

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0220.7
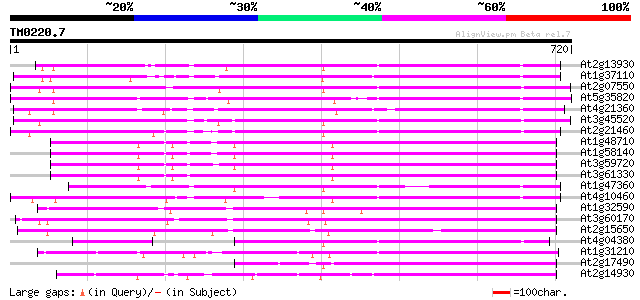
(720 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g13930 putative retroelement pol polyprotein 430 e-120
At1g37110 413 e-115
At2g07550 putative retroelement pol polyprotein 407 e-113
At5g35820 copia-like retrotransposable element 404 e-113
At4g21360 putative transposable element 401 e-112
At3g45520 copia-like polyprotein 396 e-110
At2g21460 putative retroelement pol polyprotein 391 e-109
At1g48710 hypothetical protein 380 e-105
At1g58140 hypothetical protein 380 e-105
At3g59720 copia-type reverse transcriptase-like protein 378 e-105
At3g61330 copia-type polyprotein 377 e-105
At1g47360 polyprotein, putative 366 e-101
At4g10460 putative retrotransposon 363 e-100
At1g32590 hypothetical protein, 5' partial 348 5e-96
At3g60170 putative protein 338 5e-93
At2g15650 putative retroelement pol polyprotein 306 3e-83
At4g04380 putative polyprotein 296 3e-80
At1g31210 putative reverse transcriptase 278 1e-74
At2g17490 putative retroelement pol polyprotein 272 4e-73
At2g14930 pseudogene 269 5e-72
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 430 bits (1106), Expect = e-120
Identities = 253/698 (36%), Positives = 393/698 (56%), Gaps = 38/698 (5%)
Query: 34 QPLTGKK---PNDMKQEDWDLLDR-----QALGVIRLTLSKNVAFNIVNEKTTADLMKAL 85
+PLT ++ P K+ D D + R +A VI L ++ V I KT A+ + L
Sbjct: 19 KPLTEEEEEDPEKRKKRDADEVARLERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETL 78
Query: 86 SNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSL 145
++ ++V+ + +M E + E+I+ F I++ L+ ++I +E+ + L
Sbjct: 79 DRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILL 138
Query: 146 LQSLPDSWAATVTAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRG 205
L SLP + V + S KL+ DD+ ++ + D K+ S N +RG
Sbjct: 139 LSSLPARYDGLVETMKYSNSREKLRLDDV--MVAARD---KERELSQNNRPVVEGHFARG 193
Query: 206 RGSQKSHNQ-SQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQC-KAPRKKKNYQKR* 263
R K++NQ ++G+ RS+S+ RV CW C ++GHF QC K + K+ Q+
Sbjct: 194 RPDGKNNNQGNKGKNRSRSKSADGKRV-----CWICGKEGHFKKQCYKWIERNKSKQQGS 248
Query: 264 DDDESANAATEEV---------ADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICG 314
D+ ES+ A + E D + DS + WV+D+G SFH P K+ ++
Sbjct: 249 DNGESSLAKSTEAFNPAMVLLATDETLVVTDSIANEWVLDTGCSFHMTPRKDWFKDFKEL 308
Query: 315 KFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTT 374
G V + + + GIG I IR+S+G+ L +VR++P + RNLIS+G L+D G
Sbjct: 309 SSGYVKMGNDTYSPVKGIGSIKIRNSDGSQVILTDVRYMPNMTRNLISLGTLEDRGCWFK 368
Query: 375 FGGGAWKVTKGNLVVARGKKRGSLYMV----AEEDMIAVTEAINSSSIWHQRLGHMSEKG 430
G K+ KG + +G+KR +LY++ E + + E + +++WH RLGHMS+KG
Sbjct: 369 SQDGILKIVKGCSTILKGQKRDTLYILDGVTEEGESHSSAEVKDETALWHSRLGHMSQKG 428
Query: 431 MKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPV 489
M+I+ KG + +L CE C+ GKQ +VSF+ A +K EKL VH+D+WG P
Sbjct: 429 MEILVKKGCLRREVIKELEFCEDCVYGKQHRVSFAPAQHVTK-EKLAYVHSDLWGSPHNP 487
Query: 490 KSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGE 549
SLG S+Y+++F+DD +RKVW+YFL+ K + F F +WK VENQ+ K+K L++DNG E
Sbjct: 488 ASLGNSQYFISFVDDYSRKVWIYFLRKKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLE 547
Query: 550 YDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAI 609
Y + F+KFC E GI KT TP+QNG+AER+NRT+ ++ R M +SG+ K FW +A
Sbjct: 548 YCNHYFEKFCKEEGIVRHKTCAYTPQQNGIAERLNRTIMDKVRSMLSRSGMEKKFWAEAA 607
Query: 610 NTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKC 669
+TA YLINR PS +++ LPEE W G LS L+ FGC++Y+ D + KL+P++ K
Sbjct: 608 STAVYLINRSPSTAINFDLPEEKWTGALPDLSSLRKFGCLAYIHAD---QGKLNPRSKKG 664
Query: 670 FFIGYGSDMYGYRFWDEQNRKIIRSINVTFNESVLYKD 707
F Y + GY+ W +++K + S NV F E V++KD
Sbjct: 665 IFTSYPEGVKGYKVWVLEDKKCVISRNVIFREQVMFKD 702
>At1g37110
Length = 1356
Score = 413 bits (1062), Expect = e-115
Identities = 252/747 (33%), Positives = 401/747 (52%), Gaps = 72/747 (9%)
Query: 5 KVKIERFDG-RDFGFWKMLMEDYLYQKMLYQPLTGKK--------PNDMKQEDWD----- 50
+V+I+ F+G RDF WK+ ++ L L LT ++ KQE D
Sbjct: 7 RVEIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSG 66
Query: 51 ----------LLDRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL 100
QA +I +S V + + TTADL L+ Y + N+++
Sbjct: 67 TKEVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYT 126
Query: 101 IRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSW------- 153
+L++ +M ++ ++++ F I+++L S++I D E+ + +L SLP S
Sbjct: 127 QLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTL 186
Query: 154 -----AATVTAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRGS 208
TV V++SA+ + + + DL D G+++ L T RGR
Sbjct: 187 KYGNKTLTVQDVTSSAKSLERELAEAVDL---------DKGQAA-----VLYTTERGRPL 232
Query: 209 QKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDES 268
++ NQ G+G+ +SR S+T+V CW C ++GH C + +KK + + +
Sbjct: 233 VRN-NQKGGQGKGRSRSNSKTKV----PCWYCKKEGHVKKDCYSRKKKMESEGQGE---- 283
Query: 269 ANAATEEVADTLICSLDSPV--DSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKP 326
A TE++ + S++ + D W++DSG + H ++ ++ + L D
Sbjct: 284 AGVITEKLVFSEALSVNEQMVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHS 343
Query: 327 LDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGN 386
++ G G I I + GT+ L NV++VP ++RNLIS G LD GY G G + K N
Sbjct: 344 VESQGQGTIRIDTHGGTIKILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNN 403
Query: 387 LVVARGKKRGSLYM-----VAEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMS 441
RG LY+ V E A T+ + ++ +WH RLGHMS +K++A KG +
Sbjct: 404 KTALRGSLSNGLYVLDGSTVMSELCNAETDKVKTA-LWHSRLGHMSMNNLKVLAGKGLID 462
Query: 442 NLKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVT 500
+ +L CEHC++GK +KVSF+ G+ + + L VH D+WG P S+ G +Y+++
Sbjct: 463 RKEINELEFCEHCVMGKSKKVSFN-VGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLS 521
Query: 501 FIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCS 560
IDD TRKVW+YFLKSK + F F +WK+ VENQ K+K L++DNG E+ + F +C
Sbjct: 522 IIDDKTRKVWLYFLKSKDETFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCK 581
Query: 561 ENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGP 620
E+GI +T TP+QNGVAERMNRT+ E+ RC+ +SG+ ++FW +A TAAYLINR P
Sbjct: 582 EHGIERHRTCTYTPQQNGVAERMNRTIMEKVRCLLNKSGVEEVFWAEAAATAAYLINRSP 641
Query: 621 SVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYG 680
+ +++ +PEE+W ++ HL+ FG ++YV D + KL P+A+K FF+GY + G
Sbjct: 642 ASAINHNVPEEMWLNRKPGYKHLRKFGSIAYVHQD---QGKLKPRALKGFFLGYPAGTKG 698
Query: 681 YRFWDEQNRKIIRSINVTFNESVLYKD 707
Y+ W + K + S NV F ESV+Y+D
Sbjct: 699 YKVWLLEEEKCVISRNVVFQESVVYRD 725
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 407 bits (1045), Expect = e-113
Identities = 241/750 (32%), Positives = 398/750 (52%), Gaps = 43/750 (5%)
Query: 1 LEEGKVKIERFDGR-DFGFWKMLMEDYLYQKMLYQPL-----TGKKPNDMKQEDWDLLDR 54
+ ++++E+FDGR D+ WK + ++ L L TG+K + + + D D ++
Sbjct: 1 MSTARIEVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEK 60
Query: 55 ------------QALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIR 102
+A I L+++ V I E T A ++ AL +Y N+++ +
Sbjct: 61 LEKFEALEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQ 120
Query: 103 RLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSN 162
+L++ +M E SV +I+ F II+ L ++ + +E + LL +LP ++ +
Sbjct: 121 KLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKY 180
Query: 163 SARDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRGSQKSHNQSQGRGRSK 222
S+ + L D++ I S+++ +S L K N+++G+G K
Sbjct: 181 SSGKSILTLDEVAAAIYSKELELGSVKKSIKVQAEGLYV--------KDKNENKGKGEQK 232
Query: 223 SRGRSQT-RVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDES-------ANAATE 274
+G+ + + + CW C +GHF + C K + Q + ES A AA
Sbjct: 233 GKGKGKKGKSKKKPGCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAAGY 292
Query: 275 EVADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGD 334
V++ L + D W++D+G S+H +E + G V + + + G+G
Sbjct: 293 YVSEALSSTEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGT 352
Query: 335 IDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKK 394
I +++S+G L NVR++P + RNL+S+G + GY G ++ GN V+ G++
Sbjct: 353 IRVKNSDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRR 412
Query: 395 RGSLYMV----AEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGV 450
+LY++ + +AV + + + +WHQRL HMS+K M+I+ KG + K L V
Sbjct: 413 YDTLYLLNWKPVASESLAVVKRADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDV 472
Query: 451 CEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKV 509
CE CI GK ++ SFS A +K EKLE +H+D+WG P SLG +Y+++ IDD TRKV
Sbjct: 473 CEDCIYGKAKRKSFSLAHHDTK-EKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKV 531
Query: 510 WVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKT 569
WVYF+K+K + F F +W VENQT ++K+L++DNG E+ ++ F FC GI +T
Sbjct: 532 WVYFMKTKDEAFEKFVEWVNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCESIGIHRHRT 591
Query: 570 IPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLP 629
TP+QNGVAERMNRT+ E+ R M SGLPK FW +A +T LIN+ PS L++++P
Sbjct: 592 CAYTPQQNGVAERMNRTIMEKVRSMLSDSGLPKRFWAEATHTTVLLINKTPSSALNFEIP 651
Query: 630 EEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNR 689
++ W G S+L+ +GCV++V D KL+P+A K IGY + GY+ W R
Sbjct: 652 DKKWSGNPPVYSYLRRYGCVAFVHTDD---GKLEPRAKKGVLIGYPVGVKGYKVWILDER 708
Query: 690 KIIRSINVTFNESVLYKDRSSAESMSSSKQ 719
K + S N+ F E+ +YKD + S+++
Sbjct: 709 KCVVSRNIIFQENAVYKDLMQRQENVSTEE 738
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 404 bits (1039), Expect = e-113
Identities = 238/756 (31%), Positives = 395/756 (51%), Gaps = 66/756 (8%)
Query: 1 LEEGKVKIERFDGR-DFGFWKMLMEDYLYQKMLYQPLTGKKPNDMKQEDWDLLDR----- 54
+ G+ ++E+FDG D+ WK + ++ L + L ++ ++ ++ D
Sbjct: 1 MSSGRAEVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDP 60
Query: 55 -----------------QALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANK 97
+A I L+L NV ++ +KT A ++K L ++ N+
Sbjct: 61 ETATSKLEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNR 120
Query: 98 VHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATV 157
++L +RL+ +M E ++ E++N F +IS L +VK+ +E + LL SLP +
Sbjct: 121 IYLKQRLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLK 180
Query: 158 TAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRGSQKSHNQSQG 217
+ L ++I I S+ + SG+ L + RGR + ++
Sbjct: 181 ETLKYCK--TTLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKN 238
Query: 218 RGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVA 277
+ RSKS+G +T CW C ++GHF QC K++N Q + A+ T V
Sbjct: 239 KSRSKSKGAGKT-------CWICGKEGHFKKQCYV-WKERNKQGSTSERGEASTVTARVT 290
Query: 278 DT--------LICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDI 329
D L+ + D+W++D+G SFH K+ + ++ GKV + + ++
Sbjct: 291 DAAALVVSRALLGFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEV 350
Query: 330 VGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVV 389
GIGD+ I++ +G+ L +VR++P + +NLIS+G L+D+G G + K +L V
Sbjct: 351 KGIGDVRIKNEDGSTILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTV 410
Query: 390 ARGKKRGSLYMVAEEDMIAVTEAINS----SSIWHQRLGHMSEKGMKIMASKGKMSNLKH 445
GKK +LY + + I+ +S+WH RLGH+ KG++++ SKG H
Sbjct: 411 LTGKKESTLYFLQGTTLAGEANVIDKEKDETSLWHSRLGHIGAKGLQVLVSKG------H 464
Query: 446 VDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVK-SLGGSRYYVTFIDD 504
+D K +SF A +K +KL+ VH+D+WG V S+G +Y++TFIDD
Sbjct: 465 LD----------KNIMISFGAAKHVTK-DKLDYVHSDLWGSTNVPFSIGKCQYFITFIDD 513
Query: 505 STRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGI 564
TR+ W+YF+++K + FS F +WKT++ENQ K+K L +DNG E+ +QEF FC + G+
Sbjct: 514 FTRRTWIYFIRTKDEAFSKFVEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGV 573
Query: 565 RMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPL 624
+T TP+QNGVAERMNRT+ + RCM +SGL K FW +A +TA +LIN+ PS +
Sbjct: 574 IRHRTCAYTPQQNGVAERMNRTIMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSI 633
Query: 625 DYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFW 684
++ +PEE W G LK FG V+Y+ D + KL+P+A K F+GY + ++ W
Sbjct: 634 EFDIPEEKWTGHPPDYKILKKFGSVAYIHSD---QGKLNPRAKKGIFLGYPDGVKRFKVW 690
Query: 685 DEQNRKIIRSINVTFNESVLYKDRSSAESMSSSKQL 720
++RK + S ++ F E+ +YK+ + KQL
Sbjct: 691 LLEDRKCVVSRDIVFQENQMYKELQKNDMSEEDKQL 726
>At4g21360 putative transposable element
Length = 1308
Score = 401 bits (1031), Expect = e-112
Identities = 241/746 (32%), Positives = 389/746 (51%), Gaps = 67/746 (8%)
Query: 5 KVKIERFDG-RDFGFWKMLME-------------DYLYQKMLYQPLTGKKPNDMKQEDWD 50
KV+I+ F+G RDF WK+ +E D+ K + + KK ++ + ++ D
Sbjct: 6 KVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDDETD 65
Query: 51 LLDR-------------QALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANK 97
QA I ++ V + + T A+L L+ ++ + N+
Sbjct: 66 SKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSLPNR 125
Query: 98 VHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATV 157
++ RL++ +M + S+ ++ + F I+++L S++I E+ + +L SLP S+
Sbjct: 126 IYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYIQLK 185
Query: 158 TAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSNTF----GSALNTESRGRGSQKSHN 213
+ K ++D++ S ++ E T +AL T RGR Q +
Sbjct: 186 HTLKYGN-----KTLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGR-PQTKNT 239
Query: 214 QSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAAT 273
Q QG+GR +S +S+ +TCW C ++GH C A ++K + + A T
Sbjct: 240 QGQGKGRGRSNSKSR------LTCWFCKKEGHVKKDCYAGKRKLENEGQ----GKAGVIT 289
Query: 274 EEVADTLICSL--DSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVG 331
E++ + S+ D WVIDSG ++H + S + + + L D ++ G
Sbjct: 290 EKLVYSEALSMYDQEAKDKWVIDSGCTYHMTSRMDWFSEFNENETTMILLGDDHTVESKG 349
Query: 332 IGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVAR 391
G + + + G++ L NVR VP ++RNLIS G LD GY G G + K N
Sbjct: 350 SGTVKVNTHGGSIRVLKNVRFVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALC 409
Query: 392 GKKRGSLYMVAEEDMIAVTEAINSSS----IWHQRLGHMSEKGMKIMASKGKMSNLKHVD 447
G LY++ ++ + S+ +WH RLGHMS MKI+A KG + +
Sbjct: 410 GNLVNGLYVLDGHTVVNENCNVEGSNEKTELWHCRLGHMSLNNMKILAEKGLLEKKDIKE 469
Query: 448 LGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTR 507
L CE+C++GK +K+SF+ G+ E L +H D+WG +Y+++ IDD +R
Sbjct: 470 LSFCENCVMGKSKKLSFN-VGKHITDEVLGYIHADLWG---------KQYFLSIIDDKSR 519
Query: 508 KVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMI 567
KVW+ FLK+K + F F +WK VENQ K+K L++DNG E+ + +F +FC +NGI
Sbjct: 520 KVWLMFLKTKDETFERFCEWKELVENQVNKKVKILRTDNGLEFCNLKFDEFCKQNGIERH 579
Query: 568 KTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQ 627
+T TP+QNGVA+RMNRTL E+ RC+ +SGL ++FW +A TAAYL+NR P+ +D+
Sbjct: 580 RTCTYTPQQNGVAKRMNRTLMEKVRCLLNESGLEEVFWAEAAATAAYLVNRSPASAVDHN 639
Query: 628 LPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQ 687
+PEE+W K+ HL+ FGC++YV +D + KL P+A+K F+GY GY+ W
Sbjct: 640 VPEELWLDKKPGYKHLRRFGCIAYVHLD---QGKLKPRALKGVFLGYPQGTKGYKVWLLD 696
Query: 688 NRKIIRSINVTFNESVLYKD-RSSAE 712
K + S N+ FNE+ +YKD R S+E
Sbjct: 697 EEKCVISRNIVFNENQVYKDIRESSE 722
>At3g45520 copia-like polyprotein
Length = 1363
Score = 396 bits (1018), Expect = e-110
Identities = 238/758 (31%), Positives = 391/758 (51%), Gaps = 60/758 (7%)
Query: 5 KVKIERFDGR-DFGFWKMLMEDYLYQKMLYQPLT-----------------GKKPNDMKQ 46
++++E+FDGR D+ WK + ++ L L +K K
Sbjct: 5 RIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEEREKM 64
Query: 47 EDWDLLDRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIRRLFN 106
E ++ R+A I L++S V I E + A +++AL +Y N+++L ++L++
Sbjct: 65 EAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKLYS 124
Query: 107 LRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSARD 166
+M E S+ +I+ F I++ L ++ + +E + LL SLP + + S+
Sbjct: 125 FKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSGK 184
Query: 167 NKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRGSQKSHNQSQGRG-RSKSRG 225
L D++ I S ++ +S L + + +S + +G+G RSKS+
Sbjct: 185 TVLSLDEVAAAIYSRELEFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSKSKS 244
Query: 226 RSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDD------------------- 266
+ CW C GH + C P K K K +
Sbjct: 245 KRG--------CWICGEDGHLKSTC--PNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFV 294
Query: 267 ESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKP 326
ESA E + L+ D W++D+G +H +E L ++ G V + +
Sbjct: 295 ESAGMFVSEALSSTDIHLE---DEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSI 351
Query: 327 LDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGN 386
+ G+G + I + NG TL NVR++P + RNL+S+G + G+ G ++ GN
Sbjct: 352 SRVKGVGTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGN 411
Query: 387 LVVARGKKRGSLYMV----AEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSN 442
V+ G++ +LY++ A ++ +AV A + + +WH+RL HMS+K M ++ KG +
Sbjct: 412 QVLLEGRRYDTLYILHGKPATDESLAVARANDDTVLWHRRLCHMSQKNMSLLIKKGFLDK 471
Query: 443 LKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVTF 501
K L CE CI G+ +K+ F+ A +K +KLE VH+D+WG P SLG +Y+++F
Sbjct: 472 KKVSMLDTCEDCIYGRAKKIGFNLAQHDTK-KKLEYVHSDLWGAPTVPMSLGNCQYFISF 530
Query: 502 IDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSE 561
IDD TRKVWVYFLK+K + F F W + VENQ+G ++K+L++DNG E+ ++ F FC E
Sbjct: 531 IDDYTRKVWVYFLKTKDEAFEKFVSWISLVENQSGERVKTLRTDNGLEFCNRMFDGFCEE 590
Query: 562 NGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPS 621
G + +T TP+QNGV ERMNRT+ E+ R M SGLPK FW +A +TA LIN+ P
Sbjct: 591 KGFQRHRTCAYTPQQNGVVERMNRTIMEKVRSMLCDSGLPKRFWAEATHTAVLLINKTPC 650
Query: 622 VPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGY 681
++++ P++ W GK S+L+ +GCV++V D KL+ +A K IGY S + GY
Sbjct: 651 SAINFEFPDKRWSGKAPIYSYLRRYGCVTFVHTDG---GKLNLRAKKGVLIGYPSGVKGY 707
Query: 682 RFWDEQNRKIIRSINVTFNESVLYKDRSSAESMSSSKQ 719
+ W + +K + S NV+F E+ +YKD + S ++
Sbjct: 708 KVWLIEEKKCVVSRNVSFQENAVYKDLMQRKEQVSCEE 745
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 391 bits (1004), Expect = e-109
Identities = 236/735 (32%), Positives = 390/735 (52%), Gaps = 59/735 (8%)
Query: 1 LEEGKVKIERFDGR-DFGFWKMLM---------------EDYLYQKMLYQPLTGKKPND- 43
+ ++++E+FDGR D+ WK + ED L +K+ LT ++ +
Sbjct: 1 MSAARIEVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEE 60
Query: 44 -MKQEDWDLLDRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIR 102
+++E + R+A I L+++ V I E++ A ++ L +Y N+++ +
Sbjct: 61 VLRRELLEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQ 120
Query: 103 RLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSN 162
+L++ +M E S+ +I+ F II+ L + + +E + LL SLP + +
Sbjct: 121 KLYSFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKY 180
Query: 163 SARDNKLKFDDIRDLILSEDI-----RRKDSGESSNTFGSALNTESRGRGSQKSHNQSQG 217
L D++ I S+++ ++ G++ F TE+RGR Q+ +N +
Sbjct: 181 GLGRVTLSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKE-KTETRGRTEQRGNNNNNK 239
Query: 218 RGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVA 277
+ RSKSR + CW C + ++ NY E+ E
Sbjct: 240 KSRSKSRSKKG--------CWICGESSNGSS---------NYS------EANGLYVSEAL 276
Query: 278 DTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDI 337
+ L+ D WV+D+G S+H +E + G V + + + GIG I +
Sbjct: 277 SSTDIHLE---DEWVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRV 333
Query: 338 RSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGS 397
++ G + L NVR++P + RNL+S+G + GY G + G+ V+ ++ +
Sbjct: 334 KNEAGMVVRLTNVRYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYT 393
Query: 398 LYMV----AEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEH 453
LY++ E+ ++V + + + +WH+RLGHMS+K M ++ KG + K L CE
Sbjct: 394 LYLLQWRPVTEESLSVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLETCED 453
Query: 454 CILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKVWVY 512
CI GK +++ F+ A ++ EKLE VH+D+WG P+ SLG +Y+++FIDD TRKV +Y
Sbjct: 454 CIYGKAKRIGFNLAQHDTR-EKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIY 512
Query: 513 FLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPG 572
FLK+K + F F +W VENQT +IK+L++DNG E+ ++ F +FCS+ GI +T
Sbjct: 513 FLKTKDEAFDKFVEWANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAY 572
Query: 573 TPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEV 632
TP+QNGVAERMNRTL E+ R M SGLPK FW +A +T A LIN+ PS L+Y++P++
Sbjct: 573 TPQQNGVAERMNRTLMEKVRSMLSDSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKR 632
Query: 633 WYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKII 692
W GK S+L+ FGC+++V D KL+P+A K +GY + GY+ W + +K +
Sbjct: 633 WSGKSPIYSYLRRFGCIAFVHTDD---GKLNPRAKKGILVGYPIGVKGYKIWLLEEKKCV 689
Query: 693 RSINVTFNESVLYKD 707
S NV F E+ YKD
Sbjct: 690 VSRNVIFQENASYKD 704
>At1g48710 hypothetical protein
Length = 1352
Score = 380 bits (976), Expect = e-105
Identities = 229/698 (32%), Positives = 369/698 (52%), Gaps = 60/698 (8%)
Query: 53 DRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLFN-LRM 109
D++AL +I L ++ +V + + + L Y+ KV L +R F L+M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 110 GEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSA----- 164
EG V+++ + T+ + L D+ ++ +L+SL + VT + +
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 165 ------------RDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRG----- 207
+ K K +DI + +L+ I ++++G+S G RGRG
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIIEQVLNMQITKEENGQSYQRRGGG-QVRGRGRGGYGNG 244
Query: 208 ----------SQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKK 257
+Q+ N S+GRG+ + R ++ + C+NC + GH+ ++CKAP KK
Sbjct: 245 RGWRPHEDNTNQRGENSSRGRGKGHPKSRYD---KSSVKCYNCGKFGHYASECKAPSNKK 301
Query: 258 NYQKR*DDDESANAATEEVADTLICSLDS-------PVDSWVIDSGASFHTIPSKELLSN 310
+E AN E++ + + + S W +DSGAS H K + +
Sbjct: 302 F-------EEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAE 354
Query: 311 YICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEG 370
G V L D +++ G G+I IR NG + NV ++P +K N++S+GQL ++G
Sbjct: 355 LDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKG 414
Query: 371 YHTTFGGGAWKVT--KGNLVVARGKKRGSLYMVAEEDMIAVTEAI---NSSSIWHQRLGH 425
Y + + NL+ + ++++ + IA + S +WH R GH
Sbjct: 415 YDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGH 474
Query: 426 MSEKGMKIMASKGKMSNLKHVDLG--VCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDV 483
++ G+++++ K + L ++ VCE C+LGKQ K+SF K + LEL+HTDV
Sbjct: 475 LNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKSLELIHTDV 534
Query: 484 WGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLK 543
GP KSLG S Y++ FIDD +RK WVYFLK KS+VF +FKK+K VE ++GL IK+++
Sbjct: 535 CGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMR 594
Query: 544 SDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKM 603
SD GGE+ S+EF K+C +NGIR T+P +P+QNGVAER NRT+ E AR M LPK
Sbjct: 595 SDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKE 654
Query: 604 FWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLD 663
W +A+ A YL+NR P+ + + P+E W G++ +SHL+VFG +++ + +KR KLD
Sbjct: 655 LWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSKLD 714
Query: 664 PKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNE 701
K+ K FIGY ++ GY+ ++ +K I S N+ F+E
Sbjct: 715 DKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDE 752
>At1g58140 hypothetical protein
Length = 1320
Score = 380 bits (975), Expect = e-105
Identities = 229/698 (32%), Positives = 369/698 (52%), Gaps = 60/698 (8%)
Query: 53 DRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLFN-LRM 109
D++AL +I L ++ +V + + + L Y+ KV L +R F L+M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 110 GEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSA----- 164
EG V+++ + T+ + L D+ ++ +L+SL + VT + +
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 165 ------------RDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRG----- 207
+ K K +DI + +L+ I ++++G+S G RGRG
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGG-QVRGRGRGGYGNG 244
Query: 208 ----------SQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKK 257
+Q+ N S+GRG+ + R ++ + C+NC + GH+ ++CKAP KK
Sbjct: 245 RGWRPHEDNTNQRGENSSRGRGKGHPKSRYD---KSSVKCYNCGKFGHYASECKAPSNKK 301
Query: 258 NYQKR*DDDESANAATEEVADTLICSLDS-------PVDSWVIDSGASFHTIPSKELLSN 310
+E AN E++ + + + S W +DSGAS H K + +
Sbjct: 302 F-------EEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAE 354
Query: 311 YICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEG 370
G V L D +++ G G+I IR NG + NV ++P +K N++S+GQL ++G
Sbjct: 355 LDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKG 414
Query: 371 YHTTFGGGAWKVT--KGNLVVARGKKRGSLYMVAEEDMIAVTEAI---NSSSIWHQRLGH 425
Y + + NL+ + ++++ + IA + S +WH R GH
Sbjct: 415 YDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGH 474
Query: 426 MSEKGMKIMASKGKMSNLKHVDLG--VCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDV 483
++ G+++++ K + L ++ VCE C+LGKQ K+SF K + LEL+HTDV
Sbjct: 475 LNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDV 534
Query: 484 WGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLK 543
GP KSLG S Y++ FIDD +RK WVYFLK KS+VF +FKK+K VE ++GL IK+++
Sbjct: 535 CGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMR 594
Query: 544 SDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKM 603
SD GGE+ S+EF K+C +NGIR T+P +P+QNGVAER NRT+ E AR M LPK
Sbjct: 595 SDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKE 654
Query: 604 FWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLD 663
W +A+ A YL+NR P+ + + P+E W G++ +SHL+VFG +++ + +KR KLD
Sbjct: 655 LWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLD 714
Query: 664 PKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNE 701
K+ K FIGY ++ GY+ ++ +K I S N+ F+E
Sbjct: 715 DKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDE 752
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 378 bits (970), Expect = e-105
Identities = 228/698 (32%), Positives = 370/698 (52%), Gaps = 60/698 (8%)
Query: 53 DRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLFN-LRM 109
D++AL +I L ++ +V + + + L Y+ KV L +R F L+M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 110 GEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSA----- 164
EG V+++ + T+ + L D+ ++ +L+SL + VT + +
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 165 ------------RDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRG----- 207
+ K K +DI + +L+ I ++++G+S G RGRG
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGG-QVRGRGRGGYGNG 244
Query: 208 ----------SQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKK 257
+Q+ N S+GRG+ + R ++ + C+NC + GH+ ++CKAP KK
Sbjct: 245 RGWRPHEDNTNQRGENSSRGRGKGHPKSRYD---KSSVKCYNCGKFGHYASECKAPSNKK 301
Query: 258 NYQKR*DDDESANAATEEVADTLICSLDS-------PVDSWVIDSGASFHTIPSKELLSN 310
+K AN E++ + + + S W +DSGAS H K + +
Sbjct: 302 FKEK-------ANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAE 354
Query: 311 YICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEG 370
G V L D +++ G G+I IR NG + NV ++P +K N++S+GQL ++G
Sbjct: 355 LDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKG 414
Query: 371 YHTTFGGGAWKVT--KGNLVVARGKKRGSLYMVAEEDMIAVTEAI---NSSSIWHQRLGH 425
Y + + NL+ + ++++ + IA + S +WH R GH
Sbjct: 415 YDIRLKDNNLSIRDKESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGH 474
Query: 426 MSEKGMKIMASKGKMSNLKHVDLG--VCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDV 483
++ G+++++ K + L ++ VCE C+LG Q K+SF K + LEL+HTDV
Sbjct: 475 LNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGNQFKMSFPKESSSRAQKPLELIHTDV 534
Query: 484 WGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLK 543
GP KSLG S Y++ FIDD +RK WVYFLK KS+VF +FKK+K VE ++GL IK+++
Sbjct: 535 CGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMR 594
Query: 544 SDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKM 603
SD+GGE+ S+EF K+C +NGIR T+P +P+QNGVAER NRT+ E AR M LPK
Sbjct: 595 SDSGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKE 654
Query: 604 FWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLD 663
W +A+ A YL+NR P+ + + P+E W G++ +SHL+VFG +++ + +KR+KLD
Sbjct: 655 LWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNKLD 714
Query: 664 PKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNE 701
K+ K FIGY ++ GY+ ++ +K I S N+ F+E
Sbjct: 715 DKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDE 752
>At3g61330 copia-type polyprotein
Length = 1352
Score = 377 bits (969), Expect = e-105
Identities = 225/691 (32%), Positives = 363/691 (51%), Gaps = 46/691 (6%)
Query: 53 DRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLFN-LRM 109
D++AL +I L ++ +V + + + L Y+ KV L +R F L+M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 110 GEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSA----- 164
EG V+++ + T+ + L D+ ++ +L+SL + VT + +
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 165 ------------RDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSALNTESRGRG----- 207
+ K K +DI + +L+ I ++++G+S G RGRG
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIAEQVLNMQITKEENGQSYQRRGGG-QVRGRGRGGYGNG 244
Query: 208 ----------SQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKK 257
+Q+ N S+GRG+ + R ++ + C+NC + GH+ ++CKAP KK
Sbjct: 245 RGWRPHEDNTNQRGENSSRGRGKGHPKSRYD---KSSVKCYNCGKFGHYASECKAPSNKK 301
Query: 258 NYQKR*DDDESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFG 317
+K +E + + W +DSGAS H K + + G
Sbjct: 302 FEEKAHYVEEKIQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDESVRG 361
Query: 318 KVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGG 377
V L D +++ G G+I IR NG + NV ++P +K N++S+GQL ++GY
Sbjct: 362 NVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKD 421
Query: 378 GAWKVT--KGNLVVARGKKRGSLYMVAEEDMIAVTEAI---NSSSIWHQRLGHMSEKGMK 432
+ + NL+ + ++++ + IA + S +WH R GH++ G++
Sbjct: 422 NNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLE 481
Query: 433 IMASKGKMSNLKHVDLG--VCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVK 490
+++ K + L ++ VCE C+LGKQ K+SF K + LEL+HTDV GP K
Sbjct: 482 LLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPK 541
Query: 491 SLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEY 550
SLG S Y++ FIDD +RK WVYFLK KS+VF +FKK+K VE ++GL IK+++SD GGE+
Sbjct: 542 SLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEF 601
Query: 551 DSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAIN 610
S+EF K+C +NGIR T+P +P+QNGV ER NRT+ E AR M LPK W +A+
Sbjct: 602 TSKEFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEAVA 661
Query: 611 TAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCF 670
A YL+NR P+ + + P+E W G++ +SHL+VFG +++ + +KR KLD K+ K
Sbjct: 662 CAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYI 721
Query: 671 FIGYGSDMYGYRFWDEQNRKIIRSINVTFNE 701
FIGY ++ GY+ ++ +K I S N+ F+E
Sbjct: 722 FIGYDNNSKGYKLYNPDTKKTIISRNIVFDE 752
>At1g47360 polyprotein, putative
Length = 1182
Score = 366 bits (939), Expect = e-101
Identities = 222/648 (34%), Positives = 345/648 (52%), Gaps = 59/648 (9%)
Query: 76 KTTADLMKALSNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKIT 135
+T + + L ++ ++V+ + +M E + E+I+ F I++ L+ ++I
Sbjct: 59 QTAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIE 118
Query: 136 FDNELMVLSLLQSLPDSWAATVTAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSNTF 195
+E+ + LL SLP + V + S KL+ DD+ R K+ S N
Sbjct: 119 VTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVM-----VGARDKERELSQNNR 173
Query: 196 GSALNTESRGRGSQKSHNQ-SQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQC-KAP 253
A +RGR K++NQ ++G+ RS+S+ RV CW C ++GHF QC K
Sbjct: 174 PVAEGHYARGRPEGKNNNQGNKGKNRSRSKSADGKRV-----CWICGKEGHFKKQCYKWL 228
Query: 254 RKKKNYQKR*DDDESANAATEEVADTLICSLDS---------PVDSWVIDSGASFHTIPS 304
+ K+ Q+ D ES+ A + E+ D + + + D W +D+G SFH P
Sbjct: 229 ERNKSKQQGSDVGESSLAKSSEIFDPAMVIMATGETLLVTGRDADEWFLDTGCSFHMTPR 288
Query: 305 KELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIG 364
++L ++ G V + + + GIG I IR+++GT L +VR+VP + RNLIS+G
Sbjct: 289 RDLFKDFKELSSGFVKMGNDTYSPVKGIGSIKIRNNDGTQVILTDVRYVPNMARNLISLG 348
Query: 365 QLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV---AEE-DMIAVTEAINSSSIWH 420
L+D+G G K+ KG + +G+KR +LY++ AE + + E + +S+WH
Sbjct: 349 TLEDKGCWFKSQDGVLKIVKGCSTILKGQKRETLYILEGLAENGESHSSAELKDETSLWH 408
Query: 421 QRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVH 480
RLGHMS+KGM+I+ K + +L CE C+ GK +VSF+ A +K EKL +H
Sbjct: 409 SRLGHMSQKGMEILVKKDCLQRETIKELKFCEDCVYGKNHRVSFAPAQHVTK-EKLAYIH 467
Query: 481 TDVWG-PAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKI 539
+D+WG P SLG +Y+++F+DD +RK K+
Sbjct: 468 SDLWGSPHNPASLGNCQYFISFVDDYSRK-----------------------------KV 498
Query: 540 KSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSG 599
K L++DNG EY + F+KFC + GI KT TP+QNG+AER+NRT+ ++ R M +SG
Sbjct: 499 KKLRTDNGLEYCNHYFEKFCKDEGIVRHKTCAYTPQQNGIAERLNRTIMDKVRSMLSESG 558
Query: 600 LPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKR 659
+ K FW +A +TA YLINR PS LD+ LPEE W G L L+ FGC+ Y+ D +
Sbjct: 559 MDKKFWAEAASTAVYLINRSPSTALDFDLPEEKWTGALPDLKGLRKFGCLVYIHAD---Q 615
Query: 660 DKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNESVLYKD 707
KL+P+A K F Y + GY+ W + +K + S NV F E ++YKD
Sbjct: 616 GKLNPRAKKDVFTSYLEGVKGYKVWVLEEKKCVISRNVIFREEIMYKD 663
>At4g10460 putative retrotransposon
Length = 1230
Score = 363 bits (932), Expect = e-100
Identities = 233/742 (31%), Positives = 374/742 (50%), Gaps = 67/742 (9%)
Query: 1 LEEGKVKIERFDGR-DFGFWKMLMEDYL----YQKMLYQPLTGKKPNDMKQE--DWDLLD 53
+ +V++E+FDG D+ WK + ++ L + + P + ++E + + D
Sbjct: 1 MSSARVEMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGD 60
Query: 54 RQAL---------GVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIRRL 104
++AL I L++S V EKT +++AL +Y N+++L ++L
Sbjct: 61 KEALMEEKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKL 120
Query: 105 FNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSA 164
++ +M E SV +I+ F +I+ L + + +E + LL SLP + + +
Sbjct: 121 YSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGS 180
Query: 165 RDNKLKFDDIRDLILSEDIRRKDSGESSNTFGSAL----NTESRGRGSQKSHNQSQGRGR 220
L D++ I S+++ + +S L E+RG QK ++GR R
Sbjct: 181 GRTTLSVDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKG-NKGRSR 239
Query: 221 SKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEE----- 275
S+S+G CW C +GHF C K++N K AAT +
Sbjct: 240 SRSKGWKG--------CWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSE 291
Query: 276 -----VADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIV 330
V++ L + + + WV+D+G ++H KE G V + +
Sbjct: 292 GSGYYVSEALHSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKST---- 347
Query: 331 GIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVA 390
+ V+++P + RNL+S+G L++ GY G V +G +
Sbjct: 348 ---------------SKFRVKYIPDMDRNLLSMGTLEEHGYSFESKNGVLVVKEGTRTLL 392
Query: 391 RGKKRGSLYMVAEEDMIAVTEAI----NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHV 446
G + LY++ + ++ + + + + +WH+RLGH+S+K M I+ KG + K
Sbjct: 393 IGSRHEKLYLLQGKPEVSHSMTVERRNDDTVLWHRRLGHISQKNMDILVKKGYLDGKKVS 452
Query: 447 DLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVTFIDDS 505
L +CE CI GK R++SF A ++ +KL VH+D+WG P+ SLG +Y+++FID
Sbjct: 453 KLELCEDCIYGKARRLSFVVATHNTE-DKLNYVHSDLWGAPSVPLSLGKCQYFISFIDVY 511
Query: 506 TRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIR 565
+RK WVYFLK K + F F +W VENQTG KIK L+ DNG E+ +Q+F FC E GI
Sbjct: 512 SRKTWVYFLKHKDEAFGTFAEWSVMVENQTGRKIKILRIDNGLEFCNQQFNDFCKEKGIV 571
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
+T TP+QNGVAERMN T+ E+ R M SGLPK FW +A NT LIN+ PS ++
Sbjct: 572 RHQTCAYTPQQNGVAERMNHTIMEKVRRMLSYSGLPKTFWAEATNTVVTLINKTPSSAVN 631
Query: 626 YQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWD 685
+++ ++ W GK ++LK FGCV++ D KL P+A K F+GY S GY+ W
Sbjct: 632 FEISDKRWSGKSPVYNYLKRFGCVAFTYAD---EGKLVPRAKKGVFLGYLSGEKGYKVWL 688
Query: 686 EQNRKIIRSINVTFNESVLYKD 707
+ RK S NVTF E+ +Y+D
Sbjct: 689 LEERKCSVSRNVTFQENAVYRD 710
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 348 bits (894), Expect = 5e-96
Identities = 210/691 (30%), Positives = 355/691 (50%), Gaps = 43/691 (6%)
Query: 36 LTGKKPNDMKQEDWDLLDRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAA 95
LTG + ++ ++ + D + + ++ K + I+ ++T+ DL +++ Y+
Sbjct: 10 LTGAQRTELAEKT--VKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRV 67
Query: 96 NKVHL--IRRLFN-LRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDS 152
L +RR F L M G ++T + + I + + ++ + +V +L++L +
Sbjct: 68 QSAQLQRLRRSFEVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEK 127
Query: 153 WAATVTAVSNSARDNKLKFDDIRDLIL--SEDIRRKDSGESSNTFGSALNTESR-----G 205
+ V A+ S +L D ++ ++ +++ R D E L E++ G
Sbjct: 128 FTYVVCAIEESNNIKELTVDGLQSSLMVHEQNLSRHDVEER------VLKAETQWRPDGG 181
Query: 206 RGSQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DD 265
RG S ++ +GRG + RGR R+ + C+ C + GH+ +C + K+ NY + ++
Sbjct: 182 RGRGGSPSRGRGRGGYQGRGRGYVN-RDTVECFKCHKMGHYKAECPSWEKEANYVEM-EE 239
Query: 266 DESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGK 325
D A E++ D W +DSG S H ++E G V L D +
Sbjct: 240 DLLLMAHVEQIGD-------EEKQIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDDR 292
Query: 326 PLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGA---WKV 382
+ + G G + + +G + + +V VPG+K NL S+GQL +G G W
Sbjct: 293 RMAVEGKGKLRLEV-DGRIQVISDVYFVPGLKNNLFSVGQLQQKGLRFIIEGDVCEVWHK 351
Query: 383 TKGNLVVARGKKRGSLYMV--------AEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIM 434
T+ +V+ + +++V E+ + ++++WH+R GH++ +G++ +
Sbjct: 352 TEKRMVMHSTMTKNRMFVVFAAVKKSKETEETRCLQVIGKANNMWHKRFGHLNHQGLRSL 411
Query: 435 ASKGKMSNLKHVDLG----VCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVK 490
A K + L DLG VC+ C+ GKQ + S K ++ L+LVHTD+ GP
Sbjct: 412 AEKEMVKGLPKFDLGEEEAVCDICLKGKQIRESIPKESAWKSTQVLQLVHTDICGPINPA 471
Query: 491 SLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEY 550
S G RY + FIDD +RK W Y L KS+ F FK++K EVE ++G K+ L+SD GGEY
Sbjct: 472 STSGKRYILNFIDDFSRKCWTYLLSEKSETFQFFKEFKAEVERESGKKLVCLRSDRGGEY 531
Query: 551 DSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAIN 610
+S+EF ++C E GI+ T TP+QNGVAER NR++ RCM ++ +P+ FWP+A+
Sbjct: 532 NSREFDEYCKEFGIKRQLTAAYTPQQNGVAERKNRSVMNMTRCMLMEMSVPRKFWPEAVQ 591
Query: 611 TAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCF 670
A Y++NR PS L+ PEE W + S+ HL++FG ++Y L+ KR KLD K+IKC
Sbjct: 592 YAVYILNRSPSKALNDITPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIKLDEKSIKCV 651
Query: 671 FIGYGSDMYGYRFWDEQNRKIIRSINVTFNE 701
G + YR +D KI+ S +V F+E
Sbjct: 652 MFGVSKESKAYRLYDPATGKILISRDVQFDE 682
>At3g60170 putative protein
Length = 1339
Score = 338 bits (868), Expect = 5e-93
Identities = 217/723 (30%), Positives = 355/723 (49%), Gaps = 43/723 (5%)
Query: 8 IERFDGRDFGFWKMLMEDYLYQKMLYQPL--------TGKKPNDMKQ----EDWDLLDRQ 55
I RFDG + FW M ME++L + L++ + G P Q E+ L D +
Sbjct: 12 IPRFDGY-YDFWSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDLK 70
Query: 56 ALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLFNL-RMGEG 112
+ + + + I+++ T+ + +++ Y+ + L +R+ F L M EG
Sbjct: 71 VKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKEG 130
Query: 113 NSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSARDNKLKFD 172
+ + T+++++ + + +V +L+SL + V ++ S + L D
Sbjct: 131 EKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSID 190
Query: 173 DIRDLILSEDIRRKDSGESSNTFGSALN---TESRGRGSQKSHNQSQGRGRSKSRGRSQT 229
++ +L + R + ++ RGRG + RGR + RGRS T
Sbjct: 191 ELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRGVFRG-----SRGRGRGRGRSGT 245
Query: 230 RVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVADTLICSLDSPVD 289
R + C+ C GHF +C K NY + +++E A E + +
Sbjct: 246 N-RAIVECYKCHNLGHFQYECPEWEKNANYAELEEEEELLLMAYVEQNQA------NRDE 298
Query: 290 SWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHN 349
W +DSG S H SKE S G V L + + +VG G + ++ NG +
Sbjct: 299 VWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKV-NGVTQVIPE 357
Query: 350 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKV---TKGNLVVARGKKRGSLYMVAEE-- 404
V +VP ++ NL+S+GQL + G G KV +KG ++ +++A +
Sbjct: 358 VYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASKPQ 417
Query: 405 --DMIAVTEAI--NSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG--VCEHCILGK 458
+ TE + + +WH R GH++++G+K++A K + L + +C C+ GK
Sbjct: 418 KNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKEICAICLTGK 477
Query: 459 QRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKS 518
Q + S SK S +L+LVH+D+ GP S G RY ++FIDD TRK WVYFL KS
Sbjct: 478 QHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYFLHEKS 537
Query: 519 DVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNG 578
+ F+ FK +K VE + G + L++D GGE+ S EF +FC +GI T TP+QNG
Sbjct: 538 EAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAAFTPQQNG 597
Query: 579 VAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEV 638
VAER NRT+ R M + +PKMFW +A + ++ NR P+ ++ PEE W G++
Sbjct: 598 VAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVHIQNRSPTAAVEGMTPEEAWSGRKP 657
Query: 639 SLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVT 698
+ + +VFGC+ YV I KR KLD K+ KC F+G + +R +D +KI+ S +V
Sbjct: 658 VVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKIVISKDVV 717
Query: 699 FNE 701
F+E
Sbjct: 718 FDE 720
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 306 bits (783), Expect = 3e-83
Identities = 215/743 (28%), Positives = 359/743 (47%), Gaps = 75/743 (10%)
Query: 11 FDGRDFGFWKMLMEDYLYQKMLYQPLTGKKPNDMKQ--------------EDWDLLDRQA 56
FDG + FW + M + L+ + P + Q E+ D A
Sbjct: 12 FDGEKYDFWSIKMATIFRTRKLWSVVEEGVPVEPVQAEETPETARAKTLREEAVTNDTMA 71
Query: 57 LGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHL--IRRLF-NLRMGEGN 113
L +++ ++ + I ++ + L + Y+ V L +RR + NL+M + +
Sbjct: 72 LQILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYDND 131
Query: 114 SVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSARDNKLKFDD 173
++ + + QL+ N ++ +L SLP + + V+ + + + L +
Sbjct: 132 NIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTMSE 191
Query: 174 IRDLILSEDIR------------------RKDSGESSNTFGSALNTESRGRGSQKS--HN 213
+ ++ +++ R ++SG + + +N + + G KS H
Sbjct: 192 LLGILKAQEARVTAREESTKEGAFYVRSKGRESGFKQDNTNNRVNQDKKWCGFHKSSKHT 251
Query: 214 QSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNY----QKR*DDDESA 269
+ + R + K+ + + R++I C+ C + GH+ N+C++ K++ + ++ ++D
Sbjct: 252 EEECREKPKNDDHGKNK-RSNIKCYKCGKIGHYANECRSKNKERAHVTLEEEDVNEDHML 310
Query: 270 NAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDI 329
+A+EE + TL D W++DSG + H + SN + + +G +
Sbjct: 311 FSASEEESTTL------REDVWLVDSGCTNHMTKEERYFSNINKSIKVPIRVRNGDIVMT 364
Query: 330 VGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVV 389
G GDI + + +G + NV VPG+++NL+S+ Q+ GY F + N
Sbjct: 365 AGKGDITVMTRHGKR-IIKNVFLVPGLEKNLLSVPQIISSGYWVRFQDKRCIIQDAN--- 420
Query: 390 ARGKKRGSLYMVAEEDMI--------AVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMS 441
GK+ ++ M + I A+T + + WH+RLGH+S K ++ M K ++
Sbjct: 421 --GKEIMNIEMTDKSFKIKLSSVEEEAMTANVQTEETWHKRLGHVSNKRLQQMQDKELVN 478
Query: 442 NLKH--VDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYV 499
L V C+ C LGKQ + SF K + EKLE+VHTDV GP +S+ GSRYYV
Sbjct: 479 GLPRFKVTKETCKACNLGKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYV 538
Query: 500 TFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFC 559
F+DD T WVYFLK KS+ F+ FKK+K VE Q+ IK+L+ + FC
Sbjct: 539 LFLDDYTHMCWVYFLKQKSETFATFKKFKALVEKQSNCSIKTLR----------PMEVFC 588
Query: 560 SENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRG 619
+ GI T+P +P+QNG AER NR+L E AR M ++ LP W +A+ T+AYL NR
Sbjct: 589 EDEGINRQVTLPYSPQQNGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQNRL 648
Query: 620 PSVPL-DYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDM 678
PS + D P E W G + ++SHL++FG + YV I KR KLD KA IGY +
Sbjct: 649 PSKAIEDDVTPMEKWCGHKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQT 708
Query: 679 YGYRFWDEQNRKIIRSINVTFNE 701
GYR + ++ K+ S +V F E
Sbjct: 709 KGYRVFLLEDEKVEVSRDVVFQE 731
>At4g04380 putative polyprotein
Length = 778
Score = 296 bits (758), Expect = 3e-80
Identities = 159/412 (38%), Positives = 243/412 (58%), Gaps = 11/412 (2%)
Query: 289 DSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLH 348
D W++D+G S+H +E L ++ V + + + G+G + I + NG TL
Sbjct: 197 DEWIMDTGCSYHMTHKREWLEDFDEEAGCSVRMGNKTISRVKGVGTVRIANDNGLTVTLQ 256
Query: 349 NVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV----AEE 404
NVR++ + R +S+G + G +V G V+ G++ +LY++ A +
Sbjct: 257 NVRYISDMDRIFLSLGTFEKAGRKFESENDMLRVNAGEQVLLEGRRYDTLYILHGKPATD 316
Query: 405 DMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSF 464
+ +AV +A + +WH+RL HMS+K M ++ KG + K L CE CI GK +K+ F
Sbjct: 317 ESLAVAKANDDIVLWHRRLCHMSQKNMSLLVKKGFLDKKKVSMLDTCEDCIYGKAKKIGF 376
Query: 465 SKAGRKSKSEKLELVHTDVWG-PAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSV 523
+ A +K EKLE VH D+WG P +LG +Y+++FIDD TRKV VYFLK+K + F
Sbjct: 377 NFAQHDTK-EKLEYVHYDLWGAPTMPMALGNCQYFISFIDDHTRKVCVYFLKTKDEAFEK 435
Query: 524 FKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERM 583
F W + VENQ+G ++K+L++DNG E+ ++ F FC E GI+ +T P TP+QNGV ERM
Sbjct: 436 FVSWISLVENQSGNRVKTLRTDNGLEFCNRMFDGFCEERGIQRHRTCPYTPQQNGVVERM 495
Query: 584 NRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHL 643
NRT+ ER R M S LPK FW +A +TA LIN+ P +++ P++ W GK S+L
Sbjct: 496 NRTIMERVRSMLCDSRLPKRFWAEATHTAVLLINKTPCSANNFEFPDKRWSGKAPIYSYL 555
Query: 644 KVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFW--DEQNRKIIR 693
+ +GCV++V D KL+ +A K IGY S + GY+ W +E+N ++R
Sbjct: 556 RRYGCVTFVQTDD---GKLNLRAKKGVLIGYPSGVKGYKVWLIEERNSHLVR 604
Score = 45.8 bits (107), Expect = 8e-05
Identities = 24/103 (23%), Positives = 53/103 (51%)
Query: 81 LMKALSNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNEL 140
+++A +Y N+++L ++L+ +M E S+ +I+ F I++ L ++ + +E
Sbjct: 68 VLEASDRLYMSKALPNQIYLKQKLYRFKMSENLSMEGNIDEFLHIVADLENLNVLVSDED 127
Query: 141 MVLSLLQSLPDSWAATVTAVSNSARDNKLKFDDIRDLILSEDI 183
+ LL SLP S+ + S+ L D++ I S+++
Sbjct: 128 QTILLLMSLPKSFDQLKDTLQYSSGKTVLTLDEVTAAIYSKEL 170
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 278 bits (710), Expect = 1e-74
Identities = 220/697 (31%), Positives = 331/697 (46%), Gaps = 48/697 (6%)
Query: 36 LTGKKPNDMKQEDWDLLDRQALGVIRLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAA 95
+T ++PN + E W D+ + TLS+ V ++ N T+ + +L+ + K A
Sbjct: 64 VTSEEPNPL-YESWFCTDQLVRSWLFGTLSEEVLGHVHNLSTSRQIWVSLAENFNKSSVA 122
Query: 96 NKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAA 155
+ L + L L E + + F TI LSS+ D + + L L +
Sbjct: 123 REFSLRQNLQLLSKKE-KPFSVYCREFKTICDALSSIGKPVDESMKIFGFLNGLGRDYDP 181
Query: 156 TVTAVSNSARDNKLK---FDDIRDLILSEDIRRKDSGESSN-TFGSALNTESRGRGSQKS 211
T + +S +KL F+D+ + D + + E+++ T A N E GS +
Sbjct: 182 ITTVIQSSL--SKLPTPTFNDVVSEVQGFDSKLQSYEEAASVTPHLAFNIERSESGSPQY 239
Query: 212 HNQSQGRGRS---KSRGRSQTRVRNDI-------------TCWNCDRKGHFTNQCKAPRK 255
+ +GRGRS K RG TR R C C R GH +C R
Sbjct: 240 NPNQKGRGRSGQNKGRGGYSTRGRGFSQHQSSPQVSGPRPVCQICGRTGHTALKCYN-RF 298
Query: 256 KKNYQKR*DDDESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKE-LLSNYICG 314
NYQ A + TL S D+ + W DS A+ H S L S
Sbjct: 299 DNNYQ-----------AEIQAFSTLRVSDDTGKE-WHPDSAATAHVTSSTNGLQSATEYE 346
Query: 315 KFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTT 374
V + DG L I G I+SSNG + L+ V VP I+++L+S+ +L D+ Y
Sbjct: 347 GDDAVLVGDGTYLPITHTGSTTIKSSNGKI-PLNEVLVVPNIQKSLLSVSKLCDD-YPCG 404
Query: 375 FGGGAWKVTKGNL----VVARGKKRGSLYMVAEEDMIAV---TEAINSSSIWHQRLGHMS 427
A KV +L VV G +R LY++ ++ +A+ + + +WH RLGH +
Sbjct: 405 VYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQEFVALYSNRQCAATEEVWHHRLGHAN 464
Query: 428 EKGMKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPA 487
K ++ + + + K VCE C +GK ++ F + + L+ +H D+WGP+
Sbjct: 465 SKALQHLQNSKAIQINKSRTSPVCEPCQMGKSSRLPFLISDSRVL-HPLDRIHCDLWGPS 523
Query: 488 PVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNG 547
PV S G +YY F+DD +R W Y L +KS+ SVF ++ VENQ KIK +SD G
Sbjct: 524 PVVSNQGLKYYAIFVDDYSRYSWFYPLHNKSEFLSVFISFQKLVENQLNTKIKVFQSDGG 583
Query: 548 GEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPD 607
GE+ S + K SE+GI + P TP+QNG+AER +R L E M S P+ FW +
Sbjct: 584 GEFVSNKLKTHLSEHGIHHRISCPYTPQQNGLAERKHRHLVELGLSMLFHSHTPQKFWVE 643
Query: 608 AINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAI 667
+ TA Y+INR PS L P E +G++ S L+VFG Y + ++K DP+++
Sbjct: 644 SFFTANYIINRLPSSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSL 703
Query: 668 KCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNESVL 704
+C F+GY S GYR + K+ S NV FNES L
Sbjct: 704 QCVFLGYNSQYKGYRCFYPPTGKVYISRNVIFNESEL 740
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 272 bits (696), Expect = 4e-73
Identities = 165/426 (38%), Positives = 233/426 (53%), Gaps = 22/426 (5%)
Query: 289 DSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLH 348
++W+IDSG + H P+++L + + + +G + G GDI++ + +
Sbjct: 28 NTWLIDSGCTNHMTPNEKLFTKINRDFKVPIRVGNGAVMMSEGKGDIEVMTRKDKRG-IR 86
Query: 349 NVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMV------- 401
+V VP + +NL+S+ Q+ GY T + AR KK G + MV
Sbjct: 87 DVLLVPKLGKNLLSVPQMIINGYQVTLKNNYCTIHDS----AR-KKIGEVEMVNKSFHLR 141
Query: 402 ----AEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLK--HVDLGVCEHCI 455
E M+A EA + +WH+RLGH +KI+ SK ++ L +V+ G CE CI
Sbjct: 142 WLSNEETAMVAKDEA---TELWHKRLGHTGHSNLKILQSKEMVTGLPKFNVEEGKCESCI 198
Query: 456 LGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLK 515
L K + F K KLEL+H+DV GP S+ GSRY +TFIDD+TR VWVYFLK
Sbjct: 199 LSKHSRDPFPKESETRAKHKLELIHSDVCGPMQNSSINGSRYILTFIDDATRMVWVYFLK 258
Query: 516 SKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPE 575
+KS+VF FKK+K VEN +IK L+ D G EY S+EF +F NGI T +P+
Sbjct: 259 AKSEVFQTFKKFKNLVENNANCRIKKLRIDRGTEYLSKEFSEFLEGNGIERQLTAAYSPQ 318
Query: 576 QNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYG 635
QN V+ER NR+L E AR M LP W +A++ AAY NR P+ L + P E W
Sbjct: 319 QNEVSERRNRSLVEMARAMIKAKDLPLKLWAEAVHVAAYAQNRTPTRTLKNKTPLEAWSD 378
Query: 636 KEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSI 695
+ S+SH+KVFG + YV I +KR K D K+ + F+GY S GYR + + KI S
Sbjct: 379 SKPSVSHMKVFGSICYVHIPDEKRRKWDDKSKRAIFVGYSSQTKGYRVYLLKENKIDISR 438
Query: 696 NVTFNE 701
+V F+E
Sbjct: 439 DVIFDE 444
>At2g14930 pseudogene
Length = 1149
Score = 269 bits (687), Expect = 5e-72
Identities = 200/672 (29%), Positives = 323/672 (47%), Gaps = 54/672 (8%)
Query: 61 RLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHIN 120
++ +S+++ +V KT+ ++ L+ + + ++ L RRL +L EG ++ E++
Sbjct: 84 QVVMSEDILSVVVGSKTSHEVWMNLAKHFNRISSSRIFELQRRLHSLSK-EGKTMEEYLR 142
Query: 121 SFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSN--------SARDNKLKFD 172
TI QL+SV ++ + +++ L + +T++ S D +
Sbjct: 143 YLKTICDQLASVGSPVAEKMKIFAMVHGLTREYEPLITSLEGTLDAFPGPSYEDVVYRLK 202
Query: 173 DIRDLILSEDIRRKDSGESSNTFGSALNTESRGRGSQKSHNQSQGRGRSKSRGR------ 226
+ D + + + NTF S+ +RGRG + + +G+G +RGR
Sbjct: 203 NFDDRLQGYTVTDVSPHLAFNTFRSS----NRGRGGRNN----RGKGNFSTRGRGFQQQF 254
Query: 227 ----SQTRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVADTLIC 282
S C C ++GH+ QC R DD + A L
Sbjct: 255 SSSSSSVSASEKPMCQICGKRGHYALQC---------WHRFDDSYQHSEAAAAAFSALHI 305
Query: 283 SLDSPVDSWVIDSGASFHTIPSKELLSN---YICGKFGKVYLADGKPLDIVGIGDIDIRS 339
+ S WV DS A+ H + L Y+ V +DG L I IG ++ S
Sbjct: 306 TDVSDDSGWVPDSAATAHITNNSSRLQQMQPYLGND--TVMASDGNFLPITHIGSANLPS 363
Query: 340 SNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGS-- 397
++G L L +V P I ++L+S+ +L + Y +F A V + + +GS
Sbjct: 364 TSGNL-PLKDVLVCPNIAKSLLSVSKLTKD-YPCSFTFDADGVLVKDKATCKVLTKGSST 421
Query: 398 ---LYMVAEE--DMIAVTEAINSSS-IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVC 451
LY + M T + ++ +WH RLGH + + ++++A+K K + +C
Sbjct: 422 SEGLYKLENPKFQMFYSTRQVKATDEVWHMRLGHPNPQVLQLLANK-KAIQINKSTSKMC 480
Query: 452 EHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWV 511
E C LGK ++ F A S LE VH D+WGPAPV S+ G +YYV FID+ +R W
Sbjct: 481 ESCRLGKSSRLPFI-ASDFIASRPLERVHCDLWGPAPVSSIQGFQYYVIFIDNRSRFCWF 539
Query: 512 YFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIP 571
Y LK KSD S+F K+++ VEN KI + +SD GGE+ S F + E+GI+ + P
Sbjct: 540 YPLKHKSDFCSLFMKFQSFVENLLQTKIGTFQSDGGGEFTSNRFLQHLQESGIQHYISCP 599
Query: 572 GTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQ-LPE 630
TP+QNG+AER +R L ER + QS P+ FW +A TA +L N P+ LD P
Sbjct: 600 HTPQQNGLAERKHRQLTERGLTLMFQSKAPQRFWVEAFFTANFLSNLLPTSALDSSTTPY 659
Query: 631 EVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRK 690
+V +GK S L+ FGC + + + R+K DP+++KC F+GY GYR + +
Sbjct: 660 QVLFGKAPDYSALRTFGCACFPTLRAYARNKFDPRSLKCIFLGYTEKYKGYRCFFPPTNR 719
Query: 691 IIRSINVTFNES 702
+ S +V F+ES
Sbjct: 720 VYLSRHVLFDES 731
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,347,522
Number of Sequences: 26719
Number of extensions: 727966
Number of successful extensions: 2912
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 2338
Number of HSP's gapped (non-prelim): 263
length of query: 720
length of database: 11,318,596
effective HSP length: 106
effective length of query: 614
effective length of database: 8,486,382
effective search space: 5210638548
effective search space used: 5210638548
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0220.7


