

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.19
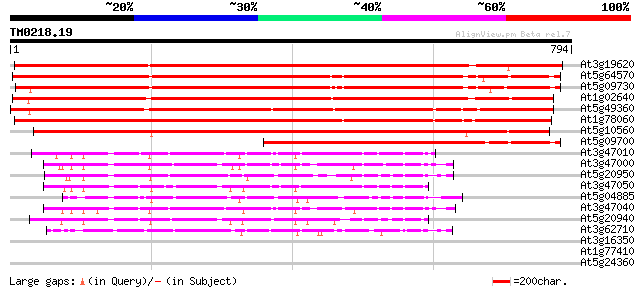
(794 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g19620 beta-xylosidase, putative 1105 0.0
At5g64570 beta-xylosidase 851 0.0
At5g09730 beta-xylosidase - like protein 809 0.0
At1g02640 beta-xylosidase, putative 794 0.0
At5g49360 xylosidase 793 0.0
At1g78060 putative protein 723 0.0
At5g10560 beta-xylosidase - like protein 711 0.0
At5g09700 beta-glucosidase - like protein 379 e-105
At3g47010 beta-D-glucan exohydrolase - like protein 133 3e-31
At3g47000 beta-D-glucan exohydrolase - like protein 130 2e-30
At5g20950 beta-D-glucan exohydrolase-like protein 126 4e-29
At3g47050 beta-D-glucan exohydrolase - like protein 124 2e-28
At5g04885 unknown protein 118 1e-26
At3g47040 beta-D-glucan exohydrolase - like protein 117 2e-26
At5g20940 beta-glucosidase - like protein 115 7e-26
At3g62710 beta-D-glucan exohydrolase-like protein 113 5e-25
At3g16350 putative MYB family transcription factor 36 0.097
At1g77410 unknown protein 31 2.4
At5g24360 unknown protein 31 3.1
>At3g19620 beta-xylosidase, putative
Length = 781
Score = 1105 bits (2858), Expect = 0.0
Identities = 542/782 (69%), Positives = 646/782 (82%), Gaps = 17/782 (2%)
Query: 7 FLSLFLSLLVLLPILTSSQKH-ACNKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKA 65
F+ L L ++ L+ L SQK+ AC+ ++P T+ + FC+ +LSYEARAKDLVSRL+L+EK
Sbjct: 6 FVRLSLLIIALVSSLCESQKNFACDISAPATAKYGFCNVSLSYEARAKDLVSRLSLKEKV 65
Query: 66 QQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNAT 125
QQLVN ++G+ RLGVP YEWWSEALHGVS++GPG FN TVPGATSFPA IL+AASFN +
Sbjct: 66 QQLVNKATGVPRLGVPPYEWWSEALHGVSDVGPGVHFNGTVPGATSFPATILTAASFNTS 125
Query: 126 LWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKY 185
LW MG+VVSTEARAM+NV LAGLT+WSPNVNVFRDPRWGRGQETPGEDPLVVS+YAV Y
Sbjct: 126 LWLKMGEVVSTEARAMHNVGLAGLTYWSPNVNVFRDPRWGRGQETPGEDPLVVSKYAVNY 185
Query: 186 VRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPF 245
V+GLQDV D K+ RLKVSSCCKHYTAYD+DNWKG+DRFHFDAKVTKQDLEDTYQ PF
Sbjct: 186 VKGLQDVHDAG--KSRRLKVSSCCKHYTAYDLDNWKGIDRFHFDAKVTKQDLEDTYQTPF 243
Query: 246 KSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIH 305
KSCV EG VSSVMCSYNRVNGIPTCADP+LL+GVIRGQW LDGYIVSDCDS+QVY+ IH
Sbjct: 244 KSCVEEGDVSSVMCSYNRVNGIPTCADPNLLRGVIRGQWRLDGYIVSDCDSIQVYFNDIH 303
Query: 306 YTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFF 365
YT T EDAVALALKAGLNMNCGD+L KYT NAVKLKK++ S VD+AL+YNYIVLMRLGFF
Sbjct: 304 YTKTREDAVALALKAGLNMNCGDFLGKYTENAVKLKKLNGSDVDEALIYNYIVLMRLGFF 363
Query: 366 D-NPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSGVLPLSKTNIKNLAVIGPNA 424
D +PKSLPF NLGPSDVC+K++Q+LALEAAKQGIVLLEN G LPL KT +K LAVIGPNA
Sbjct: 364 DGDPKSLPFGNLGPSDVCSKDHQMLALEAAKQGIVLLENRGDLPLPKTTVKKLAVIGPNA 423
Query: 425 NATTVMISNYAGIPCRYTSPLQGLQKYI-SSVTYAPGCGNVKCGDQSLFAAATKAAAPAD 483
NAT VMISNYAG+PC+YTSP+QGLQKY+ + Y PGC +VKCGDQ+L +AA KA + AD
Sbjct: 424 NATKVMISNYAGVPCKYTSPIQGLQKYVPEKIVYEPGCKDVKCGDQTLISAAVKAVSEAD 483
Query: 484 AVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKS 543
VLVVGLDQ++EAEGLDRVNLTLPG+QEKLV+DVA+A K V+LVIM+AGPIDISF K+
Sbjct: 484 VTVLVVGLDQTVEAEGLDRVNLTLPGYQEKLVRDVANAAKKTVVLVIMSAGPIDISFAKN 543
Query: 544 IRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSS 603
+ I +LWVGYPG+AGGDAIAQVIFG+YNP GR P TWYPQ +AD+V MTDMNMR NS+
Sbjct: 544 LSTIRAVLWVGYPGEAGGDAIAQVIFGDYNPSGRLPETWYPQEFADKVAMTDMNMRPNST 603
Query: 604 RNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSD 663
FPGR+YRFY G +Y+FG+GLSYS+FST++ SAPS I I+ N M +L+
Sbjct: 604 SGFPGRSYRFYTGKPIYKFGYGLSYSSFSTFVLSAPSIIHIKTNPIM---------NLNK 654
Query: 664 GQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPP---TSELVTGAPIKQLIGF 720
++DIST++C DL +VIGVKN G +GSHVVLVFW+PP S + G P+ QL+GF
Sbjct: 655 TTSVDISTVNCHDLKIRIVIGVKNHGLRSGSHVVLVFWKPPKCSKSLVGGGVPLTQLVGF 714
Query: 721 ERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQIDISLS 780
ER V MTE T++ D+C+ LS VD+ GKRKL+ G H L++GS+S+ Q+ H +++ L+
Sbjct: 715 ERVEVGRSMTEKFTVDFDVCKALSLVDTHGKRKLVTGHHKLVIGSNSDQQIYHHLNVRLA 774
Query: 781 GN 782
G+
Sbjct: 775 GD 776
>At5g64570 beta-xylosidase
Length = 784
Score = 851 bits (2198), Expect = 0.0
Identities = 440/783 (56%), Positives = 561/783 (71%), Gaps = 29/783 (3%)
Query: 5 LLFLSLFLSLLVLLPILTSSQKHACN-KASPKTSNFPFCDTTLSYEARAKDLVSRLTLQE 63
L+FL FL L SS AC+ A+P + + FC+T L E R DLV+RLTLQE
Sbjct: 23 LIFLCFFLYFLNFSNA-QSSPVFACDVAANPSLAAYGFCNTVLKIEYRVADLVARLTLQE 81
Query: 64 KAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFN 123
K LV+ ++G+ RLG+P+YEWWSEALHGVS +GPGT F+ VPGATSFP VIL+AASFN
Sbjct: 82 KIGFLVSKANGVTRLGIPTYEWWSEALHGVSYIGPGTHFSSQVPGATSFPQVILTAASFN 141
Query: 124 ATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAV 183
+L+ +G+VVSTEARAMYNV LAGLT+WSPNVN+FRDPRWGRGQETPGEDPL+ S+YA
Sbjct: 142 VSLFQAIGKVVSTEARAMYNVGLAGLTYWSPNVNIFRDPRWGRGQETPGEDPLLASKYAS 201
Query: 184 KYVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQP 243
YV+GLQ+ + +S +RLKV++CCKHYTAYDVDNWKGV+R+ F+A VT+QD++DTYQP
Sbjct: 202 GYVKGLQETDGGDS---NRLKVAACCKHYTAYDVDNWKGVERYSFNAVVTQQDMDDTYQP 258
Query: 244 PFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYES 303
PFKSCVV+G+V+SVMCSYN+VNG PTCADPDLL GVIRG+W L+GYIVSDCDSV V Y++
Sbjct: 259 PFKSCVVDGNVASVMCSYNQVNGKPTCADPDLLSGVIRGEWKLNGYIVSDCDSVDVLYKN 318
Query: 304 IHYTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLG 363
HYT TP +A A+++ AGL++NCG +L ++T AVK V+ + +D+A+ N++ LMRLG
Sbjct: 319 QHYTKTPAEAAAISILAGLDLNCGSFLGQHTEEAVKSGLVNEAAIDKAISNNFLTLMRLG 378
Query: 364 FFD-NPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSGVLPLSKTNIKNLAVIGP 422
FFD NPK+ + LGP+DVCT NQ LA +AA+QGIVLL+N+G LPLS +IK LAVIGP
Sbjct: 379 FFDGNPKNQIYGGLGPTDVCTSANQELAADAARQGIVLLKNTGCLPLSPKSIKTLAVIGP 438
Query: 423 NANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPA 482
NAN T MI NY G PC+YT+PLQGL +S+ TY PGC NV C + A ATK AA A
Sbjct: 439 NANVTKTMIGNYEGTPCKYTTPLQGLAGTVST-TYLPGCSNVACAVADV-AGATKLAATA 496
Query: 483 DAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTK 542
D VLV+G DQSIEAE DRV+L LPG Q++LV VA A KG V+LVIM+ G DI+F K
Sbjct: 497 DVSVLVIGADQSIEAESRDRVDLHLPGQQQELVIQVAKAAKGPVLLVIMSGGGFDITFAK 556
Query: 543 SIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANS 602
+ I GILWVGYPG+AGG AIA +IFG YNP G+ P TWYPQSY ++VPMT MNMR +
Sbjct: 557 NDPKIAGILWVGYPGEAGGIAIADIIFGRYNPSGKLPMTWYPQSYVEKVPMTIMNMRPDK 616
Query: 603 SRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLS 662
+ +PGRTYRFY G ++Y FG GLSY+ FS + APS + S+ N++ S S
Sbjct: 617 ASGYPGRTYRFYTGETVYAFGDGLSYTKFSHTLVKAPSLV------SLGLEENHVCRS-S 669
Query: 663 DGQAIDI------STISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQ 716
+ Q++D + +S F + I V+N G G H V +F PP + G+P K
Sbjct: 670 ECQSLDAIGPHCENAVSGGGSAFEVHIKVRNGGDREGIHTVFLFTTPPA---IHGSPRKH 726
Query: 717 LIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQID 776
L+GFE+ + V +++IC+ LS VD GKRK+ +GKH L VG ++H +
Sbjct: 727 LVGFEKIRLGKREEAVVRFKVEICKDLSVVDEIGKRKIGLGKHLLHVG-----DLKHSLS 781
Query: 777 ISL 779
I +
Sbjct: 782 IRI 784
>At5g09730 beta-xylosidase - like protein
Length = 773
Score = 809 bits (2089), Expect = 0.0
Identities = 427/782 (54%), Positives = 550/782 (69%), Gaps = 32/782 (4%)
Query: 9 SLFLSLLVLLPILTSSQKH---ACN-KASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEK 64
+LFL +V + +++Q AC+ +P + FC+ LS +AR DLV RLTL+EK
Sbjct: 13 TLFLCFIVCISEQSNNQSSPVFACDVTGNPSLAGLRFCNAGLSIKARVTDLVGRLTLEEK 72
Query: 65 AQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNA 124
L + + G++RLG+PSY+WWSEALHGVSN+G G+RF VPGATSFP VIL+AASFN
Sbjct: 73 IGFLTSKAIGVSRLGIPSYKWWSEALHGVSNVGGGSRFTGQVPGATSFPQVILTAASFNV 132
Query: 125 TLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVK 184
+L+ +G+VVSTEARAMYNV AGLTFWSPNVN+FRDPRWGRGQETPGEDP + S+YAV
Sbjct: 133 SLFQAIGKVVSTEARAMYNVGSAGLTFWSPNVNIFRDPRWGRGQETPGEDPTLSSKYAVA 192
Query: 185 YVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPP 244
YV+GLQ+ + + +RLKV++CCKHYTAYD+DNW+ V+R F+A V +QDL DT+QPP
Sbjct: 193 YVKGLQETDGGDP---NRLKVAACCKHYTAYDIDNWRNVNRLTFNAVVNQQDLADTFQPP 249
Query: 245 FKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESI 304
FKSCVV+GHV+SVMCSYN+VNG PTCADPDLL GVIRGQW L+GYIVSDCDSV V +
Sbjct: 250 FKSCVVDGHVASVMCSYNQVNGKPTCADPDLLSGVIRGQWQLNGYIVSDCDSVDVLFRKQ 309
Query: 305 HYTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGF 364
HY TPE+AVA +L AGL++NC + ++ AVK V+ + +D+A+ N+ LMRLGF
Sbjct: 310 HYAKTPEEAVAKSLLAGLDLNCDHFNGQHAMGAVKAGLVNETAIDKAISNNFATLMRLGF 369
Query: 365 FD-NPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENS-GVLPLSKTNIKNLAVIGP 422
FD +PK + LGP DVCT +NQ LA + A+QGIVLL+NS G LPLS + IK LAVIGP
Sbjct: 370 FDGDPKKQLYGGLGPKDVCTADNQELARDGARQGIVLLKNSAGSLPLSPSAIKTLAVIGP 429
Query: 423 NANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPA 482
NANAT MI NY G+PC+YT+PLQGL + +SS TY GC NV C D + +A AA A
Sbjct: 430 NANATETMIGNYHGVPCKYTTPLQGLAETVSS-TYQLGC-NVACVDADI-GSAVDLAASA 486
Query: 483 DAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTK 542
DAVVLVVG DQSIE EG DRV+L LPG Q++LV VA A +G V+LVIM+ G DI+F K
Sbjct: 487 DAVVLVVGADQSIEREGHDRVDLYLPGKQQELVTRVAMAARGPVVLVIMSGGGFDITFAK 546
Query: 543 SIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANS 602
+ + I I+WVGYPG+AGG AIA VIFG +NP G P TWYPQSY ++VPM++MNMR +
Sbjct: 547 NDKKITSIMWVGYPGEAGGLAIADVIFGRHNPSGNLPMTWYPQSYVEKVPMSNMNMRPDK 606
Query: 603 SRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLS 662
S+ +PGR+YRFY G ++Y F L+Y+ F + AP + S+S N+ S S
Sbjct: 607 SKGYPGRSYRFYTGETVYAFADALTYTKFDHQLIKAPRLV------SLSLDENHPCRS-S 659
Query: 663 DGQAIDISTISCLDLT-----FFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQL 717
+ Q++D C + F + + VKNTG GSH V +F TS V G+PIKQL
Sbjct: 660 ECQSLDAIGPHCENAVEGGSDFEVHLNVKNTGDRAGSHTVFLF---TTSPQVHGSPIKQL 716
Query: 718 IGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQIDI 777
+GFE+ + V +++C+ LS VD GKRK+ +G H L VGS ++H ++I
Sbjct: 717 LGFEKIRLGKSEEAVVRFNVNVCKDLSVVDETGKRKIALGHHLLHVGS-----LKHSLNI 771
Query: 778 SL 779
S+
Sbjct: 772 SV 773
>At1g02640 beta-xylosidase, putative
Length = 768
Score = 794 bits (2051), Expect = 0.0
Identities = 408/773 (52%), Positives = 524/773 (67%), Gaps = 27/773 (3%)
Query: 5 LLFLSLFLSLLVLLPILTSS------QKHACNKASPKTSNFPFCDTTLSYEARAKDLVSR 58
+L FL++++ I +SS + AC+ T+ FC ++ R +DL+ R
Sbjct: 2 ILHKMAFLAVILFFLISSSSVCVHSRETFACDTKDAATATLRFCQLSVPIPERVRDLIGR 61
Query: 59 LTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILS 118
LTL EK L N ++ I RLG+ YEWWSEALHGVSN+GPGT+F P ATSFP VI +
Sbjct: 62 LTLAEKVSLLGNTAAAIPRLGIKGYEWWSEALHGVSNVGPGTKFGGVYPAATSFPQVITT 121
Query: 119 AASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVV 178
ASFNA+LW ++G+VVS EARAMYN + GLT+WSPNVN+ RDPRWGRGQETPGEDP+V
Sbjct: 122 VASFNASLWESIGRVVSNEARAMYNGGVGGLTYWSPNVNILRDPRWGRGQETPGEDPVVA 181
Query: 179 SRYAVKYVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLE 238
+YA YVRGLQ + RLKV++CCKH+TAYD+DNW GVDRFHF+AKV+KQD+E
Sbjct: 182 GKYAASYVRGLQGND------RSRLKVAACCKHFTAYDLDNWNGVDRFHFNAKVSKQDIE 235
Query: 239 DTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQ 298
DT+ PF+ CV EG+V+S+MCSYN+VNG+PTCADP+LLK IR QWGL+GYIVSDCDSV
Sbjct: 236 DTFDVPFRMCVKEGNVASIMCSYNQVNGVPTCADPNLLKKTIRNQWGLNGYIVSDCDSVG 295
Query: 299 VYYESIHYTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIV 358
V Y++ HYT TPE+A A ++KAGL+++CG +L +T +AVK + S VD AL+ V
Sbjct: 296 VLYDTQHYTGTPEEAAADSIKAGLDLDCGPFLGAHTIDAVKKNLLRESDVDNALINTLTV 355
Query: 359 LMRLGFFDNP-KSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSG-VLPLSKTNIKN 416
MRLG FD + P+ +LGP+ VCT ++ LALEAA+QGIVLL+N G LPLS +
Sbjct: 356 QMRLGMFDGDIAAQPYGHLGPAHVCTPVHKGLALEAAQQGIVLLKNHGSSLPLSSQRHRT 415
Query: 417 LAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAAT 476
+AVIGPN++AT MI NYAG+ C YTSP+QG+ Y ++ + GC +V C D LF AA
Sbjct: 416 VAVIGPNSDATVTMIGNYAGVACGYTSPVQGITGYARTI-HQKGCVDVHCMDDRLFDAAV 474
Query: 477 KAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPI 536
+AA ADA VLV+GLDQSIEAE DR +L LPG Q++LV VA A KG VILV+M+ GPI
Sbjct: 475 EAARGADATVLVMGLDQSIEAEFKDRNSLLLPGKQQELVSRVAKAAKGPVILVLMSGGPI 534
Query: 537 DISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDM 596
DISF + R I I+W GYPGQ GG AIA ++FG NPGG+ P TWYPQ Y +PMT+M
Sbjct: 535 DISFAEKDRKIPAIVWAGYPGQEGGTAIADILFGSANPGGKLPMTWYPQDYLTNLPMTEM 594
Query: 597 NMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNN 656
+MR S+ PGRTYRFY+G +Y FGHGLSY+ F+ IA AP I I
Sbjct: 595 SMRPVHSKRIPGRTYRFYDGPVVYPFGHGLSYTRFTHNIADAPKVIPIAVRGR------- 647
Query: 657 IFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQ 716
G++I ++ C L+ + + V N G +G+H +LVF PP E AP KQ
Sbjct: 648 --NGTVSGKSIRVTHARCDRLSLGVHVEVTNVGSRDGTHTMLVFSAPPGGE---WAPKKQ 702
Query: 717 LIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSET 769
L+ FER HV VG + V + I +C+ LS VD G R++ IG H + +G S T
Sbjct: 703 LVAFERVHVAVGEKKRVQVNIHVCKYLSVVDRAGNRRIPIGDHGIHIGDESHT 755
>At5g49360 xylosidase
Length = 774
Score = 793 bits (2049), Expect = 0.0
Identities = 413/774 (53%), Positives = 526/774 (67%), Gaps = 18/774 (2%)
Query: 2 KSNLLFLSLFLSLLVLLPILTSSQK----HACNKASPKTSNFPFCDTTLSYEARAKDLVS 57
K+ L+ + + L+ LL ++ SS+ AC+ A+ T FC + R +DL+
Sbjct: 6 KALLIGNKVVVILVFLLCLVHSSESLRPLFACDPANGLTRTLRFCRANVPIHVRVQDLLG 65
Query: 58 RLTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVIL 117
RLTLQEK + LVN ++ + RLG+ YEWWSEALHG+S++GPG +F PGATSFP VI
Sbjct: 66 RLTLQEKIRNLVNNAAAVPRLGIGGYEWWSEALHGISDVGPGAKFGGAFPGATSFPQVIT 125
Query: 118 SAASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLV 177
+AASFN +LW +G+VVS EARAMYN +AGLT+WSPNVN+ RDPRWGRGQETPGEDP+V
Sbjct: 126 TAASFNQSLWEEIGRVVSDEARAMYNGGVAGLTYWSPNVNILRDPRWGRGQETPGEDPIV 185
Query: 178 VSRYAVKYVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDL 237
++YA YVRGLQ + +RLKV++CCKHYTAYD+DNW GVDRFHF+AKVT+QDL
Sbjct: 186 AAKYAASYVRGLQG-----TAAGNRLKVAACCKHYTAYDLDNWNGVDRFHFNAKVTQQDL 240
Query: 238 EDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSV 297
EDTY PFKSCV EG V+SVMCSYN+VNG PTCAD +LLK IRGQW L+GYIVSDCDSV
Sbjct: 241 EDTYNVPFKSCVYEGKVASVMCSYNQVNGKPTCADENLLKNTIRGQWRLNGYIVSDCDSV 300
Query: 298 QVYYESIHYTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYI 357
V++ HYT+TPE+A A ++KAGL+++CG +L +T AVK + + ++ AL
Sbjct: 301 DVFFNQQHYTSTPEEAAARSIKAGLDLDCGPFLAIFTEGAVKKGLLTENDINLALANTLT 360
Query: 358 VLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSG-VLPLSKTNIKN 416
V MRLG FD P+ANLGP DVCT ++ LALEAA QGIVLL+NS LPLS +
Sbjct: 361 VQMRLGMFDGNLG-PYANLGPRDVCTPAHKHLALEAAHQGIVLLKNSARSLPLSPRRHRT 419
Query: 417 LAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAAT 476
+AVIGPN++ T MI NYAG C YTSPLQG+ +Y ++ A GC V C F AA
Sbjct: 420 VAVIGPNSDVTETMIGNYAGKACAYTSPLQGISRYARTLHQA-GCAGVACKGNQGFGAAE 478
Query: 477 KAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPI 536
AA ADA VLV+GLDQSIEAE DR L LPG+Q+ LV VA A++G VILV+M+ GPI
Sbjct: 479 AAAREADATVLVMGLDQSIEAETRDRTGLLLPGYQQDLVTRVAQASRGPVILVLMSGGPI 538
Query: 537 DISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDM 596
D++F K+ + I+W GYPGQAGG AIA +IFG NPGG+ P TWYPQ Y +VPMT M
Sbjct: 539 DVTFAKNDPRVAAIIWAGYPGQAGGAAIANIIFGAANPGGKLPMTWYPQDYVAKVPMTVM 598
Query: 597 NMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNN 656
MRA S N+PGRTYRFY G ++ FG GLSY+TF+ +A +P + + S N
Sbjct: 599 AMRA--SGNYPGRTYRFYKGPVVFPFGFGLSYTTFTHSLAKSP-LAQLSVSLSNLNSANT 655
Query: 657 IFESLSDGQAIDISTISCLDLTFF-LVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIK 715
I S S +I +S +C L + V NTG F+G+H V VF EPP + + K
Sbjct: 656 ILNSSS--HSIKVSHTNCNSFPKMPLHVEVSNTGEFDGTHTVFVFAEPPINGIKGLGVNK 713
Query: 716 QLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSET 769
QLI FE+ HVM G + V +++D C+ L VD GKR++ +G+H L +G T
Sbjct: 714 QLIAFEKVHVMAGAKQTVQVDVDACKHLGVVDEYGKRRIPMGEHKLHIGDLKHT 767
>At1g78060 putative protein
Length = 767
Score = 723 bits (1867), Expect = 0.0
Identities = 367/763 (48%), Positives = 509/763 (66%), Gaps = 12/763 (1%)
Query: 8 LSLFLSLLVLLPILTSSQKHACNKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKAQQ 67
L L L L ++ + ++ H+C+ ++P T + FC T L RA+DLVSRLT+ EK Q
Sbjct: 5 LLLLLLLFIVHGVESAPPPHSCDPSNPTTKLYQFCRTDLPIGKRARDLVSRLTIDEKISQ 64
Query: 68 LVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLW 127
LVN + GI RLGVP+YEWWSEALHGV+ GPG RFN TV ATSFP VIL+AASF++ W
Sbjct: 65 LVNTAPGIPRLGVPAYEWWSEALHGVAYAGPGIRFNGTVKAATSFPQVILTAASFDSYEW 124
Query: 128 YNMGQVVSTEARAMYNVDLA-GLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYV 186
+ + QV+ EAR +YN A G+TFW+PN+N+FRDPRWGRGQETPGEDP++ YAV YV
Sbjct: 125 FRIAQVIGKEARGVYNAGQANGMTFWAPNINIFRDPRWGRGQETPGEDPMMTGTYAVAYV 184
Query: 187 RGLQ-DVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPF 245
RGLQ D D ++ L+ S+CCKH+TAYD+D WKG+ R+ F+A+V+ DL +TYQPPF
Sbjct: 185 RGLQGDSFDGRKTLSNHLQASACCKHFTAYDLDRWKGITRYVFNAQVSLADLAETYQPPF 244
Query: 246 KSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIH 305
K C+ EG S +MC+YNRVNGIP+CADP+LL RGQW GYI SDCD+V + Y++
Sbjct: 245 KKCIEEGRASGIMCAYNRVNGIPSCADPNLLTRTARGQWAFRGYITSDCDAVSIIYDAQG 304
Query: 306 YTATPEDAVALALKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFF 365
Y +PEDAVA LKAG+++NCG YL+K+T +A++ KKV + +D+AL+ + V +RLG F
Sbjct: 305 YAKSPEDAVADVLKAGMDVNCGSYLQKHTKSALQQKKVSETDIDRALLNLFSVRIRLGLF 364
Query: 366 D-NPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENS-GVLPLSKTNIKNLAVIGPN 423
+ +P LP+ N+ P++VC+ +Q LAL+AA+ GIVLL+N+ +LP SK ++ +LAVIGPN
Sbjct: 365 NGDPTKLPYGNISPNEVCSPAHQALALDAARNGIVLLKNNLKLLPFSKRSVSSLAVIGPN 424
Query: 424 ANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPAD 483
A+ ++ NYAG PC+ +PL L+ Y+ + Y GC +V C + ++ A A AD
Sbjct: 425 AHVVKTLLGNYAGPPCKTVTPLDALRSYVKNAVYHQGCDSVACSNAAI-DQAVAIAKNAD 483
Query: 484 AVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKS 543
VVL++GLDQ+ E E DRV+L+LPG Q++L+ VA+A K V+LV++ GP+DISF +
Sbjct: 484 HVVLIMGLDQTQEKEDFDRVDLSLPGKQQELITSVANAAKKPVVLVLICGGPVDISFAAN 543
Query: 544 IRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSS 603
IG I+W GYPG+AGG AI+++IFG++NPGGR P TWYPQS+ + + MTDM MR S+
Sbjct: 544 NNKIGSIIWAGYPGEAGGIAISEIIFGDHNPGGRLPVTWYPQSFVN-IQMTDMRMR--SA 600
Query: 604 RNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSD 663
+PGRTY+FY G +YEFGHGLSYS +S + T + N S +Q ++ +S+
Sbjct: 601 TGYPGRTYKFYKGPKVYEFGHGLSYSAYSYRFKTLAET-NLYLNQSKAQTNS---DSVRY 656
Query: 664 GQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERA 723
++ C + + V+N G G H VL+F KQL+GF+
Sbjct: 657 TLVSEMGKEGCDVAKTKVTVEVENQGEMAGKHPVLMFARHERGGEDGKRAEKQLVGFKSI 716
Query: 724 HVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSS 766
+ G + EI +C+ LS + G L GK+ L VG S
Sbjct: 717 VLSNGEKAEMEFEIGLCEHLSRANEFGVMVLEEGKYFLTVGDS 759
>At5g10560 beta-xylosidase - like protein
Length = 792
Score = 711 bits (1835), Expect = 0.0
Identities = 361/752 (48%), Positives = 496/752 (65%), Gaps = 25/752 (3%)
Query: 34 PKTSNFPFCDTTLSYEARAKDLVSRLTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGV 93
P S++PFC+ +LS + RA LVS L L EK QL N ++ + RLG+P YEWWSE+LHG+
Sbjct: 35 PHFSSYPFCNVSLSIKQRAISLVSLLMLPEKIGQLSNTAASVPRLGIPPYEWWSESLHGL 94
Query: 94 SNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWS 153
++ GPG FN ++ ATSFP VI+SAASFN TLWY +G V+ E RAMYN AGLTFW+
Sbjct: 95 ADNGPGVSFNGSISAATSFPQVIVSAASFNRTLWYEIGSAVAVEGRAMYNGGQAGLTFWA 154
Query: 154 PNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDEESIK-------------- 199
PN+NVFRDPRWGRGQETPGEDP VVS Y V++VRG Q+ + + +K
Sbjct: 155 PNINVFRDPRWGRGQETPGEDPKVVSEYGVEFVRGFQEKKKRKVLKRRFSDDVDDDRHDD 214
Query: 200 -AD-RLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKSCVVEGHVSSV 257
AD +L +S+CCKH+TAYD++ W R+ F+A VT+QD+EDTYQPPF++C+ +G S +
Sbjct: 215 DADGKLMLSACCKHFTAYDLEKWGNFTRYDFNAVVTEQDMEDTYQPPFETCIRDGKASCL 274
Query: 258 MCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATPEDAVALA 317
MCSYN VNG+P CA DLL+ R +WG +GYI SDCD+V + YT +PE+AVA A
Sbjct: 275 MCSYNAVNGVPACAQGDLLQKA-RVEWGFEGYITSDCDAVATIFAYQGYTKSPEEAVADA 333
Query: 318 LKAGLNMNCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLMRLGFFD-NPKSLPFANL 376
+KAG+++NCG Y+ ++T +A++ KV VD+AL+ + V +RLG FD +P+ + L
Sbjct: 334 IKAGVDINCGTYMLRHTQSAIEQGKVSEELVDRALLNLFAVQLRLGLFDGDPRRGQYGKL 393
Query: 377 GPSDVCTKENQLLALEAAKQGIVLLENS-GVLPLSKTNIKNLAVIGPNANATTVMISNYA 435
G +D+C+ +++ LALEA +QGIVLL+N +LPL+K ++ +LA++GP AN + M Y
Sbjct: 394 GSNDICSSDHRKLALEATRQGIVLLKNDHKLLPLNKNHVSSLAIVGPMANNISNMGGTYT 453
Query: 436 GIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSI 495
G PC+ + L +Y+ +YA GC +V C + F A A AD V++V GLD S
Sbjct: 454 GKPCQRKTLFTELLEYVKKTSYASGCSDVSCDSDTGFGEAVAIAKGADFVIVVAGLDLSQ 513
Query: 496 EAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGY 555
E E DRV+L+LPG Q+ LV VA +K VILV+ GP+D++F K+ IG I+W+GY
Sbjct: 514 ETEDKDRVSLSLPGKQKDLVSHVAAVSKKPVILVLTGGGPVDVTFAKNDPRIGSIIWIGY 573
Query: 556 PGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYN 615
PG+ GG A+A++IFG++NPGGR P TWYP+S+ D V M+DM+MRANSSR +PGRTYRFY
Sbjct: 574 PGETGGQALAEIIFGDFNPGGRLPTTWYPESFTD-VAMSDMHMRANSSRGYPGRTYRFYT 632
Query: 616 GNSLYEFGHGLSYSTFSTYIASAPSTIMIE---PNTSMSQPHNNIFESLSDGQAIDISTI 672
G +Y FG GLSY+ F I SAP + + P S + E L Q D+
Sbjct: 633 GPQVYSFGTGLSYTKFEYKILSAPIRLSLSELLPQQSSHKKQLQHGEELRYLQLDDVIVN 692
Query: 673 SCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEF 732
SC L F + + V NTG +GSHVV++F + P +++G P KQLIG++R HV
Sbjct: 693 SCESLRFNVRVHVSNTGEIDGSHVVMLFSKMP--PVLSGVPEKQLIGYDRVHVRSNEMME 750
Query: 733 VTLEIDICQLLSNVDSDGKRKLIIGKHTLLVG 764
ID C+ LS + GKR + +G H L +G
Sbjct: 751 TVFVIDPCKQLSVANDVGKRVIPLGSHVLFLG 782
>At5g09700 beta-glucosidase - like protein
Length = 411
Score = 379 bits (973), Expect = e-105
Identities = 208/424 (49%), Positives = 274/424 (64%), Gaps = 17/424 (4%)
Query: 360 MRLGFFD-NPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENS-GVLPLSKTNIKNL 417
MRLGFFD NPK+ P+ LGP DVCT EN+ LA+E A+QGIVLL+NS G LPLS + IK L
Sbjct: 1 MRLGFFDGNPKNQPYGGLGPKDVCTVENRELAVETARQGIVLLKNSAGSLPLSPSAIKTL 60
Query: 418 AVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATK 477
AVIGPNAN T MI NY G+ C+YT+PLQGL++ + + Y GC NV C + L +A T
Sbjct: 61 AVIGPNANVTKTMIGNYEGVACKYTTPLQGLERTVLTTKYHRGCFNVTCTEADLDSAKTL 120
Query: 478 AAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPID 537
AA+ ADA VLV+G DQ+IE E LDR++L LPG Q++LV VA A +G V+LVIM+ G D
Sbjct: 121 AAS-ADATVLVMGADQTIEKETLDRIDLNLPGKQQELVTQVAKAARGPVVLVIMSGGGFD 179
Query: 538 ISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMN 597
I+F K+ I I+WVGYPG+AGG AIA VIFG +NP G+ P TWYPQSY ++VPMT+MN
Sbjct: 180 ITFAKNDEKITSIMWVGYPGEAGGIAIADVIFGRHNPSGKLPMTWYPQSYVEKVPMTNMN 239
Query: 598 MRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPSTIMIEPNTSMS--QPHN 655
MR + S + GRTYRFY G ++Y FG GLSY+ FS + AP + + + S S P
Sbjct: 240 MRPDKSNGYLGRTYRFYIGETVYAFGDGLSYTNFSHQLIKAPKFVSLNLDESQSCRSPEC 299
Query: 656 NIFESLSDGQAIDISTISCLDLTFFLVIGVKNTGPFNGSHVVLVFWEPPTSELVTGAPIK 715
+++ + S F + + V+N G G+ V +F PP V G+P K
Sbjct: 300 QSLDAIGPHCEKAVGERS----DFEVQLKVRNVGDREGTETVFLFTTPPE---VHGSPRK 352
Query: 716 QLIGFERAHVMVGMTEFVTLEIDICQLLSNVDSDGKRKLIIGKHTLLVGSSSETQVRHQI 775
QL+GFE+ + V ++D+C+ L VD GKRKL +G H L VGS ++H
Sbjct: 353 QLLGFEKIRLGKKEETVVRFKVDVCKDLGVVDEIGKRKLALGHHLLHVGS-----LKHSF 407
Query: 776 DISL 779
+IS+
Sbjct: 408 NISV 411
>At3g47010 beta-D-glucan exohydrolase - like protein
Length = 609
Score = 133 bits (335), Expect = 3e-31
Identities = 157/620 (25%), Positives = 277/620 (44%), Gaps = 77/620 (12%)
Query: 32 ASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEK-------------AQQLVNPSSGIARL 78
A + S++ + + EAR KDL+SR+TL EK Q + N G +
Sbjct: 2 AETEESSWVYKNRDAPVEARVKDLLSRMTLPEKIGQMTQIERSVASPQVITNSFIGSVQS 61
Query: 79 GVPSYEW-------WSEALHGVSNLGPGTRF-------------NRTVPGATSFPAVILS 118
G S+ W++ + G +R N V GAT FP I
Sbjct: 62 GAGSWPLEDAKSSDWADMIDGFQRSALASRLGIPIIYGTDAVHGNNNVYGATVFPHNIGL 121
Query: 119 AASFNATLWYNMGQVVSTEARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQETPGEDPLV 177
A+ +A L +G + E RA +G+ + ++P V V DPRWGR E+ E +
Sbjct: 122 GATRDADLVKRIGAATALEIRA------SGVHWTFAPCVAVLGDPRWGRCYESYSEAAKI 175
Query: 178 VSRYAVKYVRGLQDVEDEE-----SIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKV 232
V ++ + GLQ EE A R V +C KH+ D KG+ +
Sbjct: 176 VCEMSL-LISGLQGEPPEEHPYGYPFLAGRNNVIACAKHFVG-DGGTEKGLS--EGNTIT 231
Query: 233 TKQDLEDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVS 292
+ +DLE + P+ +C+ +G VS+VM S++ NG +D LL V++ + G G++VS
Sbjct: 232 SYEDLEKIHVAPYLNCIAQG-VSTVMASFSSWNGSRLHSDYFLLTEVLKQKLGFKGFLVS 290
Query: 293 DCDSVQVYYESIHYTATPEDAVALALKAGLNM-----NCGDYLRKYTANAVKLKKVDVST 347
D D ++ E + + V L + AG++M +++ T + V+ ++ ++
Sbjct: 291 DWDGLETISEP--EGSNYRNCVKLGINAGIDMVMVPFKYEQFIQDMT-DLVESGEIPMAR 347
Query: 348 VDQALVYNYIVLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLEN---- 403
V+ A+ V G F++P + + LG V KE++ +A EA ++ +VLL+N
Sbjct: 348 VNDAVERILRVKFVAGLFEHPLA-DRSLLG--TVGCKEHREVAREAVRKSLVLLKNGKNA 404
Query: 404 -SGVLPLSKTNIKNLAVIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCG 462
+ LPL + N K + V+G +AN + I + + + S+ A G
Sbjct: 405 DTPFLPLDR-NAKRILVVGMHANDLGNQCGGWTKIKSGQSGRITIGTTLLDSIKAAVGDK 463
Query: 463 NVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADAT 522
+++ ++ ++ VG E +G D LT+P ++ VA+
Sbjct: 464 TEVIFEKTPTKETLASSDGFSYAIVAVGEPPYAEMKG-DNSELTIPFNGNNIITAVAE-- 520
Query: 523 KGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTW 582
K ++++ + P+ + T + ++ +PG G ++ VIFG+Y+ G+ P +W
Sbjct: 521 KIPTLVILFSGRPMVLEPT-VLEKTEALVAAWFPG-TEGQGMSDVIFGDYDFKGKLPVSW 578
Query: 583 YPQSYADQVPMTDMNMRANS 602
+ + DQ+P +N ANS
Sbjct: 579 FKR--VDQLP---LNAEANS 593
>At3g47000 beta-D-glucan exohydrolase - like protein
Length = 608
Score = 130 bits (328), Expect = 2e-30
Identities = 173/641 (26%), Positives = 283/641 (43%), Gaps = 116/641 (18%)
Query: 49 EARAKDLVSRLTLQEKAQQLV-------NPSS-------GIARLG--VPSYEW----WSE 88
EAR KDL+SR+TL EK Q+ +PS+ + G VP + W++
Sbjct: 18 EARVKDLLSRMTLPEKIGQMTQIERRVASPSAFTDFFIGSVLNAGGSVPFEDAKSSDWAD 77
Query: 89 ALHGVSNLGPGTRF-------------NRTVPGATSFPAVILSAASFNATLWYNMGQVVS 135
+ G +R N V GAT FP I A+ +A L +G +
Sbjct: 78 MIDGFQRSALASRLGIPIIYGTDAVHGNNNVYGATVFPHNIGLGATRDADLVRRIGAATA 137
Query: 136 TEARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVED 194
E RA +G+ + +SP V V RDPRWGR E+ GEDP +V V GLQ V
Sbjct: 138 LEVRA------SGVHWAFSPCVAVLRDPRWGRCYESYGEDPELVCE-MTSLVSGLQGVPP 190
Query: 195 EE-----SIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKSCV 249
EE A R V +C KH+ D KG++ + A + ++LE + PP+ C+
Sbjct: 191 EEHPNGYPFVAGRNNVVACVKHFVG-DGGTDKGINEGNTIA--SYEELEKIHIPPYLKCL 247
Query: 250 VEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTAT 309
+G VS+VM SY+ NG AD LL +++ + G G++VSD +E + +
Sbjct: 248 AQG-VSTVMASYSSWNGTRLHADRFLLTEILKEKLGFKGFLVSD-------WEGLDRLSE 299
Query: 310 PEDA-----VALALKAGLNM-----NCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVL 359
P+ + + A+ AG++M +++ T + V+ ++ ++ ++ A+ V
Sbjct: 300 PQGSNYRYCIKTAVNAGIDMVMVPFKYEQFIQDMT-DLVESGEIPMARINDAVERILRVK 358
Query: 360 MRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLE-----NSGVLPLSKTNI 414
G F +P L +L P+ C KE++ LA EA ++ +VLL+ + LPL + N
Sbjct: 359 FVAGLFGHP--LTDRSLLPTVGC-KEHRELAQEAVRKSLVLLKSGKNADKPFLPLDR-NA 414
Query: 415 KNLAVIGPNANATTVMISNYAGIPC-RYTSPLQGLQKYISSVTYAPGCGNVKCGDQS--L 471
K + V G +A+ G C +T GL I+ T GD++ +
Sbjct: 415 KRILVTGTHADD--------LGYQCGGWTKTWFGLSGRITIGTTLLDAIKEAVGDETEVI 466
Query: 472 FAAATKAAAPADA-----VVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKV 526
+ A + ++ VG E G D L +P +V VA+ V
Sbjct: 467 YEKTPSKETLASSEGFSYAIVAVGEPPYAETMG-DNSELRIPFNGTDIVTAVAEIIPTLV 525
Query: 527 ILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQS 586
IL+ + P+ + T + ++ PG G +A V+FG+Y+ G+ P +W+
Sbjct: 526 ILI--SGRPVVLEPT-VLEKTEALVAAWLPG-TEGQGVADVVFGDYDFKGKLPVSWF--K 579
Query: 587 YADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLS 627
+ + +P ++ ANS + L+ FG GL+
Sbjct: 580 HVEHLP---LDAHANSY-------------DPLFPFGFGLN 604
>At5g20950 beta-D-glucan exohydrolase-like protein
Length = 624
Score = 126 bits (317), Expect = 4e-29
Identities = 160/645 (24%), Positives = 270/645 (41%), Gaps = 128/645 (19%)
Query: 50 ARAKDLVSRLTLQEKAQQLVNPSSGIARL----------------GVPSY----EWWSEA 89
AR +DL++R+TLQEK Q+V +A VPS E W
Sbjct: 35 ARIRDLMNRMTLQEKIGQMVQIERSVATPEVMKKYFIGSVLSGGGSVPSEKATPETWVNM 94
Query: 90 LHGVSNLGPGTRF-------------NRTVPGATSFPAVILSAASFNATLWYNMGQVVST 136
++ + TR + V GAT FP + + + L +G +
Sbjct: 95 VNEIQKASLSTRLGIPMIYGIDAVHGHNNVYGATIFPHNVGLGVTRDPNLVKRIGAATAL 154
Query: 137 EARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQ-DVED 194
E RA G+ + ++P + V RDPRWGR E+ ED +V + + + GLQ D+
Sbjct: 155 EVRA------TGIPYAFAPCIAVCRDPRWGRCYESYSEDYRIVQQ-MTEIIPGLQGDLPT 207
Query: 195 EES---IKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKSCVVE 251
+ + KV++C KH+ D +G+D + + + L + P + + V +
Sbjct: 208 KRKGVPFVGGKTKVAACAKHFVG-DGGTVRGID--ENNTVIDSKGLFGIHMPGYYNAVNK 264
Query: 252 GHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATPE 311
G V+++M SY+ NG+ A+ +L+ G ++ + G+++SD + H +
Sbjct: 265 G-VATIMVSYSAWNGLRMHANKELVTGFLKNKLKFRGFVISDWQGIDRITTPPHLNYS-- 321
Query: 312 DAVALALKAGLNMNCGDYLRKYT------ANAVKLKKVDVSTVDQALVYNYIVLMRLGFF 365
+V + AG++M Y YT ++ ++ K + +S +D AL V +G F
Sbjct: 322 YSVYAGISAGIDMIMVPY--NYTEFIDEISSQIQKKLIPISRIDDALKRILRVKFTMGLF 379
Query: 366 DNP-KSLPFANLGPSDVCTKENQLLALEAAKQGIVLLEN--SGVLPLSKTNIKNLAVIGP 422
+ P L FAN + +KE++ LA EA ++ +VLL+N +G PL K+ ++
Sbjct: 380 EEPLADLSFAN----QLGSKEHRELAREAVRKSLVLLKNGKTGAKPLLPLPKKSGKILVA 435
Query: 423 NANATTVMISNYAGIPC-RYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAP 481
A+A + G C +T QGL GN ++ AA AP
Sbjct: 436 GAHADNL------GYQCGGWTITWQGLN------------GNDHTVGTTILAAVKNTVAP 477
Query: 482 A-------------------DAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADAT 522
D ++VVG E G D NLT+ ++ +V +
Sbjct: 478 TTQVVYSQNPDANFVKSGKFDYAIVVVGEPPYAEMFG-DTTNLTISDPGPSIIGNVCGSV 536
Query: 523 KGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTW 582
K ++V+++ P+ I + I ++ PG G +A +FG+Y G+ TW
Sbjct: 537 --KCVVVVVSGRPVVIQ--PYVSTIDALVAAWLPG-TEGQGVADALFGDYGFTGKLARTW 591
Query: 583 YPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLS 627
+ Q+PM N R Y + LY FG GL+
Sbjct: 592 FKS--VKQLPM-----------NVGDRHY-----DPLYPFGFGLT 618
>At3g47050 beta-D-glucan exohydrolase - like protein
Length = 612
Score = 124 bits (312), Expect = 2e-28
Identities = 151/595 (25%), Positives = 255/595 (42%), Gaps = 78/595 (13%)
Query: 49 EARAKDLVSRLTLQEKAQQLVNPSSGIA---------------RLGVPSYEW-----WSE 88
EAR KDL+SR+TL EK Q+ +A R G +E W++
Sbjct: 18 EARVKDLLSRMTLAEKIGQMTLIERSVASEAVIRDFSIGSVLNRAGGWPFEDAKSSNWAD 77
Query: 89 ALHGVSNLGPGTRF-------------NRTVPGATSFPAVILSAASFNATLWYNMGQVVS 135
+ G +R N V GAT FP I A+ +A L +G +
Sbjct: 78 MIDGFQRSALESRLGIPIIYGIDAVHGNNDVYGATIFPHNIGLGATRDADLVKRIGAATA 137
Query: 136 TEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDE 195
E RA ++P V V +DPRWGR E+ GE +VS V GLQ +
Sbjct: 138 LEVRAC-----GAHWAFAPCVAVVKDPRWGRCYESYGEVAQIVSEMT-SLVSGLQGEPSK 191
Query: 196 ESIK-----ADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKSCVV 250
+ A R V +C KH+ N K ++ + + +DLE + P+K C+
Sbjct: 192 DHTNGYPFLAGRKNVVACAKHFVGDGGTN-KAINEGNTILRY--EDLERKHIAPYKKCIS 248
Query: 251 EGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYYESIHYTATP 310
+G VS+VM SY+ NG + LL +++ + G GY+VSD +E + + P
Sbjct: 249 QG-VSTVMASYSSWNGDKLHSHYFLLTEILKQKLGFKGYVVSD-------WEGLDRLSDP 300
Query: 311 -----EDAVALALKAGLNMNCGDY----LRKYTANAVKLKKVDVSTVDQALVYNYIVLMR 361
+ V + + AG++M + R + V+ +V ++ V+ A+ V
Sbjct: 301 PGSNYRNCVKIGINAGIDMVMVPFKYEQFRNDLIDLVESGEVSMARVNDAVERILRVKFV 360
Query: 362 LGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLEN---SGVLPLSKTNIKNLA 418
G F+ P L +L P+ C KE++ LA EA ++ +VLL+N LPL+ N + +
Sbjct: 361 AGLFEFP--LTDRSLLPTVGC-KEHRELAREAVRKSLVLLKNGRYGEFLPLN-CNAERIL 416
Query: 419 VIGPNANATTVMISNYAGIPCRYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKA 478
V+G +A+ + + + + ++ A G ++S +
Sbjct: 417 VVGTHADDLGYQCGGWTKTMYGQSGRITDGTTLLDAIKAAVGDETEVIYEKSPSEETLAS 476
Query: 479 AAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDI 538
++ VG E G D L +P +++ VA+ K ++++ + P+ +
Sbjct: 477 GYRFSYAIVAVGESPYAETMG-DNSELVIPFNGSEIITTVAE--KIPTLVILFSGRPMFL 533
Query: 539 SFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPM 593
+ + ++ PG G IA VIFG+Y+ G+ P TW+ + DQ+P+
Sbjct: 534 E-PQVLEKAEALVAAWLPG-TEGQGIADVIFGDYDFRGKLPATWFKR--VDQLPL 584
>At5g04885 unknown protein
Length = 665
Score = 118 bits (296), Expect = 1e-26
Identities = 147/592 (24%), Positives = 260/592 (43%), Gaps = 90/592 (15%)
Query: 75 IARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVV 134
++RLG+P + +A+HG +N V AT FP + A+ + L +G
Sbjct: 108 VSRLGIPMI-YGIDAVHGHNN----------VYNATIFPHNVGLGATRDPDLVKRIGAAT 156
Query: 135 STEARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVE 193
+ E RA G+ + ++P + V RDPRWGR E+ ED VV + GLQ E
Sbjct: 157 AVEVRA------TGIPYTFAPCIAVCRDPRWGRCYESYSEDHKVVEDMT-DVILGLQG-E 208
Query: 194 DEESIK------ADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS 247
+ K R KV++C KHY D +GV+ + + L + P +
Sbjct: 209 PPSNYKHGVPFVGGRDKVAACAKHYVG-DGGTTRGVNENNTVTDL--HGLLSVHMPAYAD 265
Query: 248 CVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVSDCDSVQVYY--ESIH 305
V +G VS+VM SY+ NG A+ +L+ G ++G G+++SD V H
Sbjct: 266 AVYKG-VSTVMVSYSSWNGEKMHANTELITGYLKGTLKFKGFVISDWQGVDKISTPPHTH 324
Query: 306 YTATPEDAVALALKAGLNM-----NCGDYLRKYTANAVKLKKVDVSTVDQALVYNYIVLM 360
YTA +V A++AG++M N +++ T VK + V+ +D A+ +V
Sbjct: 325 YTA----SVRAAIQAGIDMVMVPFNFTEFVNDLT-TLVKNNSIPVTRIDDAVRRILLVKF 379
Query: 361 RLGFFDNP-KSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSG----VLPLSKTNIK 415
+G F+NP F+ S++ ++ ++ LA EA ++ +VLL+N +LPL + K
Sbjct: 380 TMGLFENPLADYSFS----SELGSQAHRDLAREAVRKSLVLLKNGNKTNPMLPLPRKTSK 435
Query: 416 NLAV------IGPNANATTVMISNYAGIP-CRYTSPLQGLQKYISSVTYAPGCGNVKCGD 468
L +G T+ ++G R T+ L ++ + T N
Sbjct: 436 ILVAGTHADNLGYQCGGWTITWQGFSGNKNTRGTTLLSAVKSAVDQSTEVVFRENPD--- 492
Query: 469 QSLFAAATKAAAPADAVVLVVGLDQSIEAEGLDRVNLTLPGFQEKLVKDVADATKGKVIL 528
A K+ A A++ V + A D++ + PG ++ A K ++
Sbjct: 493 ----AEFIKSNNFAYAIIAVGEPPYAETAGDSDKLTMLDPG--PAIISSTCQAV--KCVV 544
Query: 529 VIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYA 588
V+++ P+ + + +I ++ PG G I +FG++ G+ P TW+ +
Sbjct: 545 VVISGRPLVME--PYVASIDALVAAWLPG-TEGQGITDALFGDHGFSGKLPVTWFRNT-- 599
Query: 589 DQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGLSYSTFSTYIASAPS 640
+Q+PM +Y + + L+ +G GL + ++ +A + S
Sbjct: 600 EQLPM----------------SYGDTHYDPLFAYGSGLETESVASIVARSTS 635
>At3g47040 beta-D-glucan exohydrolase - like protein
Length = 636
Score = 117 bits (294), Expect = 2e-26
Identities = 166/664 (25%), Positives = 281/664 (42%), Gaps = 131/664 (19%)
Query: 49 EARAKDLVSRLTLQEKAQQLVNPSS-------------GIARLGVPSYEW-------WSE 88
EAR KDL+SR+TL EK Q+ G G S+ + W++
Sbjct: 18 EARVKDLLSRMTLPEKIGQMTQIERVVTTPPVITDNFIGSVLNGGGSWPFEDAKTSDWAD 77
Query: 89 ALHGVSNLGPGTRF-------------NRTVPGATSFPAVI-LSAASF------------ 122
+ G N +R N V GAT FP I L A S
Sbjct: 78 MIDGYQNAALASRLGIPIIYGIDAVHGNNNVYGATIFPHNIGLGATSLVMLLHIDLEPKS 137
Query: 123 ------------NATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQET 170
+A L +G + E RA ++P V RDPRWGR E+
Sbjct: 138 LGRNKVVVKCDRDADLIRRVGAATALEVRAC-----GAHWAFAPCVAALRDPRWGRSYES 192
Query: 171 PGEDPLVVSRYAVKYVRGLQDVEDEE-----SIKADRLKVSSCCKHYTAYDVDNWKGVDR 225
EDP ++ + V GLQ +E A R V +C KH+ D KG++
Sbjct: 193 YSEDPDIICELS-SLVSGLQGEPPKEHPNGYPFLAGRNNVVACAKHFVG-DGGTDKGIN- 249
Query: 226 FHFDAKVTKQDLEDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWG 285
+ V+ ++LE + P+ +C+ +G VS+VM SY+ NG +D LL +++ + G
Sbjct: 250 -EGNTIVSYEELEKIHLAPYLNCLAQG-VSTVMASYSSWNGSKLHSDYFLLTELLKQKLG 307
Query: 286 LDGYIVSDCDSVQVYYESIHYTATPEDAVALALKAGLNMNCGDY----LRKYTANAVKLK 341
G+++SD ++++ E + + + V +++ AG++M + K + V+
Sbjct: 308 FKGFVISDWEALERLSEP--FGSNYRNCVKISVNAGVDMVMVPFKYEQFIKDLTDLVESG 365
Query: 342 KVDVSTVDQALVYNYIVLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLL 401
+V +S +D A+ V G F++P + + LG V KE++ LA E+ ++ +VLL
Sbjct: 366 EVTMSRIDDAVERILRVKFVAGLFEHPLT-DRSLLG--TVGCKEHRELARESVRKSLVLL 422
Query: 402 EN-----SGVLPLSKTNIKNLAVIGPNANATTVMISNYAGIPC-RYTSPLQGLQKYISSV 455
+N LPL + N+K + V G +A+ G C +T GL I+
Sbjct: 423 KNGTNSEKPFLPLDR-NVKRILVTGTHADD--------LGYQCGGWTKAWFGLSGRITIG 473
Query: 456 TYAPGCGNVKCGDQSLFAAATKAAAPADAVV--------LVVGLDQSIEAEGL-DRVNLT 506
T GD++ P++ + +V + ++ AE L D LT
Sbjct: 474 TTLLDAIKEAVGDKT---EVIYEKTPSEETLASLQRFSYAIVAVGETPYAETLGDNSELT 530
Query: 507 LPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQ 566
+P +V +A+ K ++V+ + P+ + + ++ PG G +
Sbjct: 531 IPLNGNDIVTALAE--KIPTLVVLFSGRPLVLE-PLVLEKAEALVAAWLPG-TEGQGMTD 586
Query: 567 VIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYEFGHGL 626
VIFG+Y+ G+ P +W+ + DQ+P+T ANS + L+ G GL
Sbjct: 587 VIFGDYDFEGKLPVSWFKR--VDQLPLT---ADANSY-------------DPLFPLGFGL 628
Query: 627 SYST 630
+Y++
Sbjct: 629 NYNS 632
>At5g20940 beta-glucosidase - like protein
Length = 626
Score = 115 bits (289), Expect = 7e-26
Identities = 158/631 (25%), Positives = 259/631 (41%), Gaps = 110/631 (17%)
Query: 29 CNKASPKT--SNFPFCDTTLSYEARAKDLVSRLTLQEKAQQLVNPS-------------- 72
C A+ K +N + D R K+L+S +TL+EK Q+V
Sbjct: 18 CTVAANKVPLANAKYKDPKEPLGVRIKNLMSHMTLEEKIGQMVQVERVNATTEVMQKYFV 77
Query: 73 ----SGIARLGVPSY--EWWSEALHGVSNLGPGTRF-------------NRTVPGATSFP 113
SG + P E W ++ V TR + TV AT FP
Sbjct: 78 GSVFSGGGSVPKPYIGPEAWVNMVNEVQKKALSTRLGIPIIYGIDAVHGHNTVYNATIFP 137
Query: 114 AVILSAASFNATLWYNMGQVVSTEARAMYNVDLAGLTF-WSPNVNVFRDPRWGRGQETPG 172
+ + + L +G+ + E RA G+ + ++P + V RDPRWGR E+
Sbjct: 138 HNVGLGVTRDPGLVKRIGEATALEVRA------TGIQYVFAPCIAVCRDPRWGRCYESYS 191
Query: 173 EDPLVVSRYAVKYVRGLQ-DVEDEES---IKADRLKVSSCCKHYTAYDVDNWKGVDRFHF 228
ED +V + + + GLQ D+ + A + KV++C KH+ D +G++ +
Sbjct: 192 EDHKIVQQMT-EIIPGLQGDLPTGQKGVPFVAGKTKVAACAKHFVG-DGGTLRGMNANN- 248
Query: 229 DAKVTKQDLEDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDG 288
+ L + P + V +G V++VM SY+ +NG+ A+ L+ G ++ + G
Sbjct: 249 -TVINSNGLLGIHMPAYHDAVNKG-VATVMVSYSSINGLKMHANKKLITGFLKNKLKFRG 306
Query: 289 YIVSDCDSVQVYYESIHYTATP-----EDAVALALKAGLNMNCGD----YLRKYTANAVK 339
++SD Y + TP +V A AGL+M G L + VK
Sbjct: 307 IVISD-------YLGVDQINTPLGANYSHSVYAATTAGLDMFMGSSNLTKLIDELTSQVK 359
Query: 340 LKKVDVSTVDQALVYNYIVLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIV 399
K + +S +D A+ V +G F+NP + + + +KE++ LA EA ++ +V
Sbjct: 360 RKFIPMSRIDDAVKRILRVKFTMGLFENPIA---DHSLAKKLGSKEHRELAREAVRKSLV 416
Query: 400 LLEN-----SGVLPLSKTNIKNLAV------IGPNANATTVMISNYAGIPCRY-TSPLQG 447
LL+N +LPL K K L +G T+ G T+ L
Sbjct: 417 LLKNGENADKPLLPLPKKANKILVAGTHADNLGYQCGGWTITWQGLNGNNLTIGTTILAA 476
Query: 448 LQKYISSVTYA-----PGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAEGLDR 502
++K + T P VK GD D ++ VG E G D
Sbjct: 477 VKKTVDPKTQVIYNQNPDTNFVKAGD-------------FDYAIVAVGEKPYAEGFG-DS 522
Query: 503 VNLTLPGFQEKLVKDVADATKGKVILVIMAAGPIDISFTKSIRNIGGILWVGYPGQAGGD 562
NLT+ + +V + K ++V+++ P+ I NI ++ PG G
Sbjct: 523 TNLTISEPGPSTIGNVCASV--KCVVVVVSGRPV----VMQISNIDALVAAWLPG-TEGQ 575
Query: 563 AIAQVIFGEYNPGGRSPFTWYPQSYADQVPM 593
+A V+FG+Y G+ TW+ DQ+PM
Sbjct: 576 GVADVLFGDYGFTGKLARTWF--KTVDQLPM 604
>At3g62710 beta-D-glucan exohydrolase-like protein
Length = 650
Score = 113 bits (282), Expect = 5e-25
Identities = 151/612 (24%), Positives = 263/612 (42%), Gaps = 106/612 (17%)
Query: 53 KDLVSRLTLQEKAQQLVNPSSGIARLGVPSYEWWSEALHGVSNLGPGTRFNRTVPGATSF 112
KD+ R+ Q A + ++ S+ RLG+P + +A+HG + T AT F
Sbjct: 105 KDIAKRI-FQTNAMKKLSLST---RLGIPLL-YAVDAVHG----------HNTFIDATIF 149
Query: 113 PAVILSAASFNATLWYNMGQVVSTEARAMYNVDLAGLTFWSPNVNVFRDPRWGRGQETPG 172
P + A+ + L +G + + E RA ++P V V RDPRWGR E+
Sbjct: 150 PHNVGLGATRDPQLVKKIGAITAQEVRATGVAQA-----FAPCVAVCRDPRWGRCYESYS 204
Query: 173 EDPLVVSRYAVKYVRGLQDVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKV 232
EDP VV+ + GLQ + + ++ V+ C KH+ D G++ + A
Sbjct: 205 EDPAVVNMMTESIIDGLQG--NAPYLADPKINVAGCAKHFVG-DGGTINGINENNTVAD- 260
Query: 233 TKQDLEDTYQPPFKSCVVEGHVSSVMCSYNRVNGIPTCADPDLLKGVIRGQWGLDGYIVS 292
L + PPF+ V +G ++S+M SY+ +NG+ A+ ++ ++ G+++S
Sbjct: 261 -NATLFGIHMPPFEIAVKKG-IASIMASYSSLNGVKMHANRAMITDYLKNTLKFQGFVIS 318
Query: 293 DCDSVQVY--YESIHYTATPEDAVALALKAGLNMNC-----GDYLRKYTANAVKLKKVDV 345
D + E +YT + E ++ AG++M +YL K T N V + +
Sbjct: 319 DWLGIDKITPIEKSNYTYSIEASI----NAGIDMVMVPWAYPEYLEKLT-NLVNGGYIPM 373
Query: 346 STVDQALVYNYIVLMRLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSG 405
S +D A+ V +G F+N SL L ++ ++ ++ + EA ++ +VLL+N
Sbjct: 374 SRIDDAVRRILRVKFSIGLFEN--SLADEKLPTTEFGSEAHREVGREAVRKSMVLLKNGK 431
Query: 406 -----VLPLSKTNIKNLAVIGPNANATTVMISNYA-------------------GIPC-- 439
++PL K +K + V G +AN ++ G+P
Sbjct: 432 TDADKIVPLPK-KVKKIVVAGRHANDMGWQCGGFSLTWQGFNGTGEDMPTNTKHGLPTGK 490
Query: 440 -RYTSPLQGLQKYISSVTYAPGCGNVKCGDQSLFAAATKAAAPADAVVLVVGLDQSIEAE 498
+ T+ L+ +QK + T V+ +Q K A A ++VVG E
Sbjct: 491 IKGTTILEAIQKAVDPTTEVV---YVEEPNQD----TAKLHADAAYTIVVVGETPYAETF 543
Query: 499 G-LDRVNLTLPGFQEKLVKDVADATKG---KVILVIMAAGPIDISFTKSIRNIGGILWVG 554
G + +T PG D T G K +++++ P+ I + + + W+
Sbjct: 544 GDSPTLGITKPG------PDTLSHTCGSGMKCLVILVTGRPLVIEPYIDMLDALAVAWL- 596
Query: 555 YPGQAGGDAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFY 614
PG G +A V+FG++ G P TW + Q+PM N + Y
Sbjct: 597 -PGTEG-QGVADVLFGDHPFTGTLPRTW--MKHVTQLPM-----------NVGDKNY--- 638
Query: 615 NGNSLYEFGHGL 626
+ LY FG+G+
Sbjct: 639 --DPLYPFGYGI 648
>At3g16350 putative MYB family transcription factor
Length = 387
Score = 35.8 bits (81), Expect = 0.097
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 11/77 (14%)
Query: 30 NKASPKTSNFPFCDTTLSYEARAKDLVSRLTLQEKAQQLVNPSSGIARLGVP-------- 81
+K K S P + +L+ A+++V+ QEK+Q+ + PS+G++ + VP
Sbjct: 225 SKEPEKKSYLPSLELSLNNTTEAEEVVATAPRQEKSQEAIEPSNGVSPMLVPGGFFPPCF 284
Query: 82 --SYEWWSEA-LHGVSN 95
+Y W A LHG +
Sbjct: 285 PVTYTIWLPASLHGTEH 301
>At1g77410 unknown protein
Length = 815
Score = 31.2 bits (69), Expect = 2.4
Identities = 33/134 (24%), Positives = 54/134 (39%), Gaps = 19/134 (14%)
Query: 562 DAIAQVIFGEYNPGGRSPFTWYPQSYADQVPMTDMNMRANSSRNFPGRTYRFYNGNSLYE 621
D A+V + +Y P TWY S+ + + S G+ + NG S+
Sbjct: 590 DGSAKVQWKQYRDSKSQPLTWYKASFDTPEGEDPVALNLGSM----GKGEAWVNGQSI-- 643
Query: 622 FGHGLSYSTFSTYIASAPSTIMIEPNTSMSQPHNNIFESLSDGQ-----AIDISTISCLD 676
G + +F TY + PS I S +P++N+ L + + I I T+S +
Sbjct: 644 ---GRYWVSFHTYKGN-PSQIWYHIPRSFLKPNSNLLVILEEEREGNPLGITIDTVSVTE 699
Query: 677 LTFFLVIGVKNTGP 690
+ V NT P
Sbjct: 700 VCGH----VSNTNP 709
>At5g24360 unknown protein
Length = 881
Score = 30.8 bits (68), Expect = 3.1
Identities = 21/77 (27%), Positives = 33/77 (42%), Gaps = 4/77 (5%)
Query: 361 RLGFFDNPKSLPFANLGPSDVCTKENQLLALEAAKQGIVLLENSGVLPLSKTNIKNLAVI 420
RLGF D LPF + P+ + + L L K+ +L LPL +T I + I
Sbjct: 287 RLGFLDEALYLPFQDRKPNQLAIGDGNQLTLPGNKEAEEVLS----LPLPETVISQITDI 342
Query: 421 GPNANATTVMISNYAGI 437
+ S ++G+
Sbjct: 343 IDGSTKQAGFASKFSGL 359
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,811,805
Number of Sequences: 26719
Number of extensions: 774008
Number of successful extensions: 1917
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 1807
Number of HSP's gapped (non-prelim): 34
length of query: 794
length of database: 11,318,596
effective HSP length: 107
effective length of query: 687
effective length of database: 8,459,663
effective search space: 5811788481
effective search space used: 5811788481
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0218.19


