbjct: 402 QLLVSSKVIGCIIGKSGSIISEIRKRTKADIHISKGNNTPKCADPNDELVEISGEVSNVR 461
Query: 227 EA 228
+A
Sbjct: 462 DA 463
Score = 30.4 bits (67), Expect = 1.2
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query: 164 GFGANATA-----KISVEASLAGAIIGKGGVNSKQICRQTGAKLSI---REHESDPNLRN 215
GFG ++ + K+ +S G +IGKGG+ K I + +G+ + + R + D +
Sbjct: 307 GFGGSSRSEKLAIKVICSSSKIGRVIGKGGLTIKGIRQASGSHIEVNDSRTNHDDDCVIT 366
Query: 216 IELEGSFDQIKEASNMVKDLLLTLQ 240
+ S D +K +M + +L LQ
Sbjct: 367 VTATESPDDLK---SMAVEAVLLLQ 388
>At5g09560 putative protein
Length = 567
Score = 37.7 bits (86), Expect = 0.007
Identities = 20/79 (25%), Positives = 43/79 (54%), Gaps = 5/79 (6%)
Query: 163 TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP-----NLRNIE 217
+G ++ TA++ V ++ ++G+ G I ++TGA +++ E +P N + ++
Sbjct: 392 SGLTSSITARLVVRSNQINCLLGEEGRIKTTIQQRTGAFITVLNVEQNPKCVSENNQVVQ 451
Query: 218 LEGSFDQIKEASNMVKDLL 236
+ G F ++EA N V +L
Sbjct: 452 ISGEFPNVREAINQVTSML 470
>At5g04430 putative RNA-binding protein
Length = 313
Score = 37.0 bits (84), Expect = 0.013
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query: 167 ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH---ESDPNLRNIELEGSFD 223
A+ T I V G ++G+GG N +I + TGA++ I + S R + + G
Sbjct: 232 ASTTVTIGVADEHIGLVLGRGGRNIMEITQMTGARIKISDRGDFMSGTTDRKVSITGPQR 291
Query: 224 QIKEASNMVK 233
I++A M+K
Sbjct: 292 AIQQAETMIK 301
Score = 30.4 bits (67), Expect = 1.2
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Query: 175 VEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNL--RNIELEGSFDQIKEASNM 231
V + AG++IGKGG + ++GA++ + R E P R I + GS ++ +
Sbjct: 41 VSNAAAGSVIGKGGSTITEFQAKSGARIQLSRNQEFFPGTTDRIIMISGSIKEVVNGLEL 100
Query: 232 VKDLL 236
+ D L
Sbjct: 101 ILDKL 105
>At5g18550 zinc finger -like protein
Length = 456
Score = 36.6 bits (83), Expect = 0.017
Identities = 35/130 (26%), Positives = 45/130 (33%), Gaps = 31/130 (23%)
Query: 30 SSGVGSKSKP---------CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQA 80
S+GV +K + C F T C FG SC F H M A +
Sbjct: 278 STGVSNKEQTFPQRPEQPECQYFMRTGDCKFGTSCRF-----------HHPMEAASPEAS 326
Query: 81 PPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAH--GEWELGKHIAPSFD 138
++ P P G+ P C F CKFG C F H G L +PS
Sbjct: 327 TLSHIGLPLRP---GAVP------CTHFAQHGICKFGPACKFDHSLGSSSLSYSPSPSSL 377
Query: 139 DHRPIGHAPA 148
P+ P+
Sbjct: 378 TDMPVAPYPS 387
Score = 33.5 bits (75), Expect = 0.14
Identities = 18/62 (29%), Positives = 25/62 (40%)
Query: 17 GGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAP 76
GG + L + + K C+ F T C FG +C + H VP G A +
Sbjct: 119 GGDSVTPVSLNYMGFPLRPGEKECSYFMRTGQCKFGSTCRYHHPVPPGVQAPSQQQQQQL 178
Query: 77 SA 78
SA
Sbjct: 179 SA 180
Score = 29.3 bits (64), Expect = 2.7
Identities = 20/85 (23%), Positives = 26/85 (30%), Gaps = 19/85 (22%)
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPA 99
C + T C +G C F H N + L A P + P
Sbjct: 48 CIYYLRTGVCGYGSRCRFNH----PRNRAPVLGGLRTEAGEFPERMGQP----------- 92
Query: 100 VKTRICNKFNTAEGCKFGDKCHFAH 124
+C F CKFG C + H
Sbjct: 93 ----VCQHFMRTGTCKFGASCKYHH 113
>At2g47850 unknown protein
Length = 553
Score = 36.6 bits (83), Expect = 0.017
Identities = 31/125 (24%), Positives = 44/125 (34%), Gaps = 21/125 (16%)
Query: 40 CTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPR-NVAAPPPPVPNGSTP 98
C + T C FG SC F H P + PPR N P +P P
Sbjct: 380 CQYYLKTGDCKFGTSCKFHH----------------PRDRVPPRANCVLSPIGLP--LRP 421
Query: 99 AVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPP 158
V+ C + CKFG C F H + + + S P+ P + G +
Sbjct: 422 GVQR--CTFYVQNGFCKFGSTCKFDHPMGTIRYNPSASSLADAPVAPYPVSSLLGALAAA 479
Query: 159 PGPAT 163
P ++
Sbjct: 480 PSSSS 484
Score = 33.9 bits (76), Expect = 0.11
Identities = 25/86 (29%), Positives = 29/86 (33%), Gaps = 19/86 (22%)
Query: 39 PCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTP 98
PC + T C FG SC F H N S P N+ PV G
Sbjct: 169 PCQFYLKTGTCKFGASCKF-----------HHPKNAGGSMSHVPLNIYG--YPVREGDNE 215
Query: 99 AVKTRICNKFNTAEGCKFGDKCHFAH 124
C+ + CKFG C F H
Sbjct: 216 ------CSYYLKTGQCKFGITCKFHH 235
Score = 28.5 bits (62), Expect = 4.5
Identities = 15/49 (30%), Positives = 18/49 (36%)
Query: 16 NGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGG 64
N G S L V C+ + T C FG +C F H P G
Sbjct: 192 NAGGSMSHVPLNIYGYPVREGDNECSYYLKTGQCKFGITCKFHHPQPAG 240
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,070,744
Number of Sequences: 26719
Number of extensions: 389839
Number of successful extensions: 3064
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 2449
Number of HSP's gapped (non-prelim): 548
length of query: 296
length of database: 11,318,596
effective HSP length: 99
effective length of query: 197
effective length of database: 8,673,415
effective search space: 1708662755
effective search space used: 1708662755
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0221.10


