

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0219.11
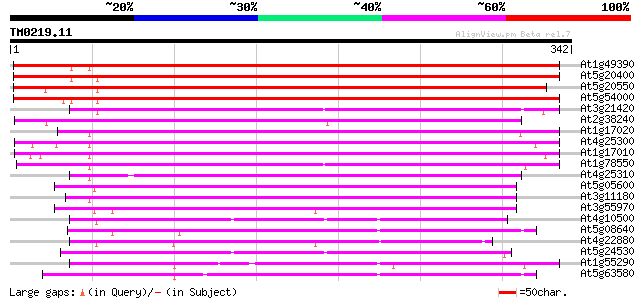
(342 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49390 hypothetical protein 322 2e-88
At5g20400 ethylene-forming-enzyme-like dioxygenase-like protein 311 3e-85
At5g20550 ethylene-forming-enzyme-like dioxygenase - like 301 3e-82
At5g54000 ethylene-forming-enzyme-like dioxygenase 298 3e-81
At3g21420 unknown protein 229 2e-60
At2g38240 putative anthocyanidin synthase 219 2e-57
At1g17020 SRG1-like protein 212 3e-55
At4g25300 SRG1-like protein 210 8e-55
At1g17010 SRG1-like protein 209 1e-54
At1g78550 unknown protein 204 4e-53
At4g25310 SRG1-like protein 201 6e-52
At5g05600 leucoanthocyanidin dioxygenase-like protein 199 1e-51
At3g11180 putative leucoanthocyanidin dioxygenase 195 3e-50
At3g55970 leucoanthocyanidin dioxygenase -like protein 192 3e-49
At4g10500 putative Fe(II)/ascorbate oxidase 180 1e-45
At5g08640 flavonol synthase (FLS) (sp|Q96330) 179 2e-45
At4g22880 putative leucoanthocyanidin dioxygenase (LDOX) 177 1e-44
At5g24530 flavanone 3-hydroxylase-like protein 176 2e-44
At1g55290 leucoanthocyanidin dioxygenase 2, putative 174 8e-44
At5g63580 flavonol synthase 171 5e-43
>At1g49390 hypothetical protein
Length = 348
Score = 322 bits (825), Expect = 2e-88
Identities = 162/342 (47%), Positives = 224/342 (65%), Gaps = 9/342 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFG-SKDSSTLIP---IPIIDVSLLSS-----EDEQGK 53
K+VQE+ P Y+ G S+ + +P IP ID+SLL S ++E K
Sbjct: 7 KTVQEVVAAGQGLPERYLHAPTGEGESQPLNGAVPEMDIPAIDLSLLFSSSVDGQEEMKK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q + HG++ +LDKI ++ K FFALP EEK K AR +GYGND ++
Sbjct: 67 LHSALSTWGVVQVMNHGITEAFLDKIYKLTKQFFALPTEEKHKCARETGNIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
S QVLDW RL L +P++KR+L WP+ P F E+L E++ K + +++ + MARSL
Sbjct: 127 SDNQVLDWIDRLFLTTYPEDKRQLKFWPQVPVGFSETLDEYTMKQRVLIEKFFKAMARSL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE FL +GE + + +RFNF+PPC RPD V+G+KPH D S IT+LL D++VEGLQ L
Sbjct: 187 ELEENCFLEMYGENAVMNSRFNFFPPCPRPDKVIGIKPHADGSAITLLLPDKDVEGLQFL 246
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D KW P +PD +++ LGDQM+IMSNGI+KSP+HRV+TN E+ R+SVA F P + E
Sbjct: 247 KDGKWYKAPIVPDTILITLGDQMEIMSNGIYKSPVHRVVTNREKERISVATFCVPGLDKE 306
Query: 294 IGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
I P +GL+ E RPRLY+ V Y D++Y+ YQ+G+ +E I
Sbjct: 307 IHPADGLVTEARPRLYKTVTKYVDLHYKYYQQGRRTIEAALI 348
>At5g20400 ethylene-forming-enzyme-like dioxygenase-like protein
Length = 348
Score = 311 bits (798), Expect = 3e-85
Identities = 158/342 (46%), Positives = 223/342 (65%), Gaps = 9/342 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFGS-KDSSTLIP---IPIIDVSLLSSEDEQG-----K 53
K+VQE+ + P Y+ G + + +P IP ID++LL S E G K
Sbjct: 7 KTVQEVVAAGEGLPERYLHAPTGDGEVQPLNAAVPEMDIPAIDLNLLLSSSEAGQQELSK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q + HG++ +LDKI ++ K FFALP EEKQK AR ++ +GYGND ++
Sbjct: 67 LHSALSTWGVVQVMNHGITKAFLDKIYKLTKEFFALPTEEKQKCAREIDSIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
QVLDW RL + +P+++R+L+ WPE P F E+L E++ K + +++ + MARSL
Sbjct: 127 WDDQVLDWIDRLYITTYPEDQRQLNFWPEVPLGFRETLHEYTMKQRIVIEQFFKAMARSL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE SFL +GE ++L RFN YPPC PD V+GVKPH D S IT+LL D++V GLQ
Sbjct: 187 ELEENSFLDMYGESATLDTRFNMYPPCPSPDKVIGVKPHADGSAITLLLPDKDVGGLQFQ 246
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D KW P +PD +++N+GDQM+IMSNGI+KSP+HRV+TN E+ R+SVA F P + E
Sbjct: 247 KDGKWYKAPIVPDTILINVGDQMEIMSNGIYKSPVHRVVTNREKERISVATFCIPGADKE 306
Query: 294 IGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
I PV L++E RPRLY+ V Y ++ ++ YQ+G+ +E I
Sbjct: 307 IQPVNELVSEARPRLYKTVKKYVELYFKYYQQGRRPIEAALI 348
>At5g20550 ethylene-forming-enzyme-like dioxygenase - like
Length = 349
Score = 301 bits (772), Expect = 3e-82
Identities = 156/335 (46%), Positives = 217/335 (64%), Gaps = 10/335 (2%)
Query: 3 KSVQEMSMDSDEPPSAYV----VERNSFGSKDSSTLIPIPIIDVSLLSSEDEQG-----K 53
K+VQE+ + P Y+ V+ N + ++ IP ID+SLL S + G K
Sbjct: 7 KTVQEVVAAGEGIPERYLQPPAVDDNGQHLNAAVPVMDIPAIDLSLLLSPSDDGREELSK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q I HG++ LDKI ++ K F ALP EEKQKYAR + +GYGND ++
Sbjct: 67 LHSALSTWGVVQVINHGITKALLDKIYKLTKEFCALPSEEKQKYAREIGSIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
QVLDW RL + +P+++R+L WP+ P F E+L E++ K + + + + MA SL
Sbjct: 127 WDDQVLDWIDRLYITTYPEDQRQLKFWPDVPVGFRETLHEYTMKQHLVFNQVFKAMAISL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE FL GE +++ RFN YPPC RPD V+GV+PH D+S T+LL D+ VEGLQ L
Sbjct: 187 ELEENCFLDMCGENATMDTRFNMYPPCPRPDKVIGVRPHADKSAFTLLLPDKNVEGLQFL 246
Query: 234 VDDKWVNVPTI-PDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPEN 292
D KW P + D +++N+GDQM+IMSNGI+KSP+HRV+TNTE+ R+SVA F P +
Sbjct: 247 KDGKWYKAPVVASDTILINVGDQMEIMSNGIYKSPVHRVVTNTEKERISVATFCIPGADK 306
Query: 293 EIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGK 327
EI PV+GL++E RPRLY+ V NY D+ + Y +G+
Sbjct: 307 EIQPVDGLVSEARPRLYKPVKNYVDLLNKYYIQGQ 341
>At5g54000 ethylene-forming-enzyme-like dioxygenase
Length = 349
Score = 298 bits (763), Expect = 3e-81
Identities = 151/343 (44%), Positives = 218/343 (63%), Gaps = 10/343 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFGSKDS--STLIP---IPIIDVSLLSSEDEQG----- 52
K+VQE+ ++ P Y+ G D + L+P I IID++LL S + G
Sbjct: 7 KTVQEVVAAGEKLPERYLYTPTGDGEGDQPFNGLLPEMKISIIDLNLLFSSSDDGREELS 66
Query: 53 KLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRV 112
KL SA+S+ G Q + HG+S LDKI E+ K FF LP +EKQKYAR ++ +G+GND +
Sbjct: 67 KLHSAISTWGVVQVMNHGISEALLDKIHELTKQFFVLPTKEKQKYAREISSFQGFGNDMI 126
Query: 113 VSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARS 172
+S QVLDW RL L +P+++R+L WPENPS F E+L E++ K + +++ + +ARS
Sbjct: 127 LSDDQVLDWVDRLYLITYPEDQRQLKFWPENPSGFRETLHEYTMKQQLVVEKFFKALARS 186
Query: 173 LNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQV 232
L LE+ FL GE ++L RFN YPPC RPD VLG+KPH+D S T++L D+ VEGLQ
Sbjct: 187 LELEDNCFLEMHGENATLETRFNIYPPCPRPDKVLGLKPHSDGSAFTLILPDKNVEGLQF 246
Query: 233 LVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPEN 292
L D KW +P +++N+GD M++MSNGI+KSP+HRV+ N ++ R+ VA F + +
Sbjct: 247 LKDGKWYKASILPHTILINVGDTMEVMSNGIYKSPVHRVVLNGKKERIYVATFCNADEDK 306
Query: 293 EIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
EI P+ GL++E RPRLY+ V + YQ+G+ +E I
Sbjct: 307 EIQPLNGLVSEARPRLYKAVKKSEKNFFDYYQQGRRPIEAAMI 349
>At3g21420 unknown protein
Length = 364
Score = 229 bits (584), Expect = 2e-60
Identities = 129/310 (41%), Positives = 182/310 (58%), Gaps = 13/310 (4%)
Query: 37 IPIIDVSLLSSEDEQG------KLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALP 90
IP+ID+S LS D KL A G FQ I HG+ ++ I EVA FF +P
Sbjct: 55 IPVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMP 114
Query: 91 VEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGES 150
+EEK+KY +GYG + S+ Q LDW +L V P + R LWP P+ F ES
Sbjct: 115 LEEKKKYPMEPGTVQGYGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFSES 174
Query: 151 LVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVK 210
L +S +++ + LL+ +A SL L+E F FGE V R N+YPPCS PDLVLG+
Sbjct: 175 LEGYSKEIRELCKRLLKYIAISLGLKEERFEEMFGEAVQAV-RMNYYPPCSSPDLVLGLS 233
Query: 211 PHTDRSGITVLLQDR-EVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMH 269
PH+D S +TVL Q + GLQ+L D+ WV V +P+ALV+N+GD ++++SNG +KS H
Sbjct: 234 PHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKSVEH 293
Query: 270 RVLTNTERLRMSVAMFNEPEPENEIGPVEGLI-NETRPRLYRNVNNYGDINYRCYQ---E 325
R +TN E+ R+++ F P E EI P+ L+ +ET P YR+ N+GD +Y +
Sbjct: 294 RAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSY-NHGDYSYHYVSNKLQ 352
Query: 326 GKIALETVQI 335
GK +L+ +I
Sbjct: 353 GKKSLDFAKI 362
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 219 bits (558), Expect = 2e-57
Identities = 119/316 (37%), Positives = 181/316 (56%), Gaps = 7/316 (2%)
Query: 4 SVQEMSMDS-DEPPSAYVV---ERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKL-RSAL 58
SVQ +S P+ YV +R F + S I IP++D++ + + E +L RSA
Sbjct: 11 SVQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIEIPVLDMNDVWGKPEGLRLVRSAC 70
Query: 59 SSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQV 118
G FQ + HG++ + ++++R + FF LP+EEK+KYA + + EGYG+ V K
Sbjct: 71 EEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPDTYEGYGSRLGVVKDAK 130
Query: 119 LDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEG 178
LDWS L P R S WP P E + ++ +V+ + + L T++ SL L+
Sbjct: 131 LDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERLTETLSESLGLKPN 190
Query: 179 SFLSQFGEQSSLVA--RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
+ G + A R NFYP C +P L LG+ H+D GIT+LL D +V GLQV D
Sbjct: 191 KLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLPDEKVAGLQVRRGD 250
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
WV + ++P+AL+VN+GDQ+QI+SNGI+KS H+V+ N+ R+S+A F P + +GP
Sbjct: 251 GWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGP 310
Query: 297 VEGLINETRPRLYRNV 312
+E L+ RP LY+ +
Sbjct: 311 IEELVTANRPALYKPI 326
>At1g17020 SRG1-like protein
Length = 358
Score = 212 bits (539), Expect = 3e-55
Identities = 116/313 (37%), Positives = 187/313 (59%), Gaps = 7/313 (2%)
Query: 30 DSSTLIPIPIIDVSLLSS----EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKH 85
D I IPIID+ L S + E KL A G FQ + HG+ S++LDK++ +
Sbjct: 46 DFDVKIEIPIIDMKRLCSSTTMDSEVEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQD 105
Query: 86 FFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPS 145
FF LP+EEK+K+ + +E EG+G VVS+ Q LDW+ V P E R+ L+P+ P
Sbjct: 106 FFNLPMEEKKKFWQRPDEIEGFGQAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPL 165
Query: 146 DFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVA-RFNFYPPCSRPD 204
F ++L +S++V+S+ L+ MAR+L ++ F + S+ + R N+YPPC +PD
Sbjct: 166 PFRDTLEMYSSEVQSVAKILIAKMARALEIKPEELEKLFDDVDSVQSMRMNYYPPCPQPD 225
Query: 205 LVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIF 264
V+G+ PH+D G+TVL+Q +VEGLQ+ D KWV V +P+A +VN+GD ++I++NG +
Sbjct: 226 QVIGLTPHSDSVGLTVLMQVNDVEGLQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTY 285
Query: 265 KSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYR--NVNNYGDINYRC 322
+S HR + N+E+ R+S+A F+ E+GP + L+ + ++ + Y D +
Sbjct: 286 RSIEHRGVVNSEKERLSIATFHNVGMYKEVGPAKSLVERQKVARFKRLTMKEYNDGLFSR 345
Query: 323 YQEGKIALETVQI 335
+GK L+ ++I
Sbjct: 346 TLDGKAYLDALRI 358
>At4g25300 SRG1-like protein
Length = 356
Score = 210 bits (535), Expect = 8e-55
Identities = 126/342 (36%), Positives = 196/342 (56%), Gaps = 10/342 (2%)
Query: 4 SVQEMSMDS--DEPPSAYVVERNSFG--SKDSSTLIPIPIIDVSLLSS----EDEQGKLR 55
SVQEM + P YV + DS IPIID+SLL S + E KL
Sbjct: 15 SVQEMVKEKMITTVPPRYVRSDQDVAEIAVDSGLRNQIPIIDMSLLCSSTSMDSEIDKLD 74
Query: 56 SALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSK 115
SA G FQ + HGM S++L+K++ + FF LP+EEK+ + +E EG+G VVS+
Sbjct: 75 SACKEWGFFQLVNHGMESSFLNKVKSEVQDFFNLPMEEKKNLWQQPDEIEGFGQVFVVSE 134
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNL 175
+Q LDW+ L + P R+ L+P+ P F ++L +S +VKS+ LL +A +L +
Sbjct: 135 EQKLDWADMFFLTMQPVRLRKPHLFPKLPLPFRDTLDMYSAEVKSIAKILLGKIAVALKI 194
Query: 176 EEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVD 235
+ F ++ R N+YP C PD V+G+ PH+D +G+T+LLQ EVEGLQ+ +
Sbjct: 195 KPEEMDKLFDDELGQRIRLNYYPRCPEPDKVIGLTPHSDSTGLTILLQANEVEGLQIKKN 254
Query: 236 DKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIG 295
KWV+V +P+ALVVN+GD ++I++NG ++S HR + N+E+ R+SVA F+ EIG
Sbjct: 255 AKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRGVVNSEKERLSVAAFHNIGLGKEIG 314
Query: 296 PVEGLINETRPRLYRNVNNYGDIN--YRCYQEGKIALETVQI 335
P+ L+ + +++V N + +GK L+ +++
Sbjct: 315 PMRSLVERHKAAFFKSVTTEEYFNGLFSRELDGKAYLDVMRL 356
>At1g17010 SRG1-like protein
Length = 361
Score = 209 bits (533), Expect = 1e-54
Identities = 124/343 (36%), Positives = 193/343 (56%), Gaps = 11/343 (3%)
Query: 4 SVQEMSMD---SDEPPS--AYVVERNSFGSKDSSTLIPIPIIDVSLLSS----EDEQGKL 54
SVQEM D + PP Y ++ DS + IPIID++ L S + E KL
Sbjct: 16 SVQEMVKDKMITTVPPRYVRYDQDKTEVVVHDSGLISEIPIIDMNRLCSSTAVDSEVEKL 75
Query: 55 RSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVS 114
A G FQ + HG+ ++LDKI+ + FF LP+EEK+K + EG+G VVS
Sbjct: 76 DFACKEYGFFQLVNHGIDPSFLDKIKSEIQDFFNLPMEEKKKLWQTPAVMEGFGQAFVVS 135
Query: 115 KKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLN 174
+ Q LDW+ L + P + R+ L+P+ P F ++L +ST+VKS+ LL MA++L
Sbjct: 136 EDQKLDWADLFFLIMQPVQLRKRHLFPKLPLPFRDTLDMYSTRVKSIAKILLAKMAKALQ 195
Query: 175 LEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLV 234
++ FG+ R N+YPPC +P+LV G+ PH+D G+T+LLQ EV+GLQ+
Sbjct: 196 IKPEEVEEIFGDDMMQSMRMNYYPPCPQPNLVTGLIPHSDAVGLTILLQVNEVDGLQIKK 255
Query: 235 DDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEI 294
+ KW V + +A +VN+GD ++I++NG ++S HR + N E+ R+S+A F+ + EI
Sbjct: 256 NGKWFFVKPLQNAFIVNVGDVLEIITNGTYRSIEHRAMVNLEKERLSIATFHNTGMDKEI 315
Query: 295 GPVEGLINETRPRLYRNVNNYGDINYRCYQE--GKIALETVQI 335
GP L+ +R++ +N +E GK L+ ++I
Sbjct: 316 GPARSLVQRQEAAKFRSLKTKDYLNGLFSRELKGKAYLDAMRI 358
>At1g78550 unknown protein
Length = 356
Score = 204 bits (520), Expect = 4e-53
Identities = 119/337 (35%), Positives = 191/337 (56%), Gaps = 7/337 (2%)
Query: 5 VQEMSMDSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSS----EDEQGKLRSALSS 60
V+E + + P V + + DSS IP+ID++ L S + E KL A
Sbjct: 21 VKEKNFTTIPPRYVRVDQEKTEILNDSSLSSEIPVIDMTRLCSVSAMDSELKKLDFACQD 80
Query: 61 AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLD 120
G FQ + HG+ S++L+K+ + FF LP++EKQK + E EG+G +VS+ Q LD
Sbjct: 81 WGFFQLVNHGIDSSFLEKLETEVQEFFNLPMKEKQKLWQRSGEFEGFGQVNIVSENQKLD 140
Query: 121 WSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSF 180
W L P R+ L+ + P F E+L +S++VKS+ L MA L ++
Sbjct: 141 WGDMFILTTEPIRSRKSHLFSKLPPPFRETLETYSSEVKSIAKILFAKMASVLEIKHEEM 200
Query: 181 LSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVN 240
F + + + N+YPPC +PD V+G+ H+D +G+T+LLQ +VEGLQ+ D KWV
Sbjct: 201 EDLFDDVWQSI-KINYYPPCPQPDQVMGLTQHSDAAGLTILLQVNQVEGLQIKKDGKWVV 259
Query: 241 VPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGL 300
V + DALVVN+G+ ++I++NG ++S HR + N+E+ R+SVAMF+ P E I P + L
Sbjct: 260 VKPLRDALVVNVGEILEIITNGRYRSIEHRAVVNSEKERLSVAMFHSPGKETIIRPAKSL 319
Query: 301 INETRPRLYRNVN--NYGDINYRCYQEGKIALETVQI 335
++ + L+++++ Y D + GK L+ ++I
Sbjct: 320 VDRQKQCLFKSMSTQEYFDAFFTQKLNGKSHLDLMRI 356
>At4g25310 SRG1-like protein
Length = 353
Score = 201 bits (510), Expect = 6e-52
Identities = 110/280 (39%), Positives = 168/280 (59%), Gaps = 7/280 (2%)
Query: 37 IPIIDVSLLSS----EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVE 92
IPIID+SLLSS + E KL A G FQ + HGM LDK + + FF LP+E
Sbjct: 52 IPIIDMSLLSSSTSMDSEIDKLDFACKEWGFFQLVNHGMD---LDKFKSDIQDFFNLPME 108
Query: 93 EKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLV 152
EK+K + + EG+G V S++Q LDW+ L + P R+ L+P+ P F ++L
Sbjct: 109 EKKKLWQQPGDIEGFGQAFVFSEEQKLDWADVFFLTMQPVPLRKPHLFPKLPLPFRDTLD 168
Query: 153 EFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPH 212
+S ++KS+ L +A +L ++ F ++ R N+YPPC PD +G+ PH
Sbjct: 169 TYSAELKSIAKVLFAKLASALKIKPEEMEKLFDDELGQRIRMNYYPPCPEPDKAIGLTPH 228
Query: 213 TDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVL 272
+D +G+T+LLQ EVEGLQ+ D KWV+V +P+ALVVN+GD ++I++NG ++S HR +
Sbjct: 229 SDATGLTILLQVNEVEGLQIKKDGKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRGV 288
Query: 273 TNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNV 312
N+E+ R+SVA F+ EIGP+ L+ + L++ +
Sbjct: 289 VNSEKERLSVASFHNTGFGKEIGPMRSLVERHKGALFKTL 328
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 199 bits (507), Expect = 1e-51
Identities = 105/289 (36%), Positives = 160/289 (55%), Gaps = 7/289 (2%)
Query: 28 SKDSSTLIPIPIIDVSLLSSEDE------QGKLRSALSSAGCFQAIGHGMSSTYLDKIRE 81
++D+ T IPIID+ L SE+ ++ A G FQ + HG+ +D RE
Sbjct: 53 TEDAPTATNIPIIDLEGLFSEEGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARE 112
Query: 82 VAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWP 141
+ FF +PV K+ Y+ + EGYG+ V K LDWS L + P + + WP
Sbjct: 113 NWREFFHMPVNAKETYSNSPRTYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWP 172
Query: 142 ENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG-EQSSLVARFNFYPPC 200
P E + E+ ++ + ++R ++ +L L+E F FG E R N+YP C
Sbjct: 173 SFPPTIREVIDEYGEELVKLSGRIMRVLSTNLGLKEDKFQEAFGGENIGACLRVNYYPKC 232
Query: 201 SRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMS 260
RP+L LG+ PH+D G+T+LL D +V GLQV DD W+ V P A +VN+GDQ+QI+S
Sbjct: 233 PRPELALGLSPHSDPGGMTILLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILS 292
Query: 261 NGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLY 309
N +KS HRV+ N+++ R+S+A F P+ + I P++ L++ P LY
Sbjct: 293 NSTYKSVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELVSTHNPPLY 341
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 195 bits (496), Expect = 3e-50
Identities = 106/278 (38%), Positives = 155/278 (55%), Gaps = 3/278 (1%)
Query: 35 IPIPIIDVSLLSS--EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVE 92
I IPIID+ L S ED++ ++ A G FQ I HG+ +D RE K FF LPVE
Sbjct: 93 INIPIIDLDSLFSGNEDDKKRISEACREWGFFQVINHGVKPELMDAARETWKSFFNLPVE 152
Query: 93 EKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLV 152
K+ Y+ + EGYG+ V K +LDW+ L P + + WP PS+ E
Sbjct: 153 AKEVYSNSPRTYEGYGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIREMND 212
Query: 153 EFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG-EQSSLVARFNFYPPCSRPDLVLGVKP 211
E+ ++ + L+ ++ +L L FG E R N+YP C +P+L LG+ P
Sbjct: 213 EYGKELVKLGGRLMTILSSNLGLRAEQLQEAFGGEDVGACLRVNYYPKCPQPELALGLSP 272
Query: 212 HTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRV 271
H+D G+T+LL D +V GLQV D W+ V + A +VN+GDQ+QI+SN +KS HRV
Sbjct: 273 HSDPGGMTILLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRV 332
Query: 272 LTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLY 309
+ N+E+ R+S+A F P+ + I P++ L+ T P LY
Sbjct: 333 IVNSEKERVSLAFFYNPKSDIPIQPMQQLVTSTMPPLY 370
>At3g55970 leucoanthocyanidin dioxygenase -like protein
Length = 363
Score = 192 bits (487), Expect = 3e-49
Identities = 106/291 (36%), Positives = 162/291 (55%), Gaps = 9/291 (3%)
Query: 28 SKDSSTLIPIPIIDVSLLSSEDE--QGKLRSALSSA----GCFQAIGHGMSSTYLDKIRE 81
+K + IPIID+ L ++D Q K +S A G FQ + HGMS +D+ +
Sbjct: 43 NKHNPQTTTIPIIDLGRLYTDDLTLQAKTLDEISKACRELGFFQVVNHGMSPQLMDQAKA 102
Query: 82 VAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWP 141
+ FF LP+E K +A + EGYG+ V K +LDWS L P + + WP
Sbjct: 103 TWREFFNLPMELKNMHANSPKTYEGYGSRLGVEKGAILDWSDYYYLHYQPSSLKDYTKWP 162
Query: 142 ENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG--EQSSLVARFNFYPP 199
P E L ++ ++ + ++L++ ++++L L+E + FG E+S R N+YP
Sbjct: 163 SLPLHCREILEDYCKEMVKLCENLMKILSKNLGLQEDRLQNAFGGKEESGGCLRVNYYPK 222
Query: 200 CSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL-VDDKWVNVPTIPDALVVNLGDQMQI 258
C +P+L LG+ PH+D G+T+LL D +V LQV DD W+ V P A +VN+GDQ+Q+
Sbjct: 223 CPQPELTLGISPHSDPGGLTILLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQM 282
Query: 259 MSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLY 309
+SN I+KS HRV+ N E R+S+A F P+ I P++ L+ P LY
Sbjct: 283 LSNSIYKSVEHRVIVNPENERLSLAFFYNPKGNVPIEPLKELVTVDSPALY 333
>At4g10500 putative Fe(II)/ascorbate oxidase
Length = 349
Score = 180 bits (456), Expect = 1e-45
Identities = 106/271 (39%), Positives = 153/271 (56%), Gaps = 7/271 (2%)
Query: 37 IPIIDVSLLSSEDEQ---GKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEE 93
IP+ID+ L + +L SA S+ G FQ HG+ T ++K++ VA+ FF P E
Sbjct: 44 IPLIDLRDLHGPNRAVIVQQLASACSTYGFFQIKNHGVPDTTVNKMQTVAREFFHQPESE 103
Query: 94 KQK-YARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLV 152
+ K Y+ + V +VL+W L L FP E + WP +P F E
Sbjct: 104 RVKHYSADPTKTTRLSTSFNVGADKVLNWRDFLRLHCFPIEDF-IEEWPSSPISFREVTA 162
Query: 153 EFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPH 212
E++T V++++ LL ++ SL LE + G+ + +A FN+YPPC P+L G+ H
Sbjct: 163 EYATSVRALVLRLLEAISESLGLESDHISNILGKHAQHMA-FNYYPPCPEPELTYGLPGH 221
Query: 213 TDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVL 272
D + ITVLLQD +V GLQV DDKWV V IP+ +VN+GDQMQ++SN +KS +HR +
Sbjct: 222 KDPTVITVLLQD-QVSGLQVFKDDKWVAVSPIPNTFIVNIGDQMQVISNDKYKSVLHRAV 280
Query: 273 TNTERLRMSVAMFNEPEPENEIGPVEGLINE 303
NTE R+S+ F P + IGP L+NE
Sbjct: 281 VNTENERLSIPTFYFPSTDAVIGPAHELVNE 311
>At5g08640 flavonol synthase (FLS) (sp|Q96330)
Length = 336
Score = 179 bits (454), Expect = 2e-45
Identities = 101/292 (34%), Positives = 160/292 (54%), Gaps = 8/292 (2%)
Query: 36 PIPIIDVSLLSSEDEQGKLRSALSSA---GCFQAIGHGMSSTYLDKIREVAKHFFALPVE 92
P P I V LS DE+ R+ + ++ G FQ + HG+ + + ++++V + FF LP
Sbjct: 39 PTPAIPVVDLSDPDEESVRRAVVKASEEWGLFQVVNHGIPTELIRRLQDVGRKFFELPSS 98
Query: 93 EKQKYARAVN--EAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGES 150
EK+ A+ + + EGYG + W L R++P WP+NP ++ E
Sbjct: 99 EKESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRIWPPSCVNYRFWPKNPPEYREV 158
Query: 151 LVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG-EQSSLVARFNFYPPCSRPDLVLGV 209
E++ VK + + LL ++ L L+ + G E + + + N+YPPC RPDL LGV
Sbjct: 159 NEEYAVHVKKLSETLLGILSDGLGLKRDALKEGLGGEMAEYMMKINYYPPCPRPDLALGV 218
Query: 210 KPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMH 269
HTD SGIT+L+ + EV GLQV DD W + IP A++V++GDQ+ +SNG +K+ +H
Sbjct: 219 PAHTDLSGITLLVPN-EVPGLQVFKDDHWFDAEYIPSAVIVHIGDQILRLSNGRYKNVLH 277
Query: 270 RVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGDINYR 321
R + E+ RMS +F EP E +GP+ L + P ++ + D +YR
Sbjct: 278 RTTVDKEKTRMSWPVFLEPPREKIVGPLPELTGDDNPPKFKPF-AFKDYSYR 328
>At4g22880 putative leucoanthocyanidin dioxygenase (LDOX)
Length = 356
Score = 177 bits (448), Expect = 1e-44
Identities = 93/268 (34%), Positives = 162/268 (59%), Gaps = 12/268 (4%)
Query: 37 IPIIDVSLLSSEDEQ------GKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALP 90
+P ID+ + S+DE+ +L+ A G I HG+ + ++++++ + FF+L
Sbjct: 47 VPTIDLKNIESDDEKIRENCIEELKKASLDWGVMHLINHGIPADLMERVKKAGEEFFSLS 106
Query: 91 VEEKQKYA--RAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFG 148
VEEK+KYA +A + +GYG+ + L+W +P+EKR LS+WP+ PSD+
Sbjct: 107 VEEKEKYANDQATGKIQGYGSKLANNASGQLEWEDYFFHLAYPEEKRDLSIWPKTPSDYI 166
Query: 149 ESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG--EQSSLVARFNFYPPCSRPDLV 206
E+ E++ ++ + + + ++ L LE + G E+ L + N+YP C +P+L
Sbjct: 167 EATSEYAKCLRLLATKVFKALSVGLGLEPDRLEKEVGGLEELLLQMKINYYPKCPQPELA 226
Query: 207 LGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKS 266
LGV+ HTD S +T +L + V GLQ+ + KWV +PD++V+++GD ++I+SNG +KS
Sbjct: 227 LGVEAHTDVSALTFILHNM-VPGLQLFYEGKWVTAKCVPDSIVMHIGDTLEILSNGKYKS 285
Query: 267 PMHRVLTNTERLRMSVAMFNEPEPENEI 294
+HR L N E++R+S A+F EP P+++I
Sbjct: 286 ILHRGLVNKEKVRISWAVFCEP-PKDKI 312
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 176 bits (445), Expect = 2e-44
Identities = 102/280 (36%), Positives = 156/280 (55%), Gaps = 7/280 (2%)
Query: 32 STLIPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPV 91
S L P+ID+S ++ A + G FQ I HG++ +D++ VA+ FF++ +
Sbjct: 33 SQLEDFPLIDLSSTDRSFLIQQIHQACARFGFFQVINHGVNKQIIDEMVSVAREFFSMSM 92
Query: 92 EEKQK-YARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGES 150
EEK K Y+ + V K++V +W L L +P K ++ WP NP F E
Sbjct: 93 EEKMKLYSDDPTKTTRLSTSFNVKKEEVNNWRDYLRLHCYPIHKY-VNEWPSNPPSFKEI 151
Query: 151 LVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVK 210
+ ++S +V+ + + ++ SL LE+ GEQ +A N+YPPC P+L G+
Sbjct: 152 VSKYSREVREVGFKIEELISESLGLEKDYMKKVLGEQGQHMA-VNYYPPCPEPELTYGLP 210
Query: 211 PHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHR 270
HTD + +T+LLQD V GLQ+L+D +W V PDA V+N+GDQ+Q +SNG++KS HR
Sbjct: 211 AHTDPNALTILLQDTTVCGLQILIDGQWFAVNPHPDAFVINIGDQLQALSNGVYKSVWHR 270
Query: 271 VLTNTERLRMSVAMFNEPEPENEIGPVEGL----INETRP 306
+TNTE R+SVA F P + P + L +ET+P
Sbjct: 271 AVTNTENPRLSVASFLCPADCAVMSPAKPLWEAEDDETKP 310
>At1g55290 leucoanthocyanidin dioxygenase 2, putative
Length = 361
Score = 174 bits (440), Expect = 8e-44
Identities = 107/305 (35%), Positives = 165/305 (54%), Gaps = 11/305 (3%)
Query: 37 IPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQK 96
IP+ID+S L + + A G FQ I HG+S L+ ++ FF LPVEEK+K
Sbjct: 62 IPVIDISNLDEKSVSKAVCDAAEEWGFFQVINHGVSMEVLENMKTATHRFFGLPVEEKRK 121
Query: 97 YAR--AVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEF 154
++R +++ +G ++ L+W LSL F E LWP++ +E+
Sbjct: 122 FSREKSLSTNVRFGTSFSPHAEKALEWKDYLSL-FFVSEAEASQLWPDSCRS---ETLEY 177
Query: 155 STKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTD 214
+ K ++ LLR + +LN++E + S N+YP C P+L +GV H+D
Sbjct: 178 MNETKPLVKKLLRFLGENLNVKELDKTKESFFMGSTRINLNYYPICPNPELTVGVGRHSD 237
Query: 215 RSGITVLLQDREVEGLQV--LVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVL 272
S +T+LLQD E+ GL V L +WV+VP I +LV+N+GD MQIMSNG +KS HRVL
Sbjct: 238 VSSLTILLQD-EIGGLHVRSLTTGRWVHVPPISGSLVINIGDAMQIMSNGRYKSVEHRVL 296
Query: 273 TNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNV--NNYGDINYRCYQEGKIAL 330
N R+SV +F P+PE+ IGP+ +I +Y+++ +Y +R +GK +
Sbjct: 297 ANGSYNRISVPIFVSPKPESVIGPLLEVIENGEKPVYKDILYTDYVKHFFRKAHDGKKTI 356
Query: 331 ETVQI 335
+ I
Sbjct: 357 DFANI 361
>At5g63580 flavonol synthase
Length = 310
Score = 171 bits (433), Expect = 5e-43
Identities = 101/304 (33%), Positives = 158/304 (51%), Gaps = 7/304 (2%)
Query: 21 VERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIR 80
VER+ S S IPIID+S L E + G F + HG+ + +++
Sbjct: 3 VERDQHISPPSLMAKTIPIIDLSNLDEELVAHAVVKGSEEWGIFHVVNHGIPMDLIQRLK 62
Query: 81 EVAKHFFALPVEEKQKYAR--AVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLS 138
+V FF LP EK+ A+ + EGY + K +V W+ L R++P
Sbjct: 63 DVGTQFFELPETEKKAVAKQDGSKDFEGYTTNLKYVKGEV--WTENLFHRIWPPTCINFD 120
Query: 139 LWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFG-EQSSLVARFNFY 197
WP+NP + E + E++ + K + + +L ++ L L + + G E + V R N Y
Sbjct: 121 YWPKNPPQYREVIEEYTKETKKLSERILGYLSEGLGLPSEALIQGLGGESTEYVMRINNY 180
Query: 198 PPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQ 257
PP +PDL LGV HTD GIT+++ + EV GLQ+ DD W++V IP ++ VN+GDQ+
Sbjct: 181 PPDPKPDLTLGVPEHTDIIGITIIITN-EVPGLQIFKDDHWLDVHYIPSSITVNIGDQIM 239
Query: 258 IMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYRNVNNYGD 317
+SNG +K+ +HR + + E RMS +F + P+ IGP+ LI P ++ + D
Sbjct: 240 RLSNGKYKNVLHRAIVDKENQRMSWPVFVDANPDVVIGPLPELITGDNPSKFKPI-TCKD 298
Query: 318 INYR 321
YR
Sbjct: 299 FKYR 302
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,574,624
Number of Sequences: 26719
Number of extensions: 315435
Number of successful extensions: 1094
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 804
Number of HSP's gapped (non-prelim): 123
length of query: 342
length of database: 11,318,596
effective HSP length: 100
effective length of query: 242
effective length of database: 8,646,696
effective search space: 2092500432
effective search space used: 2092500432
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0219.11


