

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.25
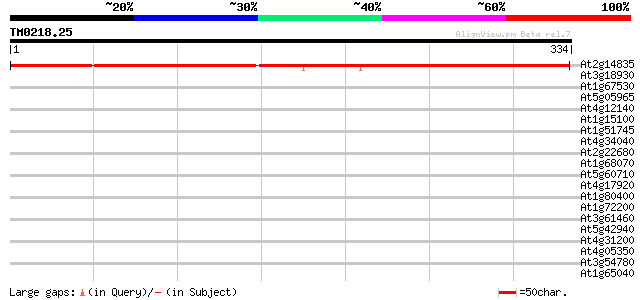
(334 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g14835 unknown protein 489 e-139
At3g18930 unknown protein 35 0.057
At1g67530 unknown protein 35 0.075
At5g05965 putative protein 34 0.13
At4g12140 putative protein 34 0.13
At1g15100 RING-H2 zinc finger like protein 34 0.13
At1g51745 unknown protein 33 0.17
At4g34040 putative protein 32 0.37
At2g22680 copia-like retroelement pol polyprotein 32 0.37
At1g68070 RING zinc finger like protein 32 0.37
At5g60710 unknown protein 32 0.49
At4g17920 putative protein 32 0.49
At1g80400 putative RING zinc finger protein 32 0.49
At1g72200 RING-H2 zinc finger protein ATL3, putative 32 0.49
At3g61460 RING finger protein 32 0.63
At5g42940 unknown protein 31 1.1
At4g31200 predicted protein 31 1.1
At4g05350 putative protein 31 1.1
At3g54780 unknown protein 31 1.1
At1g65040 31 1.1
>At2g14835 unknown protein
Length = 343
Score = 489 bits (1259), Expect = e-139
Identities = 240/344 (69%), Positives = 273/344 (78%), Gaps = 13/344 (3%)
Query: 1 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQAV 60
MVVCKC+KAT+LYCFVHK PVCGECICFPEHQ CV+RTYSEWVIDGEYD PKCCQCQA
Sbjct: 1 MVVCKCKKATRLYCFVHKAPVCGECICFPEHQTCVVRTYSEWVIDGEYD-QPKCCQCQAT 59
Query: 61 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSR 120
+EG G Q TRLGCLH IHT+CLVS IKSFPPHTAPAGY CP CST IWPP VKD+GSR
Sbjct: 60 FDEGAGHQVTRLGCLHAIHTSCLVSLIKSFPPHTAPAGYVCPACSTPIWPPMMVKDAGSR 119
Query: 121 LHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD------- 173
LH+ L+E I QTG+EKN+ GNHPVS S TESRSPPPAFAS+ LI + ++
Sbjct: 120 LHALLREVITQTGLEKNLLGNHPVSRS-TESRSPPPAFASDALINISSSSHTQEGNNLPD 178
Query: 174 --SPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSP--VGPATRKGAFNVERQNSEISYY 229
S A E K +V++IVEI+ SAG+++K SSP A RKG V+RQNSE YY
Sbjct: 179 GYSVAGNGEYSKSAVSEIVEIDVPASAGSYMKSSSPGLAAAAARKGVPAVDRQNSETLYY 238
Query: 230 ADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQ 289
ADDEDGNRKKY++RGP HKFLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQ
Sbjct: 239 ADDEDGNRKKYSRRGPLRHKFLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQ 298
Query: 290 RSSRMDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPNEEQ 333
RSS+MD RKIL+ IA++ACMATMGILYYRL + G+ELP+EEQ
Sbjct: 299 RSSKMDIRKILIFIALIACMATMGILYYRLALQAIGQELPDEEQ 342
>At3g18930 unknown protein
Length = 411
Score = 35.0 bits (79), Expect = 0.057
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 9/55 (16%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
C C EEGD +T L C H H C+ ++S P CP C T+I
Sbjct: 156 CAVCLLEFEEGDYVRTLPL-CFHAFHLECIDEWLRSHP--------NCPLCRTAI 201
>At1g67530 unknown protein
Length = 782
Score = 34.7 bits (78), Expect = 0.075
Identities = 17/51 (33%), Positives = 28/51 (54%)
Query: 184 LSVTDIVEIEGANSAGNFVKGSSPVGPATRKGAFNVERQNSEISYYADDED 234
LS ++ + + NS G++ + P G VERQN+E S+ +DD+D
Sbjct: 363 LSDSESTKSQSVNSIGSYKLKGVKIVPLEENGTTVVERQNTEESFVSDDDD 413
>At5g05965 putative protein
Length = 131
Score = 33.9 bits (76), Expect = 0.13
Identities = 36/132 (27%), Positives = 52/132 (39%), Gaps = 26/132 (19%)
Query: 85 SHIKSFPPHTAPAGYA----CPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFG 140
SH SF P G A C T SI+PP + GS +++ I Q E++
Sbjct: 12 SHSSSFDNLFGPKGSASASSCSTILDSIFPPPVARKKGSHTAPEVQGNISQATDERS--- 68
Query: 141 NHPVSLSVTESRSPPPAFASEPLIGRENH-------GNSDSPATGSEPPKLSVTDIVEIE 193
+ + ++S G + H G+S SP S PK D +I
Sbjct: 69 --------SHEKRESSYYSSSIYYGGQQHYSPPRTDGSSTSP---SHQPK-ETNDRTDIT 116
Query: 194 GANSAGNFVKGS 205
+ S GN+ KGS
Sbjct: 117 TSTSRGNWWKGS 128
>At4g12140 putative protein
Length = 202
Score = 33.9 bits (76), Expect = 0.13
Identities = 17/56 (30%), Positives = 24/56 (42%), Gaps = 8/56 (14%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIW 109
C C L + TR+ C HV H++CLV +K CP C T ++
Sbjct: 153 CSICLQSLVSSSKTGPTRMSCSHVFHSSCLVEWLK--------RKNTCPMCRTVLY 200
>At1g15100 RING-H2 zinc finger like protein
Length = 155
Score = 33.9 bits (76), Expect = 0.13
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
C C + L+EG+ + +L C HV H CL + F + CP C +++
Sbjct: 86 CVVCLSKLKEGE--EVRKLECRHVFHKKCLEGWLHQF-------NFTCPLCRSAL 131
>At1g51745 unknown protein
Length = 597
Score = 33.5 bits (75), Expect = 0.17
Identities = 24/68 (35%), Positives = 28/68 (40%), Gaps = 1/68 (1%)
Query: 145 SLSVTESRSPPPAFASEPLIGRENHGNSDSPATG-SEPPKLSVTDIVEIEGANSAGNFVK 203
S+SV+ PL G ENH A S P K VTD+ G NS FVK
Sbjct: 357 SISVSAEDDSSDRLFDVPLTGEENHSEGFPAACRISSPRKALVTDLTRRCGRNSHNVFVK 416
Query: 204 GSSPVGPA 211
+ G A
Sbjct: 417 NEASNGSA 424
>At4g34040 putative protein
Length = 666
Score = 32.3 bits (72), Expect = 0.37
Identities = 19/53 (35%), Positives = 21/53 (38%), Gaps = 10/53 (18%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCST 106
CC CQ EGD T LGC H HT C+ + CP C T
Sbjct: 619 CCVCQEEYAEGDDLGT--LGCGHEFHTACVKQWLM--------LKNLCPICKT 661
>At2g22680 copia-like retroelement pol polyprotein
Length = 683
Score = 32.3 bits (72), Expect = 0.37
Identities = 20/94 (21%), Positives = 39/94 (41%), Gaps = 5/94 (5%)
Query: 53 KCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPP- 111
KC C ++ G G+ C H H C+ S + A +CP C +S+ P
Sbjct: 129 KCGICLQSVKSGQGTAIFTAECSHTFHFPCVTSRAAANHNRLA----SCPVCGSSLLPEI 184
Query: 112 KSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVS 145
++ S++ ++K ++ + + P+S
Sbjct: 185 RNYAKPESQIKPEIKNKSLRVYNDDEALISSPIS 218
>At1g68070 RING zinc finger like protein
Length = 343
Score = 32.3 bits (72), Expect = 0.37
Identities = 17/55 (30%), Positives = 24/55 (42%), Gaps = 10/55 (18%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
CC C + E DG++ L C H H+ C+V +K CP C +I
Sbjct: 292 CCICLSSYE--DGAELVSLPCNHHFHSTCIVKWLK--------MNATCPLCKFNI 336
>At5g60710 unknown protein
Length = 704
Score = 32.0 bits (71), Expect = 0.49
Identities = 29/146 (19%), Positives = 50/146 (33%), Gaps = 17/146 (11%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKS 113
C C ++ G G C H H C+ +++K CP C W
Sbjct: 72 CAICLTAMKAGQGHAIFTAECSHSFHFQCITTNVKH-------GNQICPVCRAK-WNEIP 123
Query: 114 VKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD 173
++ ++K K + G ++ +S+ RS P + S P R + +
Sbjct: 124 IQSP----NAKPKSGVKPIGRPRD-----DAWMSIPPRRSSPIQYTSRPDCLRVSSIFNT 174
Query: 174 SPATGSEPPKLSVTDIVEIEGANSAG 199
PA ++ L D G + G
Sbjct: 175 EPAVFNDDEALEHQDRSAESGLDKPG 200
>At4g17920 putative protein
Length = 289
Score = 32.0 bits (71), Expect = 0.49
Identities = 20/79 (25%), Positives = 31/79 (38%), Gaps = 10/79 (12%)
Query: 56 QCQAVLEEGDGSQTTRL--GCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKS 113
+C L E DG RL C HV H C+ +S CP C + PP
Sbjct: 109 ECAICLLEFDGDHVLRLLTTCYHVFHQECIDLWFESHR--------TCPVCRRDLDPPPP 160
Query: 114 VKDSGSRLHSKLKEAIMQT 132
+++ + + + I +T
Sbjct: 161 PENTKPTVDEMIIDVIQET 179
>At1g80400 putative RING zinc finger protein
Length = 407
Score = 32.0 bits (71), Expect = 0.49
Identities = 16/55 (29%), Positives = 21/55 (38%), Gaps = 10/55 (18%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
CC C + GD Q L C HV H +C+ +K CP C +
Sbjct: 355 CCIC--LTRYGDDEQVRELPCSHVFHVDCVDKWLK--------INATCPLCKNEV 399
>At1g72200 RING-H2 zinc finger protein ATL3, putative
Length = 404
Score = 32.0 bits (71), Expect = 0.49
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 10/57 (17%)
Query: 56 QCQAVLEEGDGSQTTRL--GCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP 110
+C L E + +T RL C HV H C+ + ++S HT CP C + P
Sbjct: 143 ECSVCLNEFEDDETLRLIPKCCHVFHPGCIDAWLRS---HT-----TCPLCRADLIP 191
>At3g61460 RING finger protein
Length = 170
Score = 31.6 bits (70), Expect = 0.63
Identities = 26/95 (27%), Positives = 37/95 (38%), Gaps = 19/95 (20%)
Query: 32 QICVIRTYSEWVIDGEYDWPPKCCQCQAVLEEGDGSQTTRL--GCLHVIHTNCL---VSH 86
+I + + E GE D P C C L E +G Q R C H+ H +CL + H
Sbjct: 73 EILPVIKFEELTNSGE-DLPENCAVC---LYEFEGEQEIRWLRNCRHIFHRSCLDRWMDH 128
Query: 87 IKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRL 121
+ CP C T P + ++ RL
Sbjct: 129 DQK----------TCPLCRTPFVPDEMQEEFNQRL 153
>At5g42940 unknown protein
Length = 691
Score = 30.8 bits (68), Expect = 1.1
Identities = 17/54 (31%), Positives = 22/54 (40%), Gaps = 10/54 (18%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTS 107
CC CQ EG+ T L C H H+ C+ +K CP C T+
Sbjct: 637 CCVCQEEYTEGEDMGT--LECGHEFHSQCIKEWLKQ--------KNLCPICKTT 680
>At4g31200 predicted protein
Length = 650
Score = 30.8 bits (68), Expect = 1.1
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 10/85 (11%)
Query: 111 PKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPA--FASEPLIGREN 168
P++ +++ SRL E I+Q K +F +S E +S PPA F+ P+I
Sbjct: 369 PQNKEENQSRL-----EKILQFWASKEVFDQDTISSLEKEMKSGPPANTFSHSPIIAAH- 422
Query: 169 HGNSDSPATGSEPPKLSVTDIVEIE 193
P +PP +V+ + +E
Sbjct: 423 --ALQRPGMLQQPPNSNVSSTMNLE 445
>At4g05350 putative protein
Length = 206
Score = 30.8 bits (68), Expect = 1.1
Identities = 18/57 (31%), Positives = 23/57 (39%), Gaps = 9/57 (15%)
Query: 54 CCQCQAVLEEGDGSQ-TTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIW 109
C C L G + TR+ C HV H CL+ +K CP C T I+
Sbjct: 157 CSICLESLVSGPKPRDVTRMTCSHVFHNGCLLEWLK--------RKNTCPLCRTEIY 205
>At3g54780 unknown protein
Length = 676
Score = 30.8 bits (68), Expect = 1.1
Identities = 14/51 (27%), Positives = 21/51 (40%), Gaps = 7/51 (13%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTC 104
C C ++EG G C H+ H +C+ S++K CP C
Sbjct: 75 CSICLNKMKEGCGHAIFTAECSHMFHFHCIASNVKH-------GNQVCPVC 118
>At1g65040
Length = 537
Score = 30.8 bits (68), Expect = 1.1
Identities = 21/84 (25%), Positives = 37/84 (44%), Gaps = 9/84 (10%)
Query: 71 RLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIM 130
+L C H+ H +CL S ++ CPTC + P ++ + S +E++
Sbjct: 304 KLVCGHLFHVHCLRSWLER--------QNTCPTCRALVVPAENATSTASGNRGPHQESLQ 355
Query: 131 Q-TGMEKNIFGNHPVSLSVTESRS 153
Q TG + VS + +E+ S
Sbjct: 356 QGTGTSSSDGQGSSVSAAASENMS 379
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,540,616
Number of Sequences: 26719
Number of extensions: 385827
Number of successful extensions: 1150
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 1116
Number of HSP's gapped (non-prelim): 87
length of query: 334
length of database: 11,318,596
effective HSP length: 100
effective length of query: 234
effective length of database: 8,646,696
effective search space: 2023326864
effective search space used: 2023326864
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0218.25


