

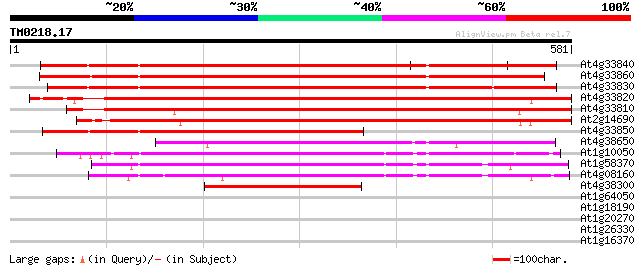
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.17
(581 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33840 putative protein 562 e-160
At4g33860 unknown protein 540 e-154
At4g33830 putative protein 516 e-146
At4g33820 putative glycosyl hydrolase family 10 protein 499 e-141
At4g33810 beta-xylan endohydrolase -like protein 467 e-132
At2g14690 1,4-beta-xylan endohydrolase 454 e-128
At4g33850 putative protein 365 e-101
At4g38650 putative protein 310 2e-84
At1g10050 putative xylan endohydrolase 199 4e-51
At1g58370 unknown protein 197 1e-50
At4g08160 xylan endohydrolase like proteinchimeric 192 3e-49
At4g38300 unknown protein 175 5e-44
At1g64050 unknown protein 30 2.8
At1g18190 hypothetical protein 30 2.8
At1g20270 unknown protein 30 4.8
At1g26330 hypothetical protein 29 6.3
At1g16370 putative transport protein 29 6.3
>At4g33840 putative protein
Length = 669
Score = 562 bits (1448), Expect = e-160
Identities = 270/537 (50%), Positives = 361/537 (66%), Gaps = 7/537 (1%)
Query: 33 DYSATTECLAEPHREHYGGGIIVNPGFDHNLEGWTVFGNGAIEERRSNEGNAFIVARNRT 92
D++A +CL P++ Y GGIIVNP + +GW+ FGN ++ R GN F+VA R
Sbjct: 119 DFAAKQQCLENPYKPQYNGGIIVNPDLQNGSQGWSQFGNAKVDFREFG-GNKFVVATQRN 177
Query: 93 QPLDSFSQKIQLKKGMLYTFSAWLQLSEGSDTVSVVFKINGRELIRGGHVIAKHGCWTLM 152
Q DS SQK+ L+KG+LYTFSAWLQ+S G VS VFK NG E G V+A+ CW+++
Sbjct: 178 QSSDSISQKVYLEKGILYTFSAWLQVSIGKSPVSAVFKKNG-EYKHAGSVVAESKCWSML 236
Query: 153 KGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQV 212
KGGL + + PAE+ FES+N VEIW DSVSLQPFT+E+W SH E+SI +VRK VR +V
Sbjct: 237 KGGLTVDESGPAELFFESENTMVEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRV 296
Query: 213 THINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYST 272
+ + ATI+ +Q KL +PFGC + NNIL N YQ WF RF TTF N+MKWYST
Sbjct: 297 MNNKGETIPNATISIEQKKLGYPFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYST 356
Query: 273 EIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRIN 332
E ++GQE+Y+T DAM+S K++GI+VRGHN+ WDDPK QP WV SLS +L A +R+
Sbjct: 357 ERIRGQEDYSTADAMLSFFKSHGIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVY 416
Query: 333 SVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEY 392
SV SRY G+L+ WDVVNENLHF+FFE G AS Y+ AH +DP T MF+NE+NT+E
Sbjct: 417 SVVSRYKGQLLGWDVVNENLHFSFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQ 476
Query: 393 SGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFP 452
D+ +SP Y+ K++++ + IPLAIGL+ HF++ PN+ YMR+ LD GATG P
Sbjct: 477 PKDLTSSPARYLGKLRELQSIRVAGKIPLAIGLESHFST--PNIPYMRSALDTFGATGLP 534
Query: 453 IWLTETSIDPQPH-QEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTP 511
IWLTE +D P+ + +YF+ +LREG+AHP V G++M+ G +G + L D NFKN P
Sbjct: 535 IWLTEIDVDAPPNVRANYFEQVLREGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLP 594
Query: 512 IGDVVDMLIREWG--TGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQISKTLNLSV 566
GDVVD L+REWG D+ G+ + L HGDYD+ ++HP + N ++
Sbjct: 595 TGDVVDKLLREWGGLRSQTTGVTDANGLFEAPLFHGDYDLRISHPLTNSKASYNFTL 651
Score = 94.0 bits (232), Expect = 2e-19
Identities = 48/101 (47%), Positives = 67/101 (65%), Gaps = 3/101 (2%)
Query: 416 SAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQPH-QEDYFDWIL 474
S I LAIGL+ HF + PN+ YMR+ LD+L ATG IWLTE ++ P Q YF+ +L
Sbjct: 11 SGYIRLAIGLESHFKT--PNIPYMRSALDILAATGLLIWLTEIDVEAPPSVQAKYFEQVL 68
Query: 475 REGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDV 515
R+G+AHP V+G++++ G +G + L D NF+N P GDV
Sbjct: 69 RDGHAHPQVKGMVVWGGYSPSGCYRMCLTDGNFRNLPTGDV 109
>At4g33860 unknown protein
Length = 576
Score = 540 bits (1391), Expect = e-154
Identities = 265/526 (50%), Positives = 348/526 (65%), Gaps = 7/526 (1%)
Query: 32 YDYSATTECLAEPHREHYGGGIIVNPGFDHNLEGWTVFGNGAIEERRSNEGNAFIVARNR 91
YDYSAT ECL P + Y GGIIV+P GWT FGN ++ R+ N F VAR+R
Sbjct: 25 YDYSATIECLEIPLKPQYNGGIIVSPDVRDGTLGWTPFGNAKVDFRKIGNHN-FFVARDR 83
Query: 92 TQPLDSFSQKIQLKKGMLYTFSAWLQLSEGSDTVSVVFKINGRELIRGGHVIAKHGCWTL 151
QP DS SQK+ L+KG+LYTFSAWLQ+S+G V VFK NG E G V+A+ CW++
Sbjct: 84 KQPFDSVSQKVYLEKGLLYTFSAWLQVSKGKAPVKAVFKKNG-EYKLAGSVVAESKCWSM 142
Query: 152 MKGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQ 211
+KGGL + + PAE+ FES++ TVEIW DSVSLQPFT+E+W SH E+SI++ RK VR +
Sbjct: 143 LKGGLTVDESGPAELYFESEDTTVEIWVDSVSLQPFTQEEWNSHHEQSIQKERKRTVRIR 202
Query: 212 VTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYS 271
+ + ATI+ +Q KL FPFGC + NIL N YQ WF RF TTF N+MKWYS
Sbjct: 203 AVNSKGEPIPKATISIEQRKLGFPFGCEVEKNILGNKAYQNWFTQRFTVTTFANEMKWYS 262
Query: 272 TEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRI 331
TE+V+G+E+Y+T DAM+ K +G++VRGHNI W+DPK QP+WV +LS +L A +R+
Sbjct: 263 TEVVRGKEDYSTADAMLRFFKQHGVAVRGHNILWNDPKYQPKWVNALSGNDLYNAVKRRV 322
Query: 332 NSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIE 391
SV SRY G+L WDVVNENLHF++FE +G AS + A DP T MF+NE+NT+E
Sbjct: 323 FSVVSRYKGQLAGWDVVNENLHFSYFEDKMGPKASYNIFKMAQAFDPTTTMFMNEYNTLE 382
Query: 392 YSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGF 451
S D +S Y++K+++I I L IGL+ HF + PN+ YMR+ LD L ATG
Sbjct: 383 ESSDSDSSLARYLQKLREIRSIRVCGNISLGIGLESHFKT--PNIPYMRSALDTLAATGL 440
Query: 452 PIWLTETSIDPQPH-QEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNT 510
PIWLTE ++ P+ Q YF+ +LREG+AHP V+GI+ + G +G + L D NFKN
Sbjct: 441 PIWLTEVDVEAPPNVQAKYFEQVLREGHAHPQVKGIVTWSGYSPSGCYRMCLTDGNFKNV 500
Query: 511 PIGDVVDMLIREWGTGPHEAK--ADSRGIVDISLHHGDYDVIVTHP 554
P GDVVD L+ EWG + D+ G + SL HGDYD+ + HP
Sbjct: 501 PTGDVVDKLLHEWGGFRRQTTGVTDADGYFEASLFHGDYDLKIAHP 546
>At4g33830 putative protein
Length = 544
Score = 516 bits (1328), Expect = e-146
Identities = 259/530 (48%), Positives = 340/530 (63%), Gaps = 8/530 (1%)
Query: 40 CLAEPHREHYGGGIIVNPGFDHNLEGWTVFGNGAIEERRSNEGNAFIVARNRTQPLDSFS 99
CL P++ Y GGIIVNP + +GW+ F N + R GN F+VA R Q DS S
Sbjct: 2 CLEIPYKPQYNGGIIVNPDMQNGSQGWSQFENAKVNFREFG-GNKFVVATQRNQSSDSVS 60
Query: 100 QKIQLKKGMLYTFSAWLQLSEGSDTVSVVFKINGRELIRGGHVIAKHGCWTLMKGGLVAN 159
QK+ L+KG+LYTFSAWLQ+S G VS VFK NG E G V+A+ CW+++KGGL +
Sbjct: 61 QKVYLEKGILYTFSAWLQVSTGKAPVSAVFKKNG-EYKHAGSVVAESKCWSMLKGGLTVD 119
Query: 160 FTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTHINETA 219
+ PAE+ ES++ TVEIW DSVSLQPFT+++W +HQE+SI+ RK VR +V +
Sbjct: 120 ESGPAELFVESEDTTVEIWVDSVSLQPFTQDEWNAHQEQSIDNSRKGPVRIRVVNNKGEK 179
Query: 220 LEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQE 279
+ A+IT +Q +L FPFG + NIL N YQ WF RF TTF N+MKWYSTE V+G E
Sbjct: 180 IPNASITIEQKRLGFPFGSAVAQNILGNQAYQNWFTQRFTVTTFENEMKWYSTESVRGIE 239
Query: 280 NYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYS 339
NYT DAM+ +GI+VRGHN+ WD PK Q +WV SLS +L A +R+ SV SRY
Sbjct: 240 NYTVADAMLRFFNQHGIAVRGHNVVWDHPKYQSKWVTSLSRNDLYNAVKRRVFSVVSRYK 299
Query: 340 GELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAAS 399
G+L WDVVNENLH +FFE G NAS ++ AH +DP+T MF+NEF T+E D+ AS
Sbjct: 300 GQLAGWDVVNENLHHSFFESKFGPNASNNIFAMAHAIDPSTTMFMNEFYTLEDPTDLKAS 359
Query: 400 PTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETS 459
P Y++K++++ IPL IGL+ HF++ PN+ YMR+ LD LGATG PIWLTE
Sbjct: 360 PAKYLEKLRELQSIRVRGNIPLGIGLESHFST--PNIPYMRSALDTLGATGLPIWLTEID 417
Query: 460 ID-PQPHQEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDM 518
+ P Q YF+ +LREG+AHP V+G++ + + L D NFKN P GDVVD
Sbjct: 418 VKAPSSDQAKYFEQVLREGHAHPHVKGMVTWTAYAPNCYHMC-LTDGNFKNLPTGDVVDK 476
Query: 519 LIREWG--TGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQISKTLNLSV 566
LIREWG D+ G + SL HGDYD+ ++HP S + N ++
Sbjct: 477 LIREWGGLRSQTTEVTDADGFFEASLFHGDYDLNISHPLTNSSVSHNFTL 526
>At4g33820 putative glycosyl hydrolase family 10 protein
Length = 570
Score = 499 bits (1285), Expect = e-141
Identities = 261/575 (45%), Positives = 366/575 (63%), Gaps = 40/575 (6%)
Query: 21 SGLTVHSLSHSYDYSATTECLAEPHREHYGGGIIVNPGFDHNLEG-----WTVFGNGAIE 75
SG+++ SHS+ S TEC+ +P R G++ F +LE W + GNG I
Sbjct: 22 SGVSIDPFSHSH--SLNTECVMKPPRSSETKGLLQ---FSRSLEDDSDEEWKIDGNGFIR 76
Query: 76 ERRSNEGNAFIVARNRTQPLDSFSQKIQLKKGMLYTFSAWLQLSEGSDT-VSVVFKINGR 134
E +Q+IQL +G +Y+FSAW++L EG+D V VVF+
Sbjct: 77 E---------------------MAQRIQLHQGNIYSFSAWVKLREGNDKKVGVVFRTENG 115
Query: 135 ELIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRS 194
L+ GG V A CWTL+KGG+V +F+ P +I FES+N +I A +V L+ F+KE+W+
Sbjct: 116 RLVHGGEVRANQECWTLLKGGIVPDFSGPVDIFFESENRGAKISAHNVLLKQFSKEEWKL 175
Query: 195 HQEESIERVRKSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWF 254
Q++ IE++RKSKVRF+VT+ N+TA++G I+ KQTK F GCG+N IL + Y+KWF
Sbjct: 176 KQDQLIEKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWF 235
Query: 255 LSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQW 314
SRFK T+FTN+MKWY+TE +GQENYT D+M+ A++NGI VRGH + WD+PK QP W
Sbjct: 236 ASRFKITSFTNEMKWYATEKARGQENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSW 295
Query: 315 VKSL-SPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTA 373
VK++ P ++ RINSV RY G+L WDVVNENLH+++FE LG NAS ++Y+ A
Sbjct: 296 VKNIKDPNDVMNVTLNRINSVMKRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLA 355
Query: 374 HHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQ 433
+DP+ R+F+NE+NTIE + + A+P K +++IL +PG+ + AIG Q HF Q
Sbjct: 356 FKIDPDVRLFVNEYNTIENTKEFTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQ 415
Query: 434 PNLAYMRAGLDLLGATGFPIWLTETSIDPQPHQEDYFDWILREGYAHPAVQGIIMFMGPV 493
PNLAY+R+ LD LG+ G PIWLTE + P+Q Y + ILRE Y+HPAV+GII+F GP
Sbjct: 416 PNLAYIRSALDTLGSLGLPIWLTEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPE 475
Query: 494 QAGFKKAPLADENFKNTPIGDVVDMLIREWGTGPHEAKADSRGIVD-----ISLHHGDYD 548
+GF K LAD++F NT GDV+D L++EW E + + D +SL HG Y+
Sbjct: 476 VSGFDKLTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQTNFTADSDNEEEEVSLLHGHYN 535
Query: 549 VIVTHPQ-KQISKTLNLSVRKGSPQ-QTIQVKMHA 581
V V+HP +S + +L V K Q Q I+V + A
Sbjct: 536 VNVSHPWIANLSTSFSLEVTKEMDQDQVIRVVISA 570
>At4g33810 beta-xylan endohydrolase -like protein
Length = 536
Score = 467 bits (1201), Expect = e-132
Identities = 245/537 (45%), Positives = 341/537 (62%), Gaps = 36/537 (6%)
Query: 60 DHNLEGWTVFGNGAIEERRSNEGNAFIVARNRTQPLDSFSQKIQLKKGMLYTFSAWLQLS 119
D + E W + G+G+I E +Q+IQL +G +Y+FSAW++L
Sbjct: 21 DDSDEEWKIDGSGSIRE---------------------MTQRIQLHEGNIYSFSAWVKLR 59
Query: 120 EGSDT-VSVVFKINGRELIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFE-------SK 171
EG++ V VVF+ + GG V AK CWTL+KGG+V + + +I FE S
Sbjct: 60 EGNNKKVGVVFRTENGRFVHGGEVRAKKRCWTLLKGGIVPDVSGSVDIFFEVQQLAIYSD 119
Query: 172 NPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTHINETALEGATITTKQTK 231
+ +I A VSL+ F+K++W+ Q++ IE++RKSKVRF+VT+ N+TA++GA I+ +QTK
Sbjct: 120 DKEAKISASDVSLKQFSKQEWKLKQDQLIEKIRKSKVRFEVTYQNKTAVKGAVISIEQTK 179
Query: 232 LDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLA 291
F GC +N IL + Y+ WF SRFK T+FTN+MKWY+TE +G ENYT D+M+ A
Sbjct: 180 PSFLLGCAMNFRILQSEGYRNWFASRFKITSFTNEMKWYTTEKERGHENYTAADSMLKFA 239
Query: 292 KNNGISVRGHNIFWDDPKQQPQWVKSLS-PEELRKAAAKRINSVASRYSGELIAWDVVNE 350
+ NGI VRGH + WDDP QP WV + P +L RINSV +RY G+L WDVVNE
Sbjct: 240 EENGILVRGHTVLWDDPLMQPTWVPKIEDPNDLMNVTLNRINSVMTRYKGKLTGWDVVNE 299
Query: 351 NLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQI 410
N+H+++FE LG NAS+++Y+ A LDP+ MF+NE+NTIE +V A+P +K+++I
Sbjct: 300 NVHWDYFEKMLGANASSSFYNLAFKLDPDVTMFVNEYNTIENRVEVTATPVKVKEKMEEI 359
Query: 411 LQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQPHQEDYF 470
L +PG+ I AIG Q HF QPNLAYMR+ LD LG+ G PIWLTE + P+QE Y
Sbjct: 360 LAYPGNMNIKGAIGAQGHFRPTQPNLAYMRSALDTLGSLGLPIWLTEVDMPKCPNQEVYI 419
Query: 471 DWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDMLIREWGTG---P 527
+ ILRE Y+HPAV+GII+F GP +GF K LAD+ F NT GDV+D L++EW P
Sbjct: 420 EEILREAYSHPAVKGIIIFAGPEVSGFDKLTLADKYFNNTATGDVIDKLLKEWQQSSEIP 479
Query: 528 HEAKADSRG-IVDISLHHGDYDVIVTHP-QKQISKTLNLSVRKGSPQ-QTIQVKMHA 581
DS ++SL HG Y+V V+HP K +S + +L V K Q Q ++V ++A
Sbjct: 480 KIFMTDSENDEEEVSLLHGHYNVNVSHPWMKNMSTSFSLEVTKEMGQRQVVRVVINA 536
>At2g14690 1,4-beta-xylan endohydrolase
Length = 552
Score = 454 bits (1167), Expect = e-128
Identities = 244/552 (44%), Positives = 334/552 (60%), Gaps = 50/552 (9%)
Query: 70 GNGAIEERRSNEGNAFIVARNRTQPLDSFSQKIQLKKGMLYTFSAWLQL-SEGSDTVSVV 128
G ++E + NE RN + +D L++G +Y SAW++L +E V +
Sbjct: 11 GGEGVKELKINENGGI---RNVVEGVD-------LREGNIYITSAWVKLRNESQRKVGMT 60
Query: 129 FKINGRELIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFESKNPTV------------- 175
F + GG V+AK GCW+L+KGG+ A+F+ P +I FE K V
Sbjct: 61 FSEKNGRNVFGGEVMAKRGCWSLLKGGITADFSGPIDIFFEVKELIVCSFVEISSTNVGI 120
Query: 176 -----------EIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTHINETALEGAT 224
EI +V +Q F K QWR Q++ IE++RK+KVRFQ++ N++ALEG+
Sbjct: 121 YTKQSDGLAGLEISVQNVRMQRFHKTQWRLQQDQVIEKIRKNKVRFQMSFKNKSALEGSV 180
Query: 225 ITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTP 284
I+ +Q K F GC +N IL + Y++WF+SRF+ T+FTN+MKWY+TE V+GQENY
Sbjct: 181 ISIEQIKPSFLLGCAMNYRILESDSYREWFVSRFRLTSFTNEMKWYATEAVRGQENYKIA 240
Query: 285 DAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLS-PEELRKAAAKRINSVASRYSGELI 343
D+MM LA+ N I V+GH + WDD QP WVK+++ PE+L+ R+NSV RY G LI
Sbjct: 241 DSMMQLAEENAILVKGHTVLWDDKYWQPNWVKTITDPEDLKNVTLNRMNSVMKRYKGRLI 300
Query: 344 AWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNY 403
WDV+NEN+HFN+FE LG NASA YS A LDP+ +FLNEFNT+EY D SP N
Sbjct: 301 GWDVMNENVHFNYFENMLGGNASAIVYSLASKLDPDIPLFLNEFNTVEYDKDRVVSPVNV 360
Query: 404 IKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQ 463
+KK+Q+I+ FPG+ I IG Q HFA QPNLAYMR LD LG+ FP+WLTE +
Sbjct: 361 VKKMQEIVSFPGNNNIKGGIGAQGHFAPVQPNLAYMRYALDTLGSLSFPVWLTEVDMFKC 420
Query: 464 PHQEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDMLIREW 523
P Q Y + ILRE Y+HPAV+ II++ GP +GF K LAD++FKNT GD++D L++EW
Sbjct: 421 PDQVKYMEDILREAYSHPAVKAIILYGGPEVSGFDKLTLADKDFKNTQAGDLIDKLLQEW 480
Query: 524 GTGP-------HEAKADSRGIV-----DISLHHGDYDVIVTHP-QKQISKTLNLSVRKGS 570
P HE + G + +ISL HG Y V VT+P K +S ++ V K S
Sbjct: 481 KQEPVEIPIQHHEHNDEEGGRIIGFSPEISLLHGHYRVTVTNPSMKNLSTRFSVEVTKES 540
Query: 571 PQ-QTIQVKMHA 581
Q +Q+ + A
Sbjct: 541 GHLQEVQLVIDA 552
>At4g33850 putative protein
Length = 371
Score = 365 bits (938), Expect = e-101
Identities = 171/332 (51%), Positives = 224/332 (66%), Gaps = 2/332 (0%)
Query: 35 SATTECLAEPHREHYGGGIIVNPGFDHNLEGWTVFGNGAIEERRSNEGNAFIVARNRTQP 94
S + ECL P++ Y GGIIVNP + +GW+ FGN +E R + N ++VAR R Q
Sbjct: 39 STSLECLENPYKPQYNGGIIVNPELQNGSQGWSKFGNAKVEFREFGD-NHYVVARQRNQS 97
Query: 95 LDSFSQKIQLKKGMLYTFSAWLQLSEGSDTVSVVFKINGRELIRGGHVIAKHGCWTLMKG 154
DS SQ + L+K +LYTFSAWLQ+SEG V +FK NG E G V+A+ CW+++KG
Sbjct: 98 FDSVSQTVYLEKELLYTFSAWLQVSEGKAPVRAIFKKNG-EYKNAGSVVAESKCWSMLKG 156
Query: 155 GLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTH 214
GL + + PAE+ FES + VEIW DSVSLQPFT+++W HQE+SI + RK VR +
Sbjct: 157 GLTVDESGPAELYFESDDTMVEIWVDSVSLQPFTQKEWNFHQEQSIYKARKGAVRIRAVD 216
Query: 215 INETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEI 274
+ ATI+ +Q +L FPFGC + NIL N Y+ WF RF TTF N+MKWYSTE+
Sbjct: 217 SEGQPIPNATISIQQKRLGFPFGCEVEKNILGNQAYENWFTQRFTVTTFANEMKWYSTEV 276
Query: 275 VKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSV 334
V+G+E+Y+T DAM+ K +G++VRGHN+ WDDPK QP WV SLS +L A +R+ SV
Sbjct: 277 VRGKEDYSTADAMLRFFKQHGVAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVFSV 336
Query: 335 ASRYSGELIAWDVVNENLHFNFFEGNLGENAS 366
SRY G+L WDVVNENLHF+FFE +G AS
Sbjct: 337 VSRYKGQLAGWDVVNENLHFSFFENKMGPKAS 368
>At4g38650 putative protein
Length = 433
Score = 310 bits (793), Expect = 2e-84
Identities = 166/425 (39%), Positives = 242/425 (56%), Gaps = 15/425 (3%)
Query: 152 MKGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERV-------- 203
M GL + S +L + +++ S SLQPFT+EQWR++Q+ I V
Sbjct: 1 MVAGLSSKEDSFLILLTSEDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTVIHTLISNA 60
Query: 204 RKSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTF 263
RK V V+ N ++EGA +T +Q DF G I+ IL N YQ+WF+ RF T F
Sbjct: 61 RKRAVTIHVSKENGESVEGAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVF 120
Query: 264 TNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEEL 323
N++KWY+TE +G+ NYT D MM+ + N I RGHNIFW+DPK P WV++L+ E+L
Sbjct: 121 ENELKWYATEPDQGKLNYTLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDL 180
Query: 324 RKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMF 383
R A +RI S+ +RY GE + WDV NE LHF+F+E LG+NAS +++ A +D +F
Sbjct: 181 RSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLF 240
Query: 384 LNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMRAGL 443
N+FN +E D ++ YI +++++ ++ G IGL+ HF + PN+A MRA L
Sbjct: 241 FNDFNVVETCSDEKSTVDEYIARVRELQRYDGVR--MDGIGLEGHFTT--PNVALMRAIL 296
Query: 444 DLLGATGFPIWLTETSID---PQPHQEDYFDWILREGYAHPAVQGIIMFMGPVQAGFKKA 500
D L PIWLTE I Q Y + +LREG++HP+V GI+++ G +
Sbjct: 297 DKLATLQLPIWLTEIDISSSLDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQM 356
Query: 501 PLADENFKNTPIGDVVDMLIREWGTGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQISK 560
L D+ F+N P GDVVD + EW TG +A D G G+Y V + + K ++
Sbjct: 357 CLTDDKFRNLPAGDVVDQKLLEWKTGEVKATTDDHGSFSFFGFLGEYRVGIMYQGKTVNS 416
Query: 561 TLNLS 565
+ +LS
Sbjct: 417 SFSLS 421
>At1g10050 putative xylan endohydrolase
Length = 1063
Score = 199 bits (506), Expect = 4e-51
Identities = 149/554 (26%), Positives = 260/554 (46%), Gaps = 45/554 (8%)
Query: 49 YGGGIIVNPGF-DHNLEGWTVFGNG---------------AIEERRSNEG---NAFIVAR 89
+G I+ N D +EGW G+ A + R +G +++A
Sbjct: 514 FGMNIVSNSHLSDGTIEGWFPLGDCHLKVGDGSPRILPPLARDSLRKTQGYLSGRYVLAT 573
Query: 90 NRTQ----PLDSFSQKIQLKKGMLYTFSAWLQLSEGSDT----VSVVFKINGRELIRGGH 141
NR+ P + + K++L + Y SAW+++ G T V++ ++G + GG
Sbjct: 574 NRSGTWMGPAQTITDKVKLF--VTYQVSAWVKIGSGGRTSPQDVNIALSVDGN-WVNGGK 630
Query: 142 VIAKHGCWTLMKGGL-VANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESI 200
V G W + G + + + +P V++ + + ++ S+
Sbjct: 631 VEVDDGDWHEVVGSFRIEKEAKEVMLHVQGPSPGVDLMVAGLQIFAVDRKARLSYLRGQA 690
Query: 201 ERVRKSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKY 260
+ VRK V + + ++ + L GAT+ +QT+ FP G I+ + + N ++ +FL+ F +
Sbjct: 691 DVVRKRNVCLKFSGLDPSELSGATVKIRQTRNSFPLGSCISRSNIDNEDFVDFFLNNFDW 750
Query: 261 TTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSP 320
F ++KWY TE +G NY + M+ + I RGH IFW+ WV+ L+
Sbjct: 751 AVFGYELKWYWTEPEQGNFNYRDANEMIEFCERYNIKTRGHCIFWEVESAIQPWVQQLTG 810
Query: 321 EELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDPNT 380
+L A R+ + +RY+G+ +DV NE LH +F+ L +A A + TAH LDP
Sbjct: 811 SKLEAAVENRVTDLLTRYNGKFRHYDVNNEMLHGSFYRDRLDSDARANMFKTAHELDPLA 870
Query: 381 RMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAYMR 440
+FLNE++ IE D +SP YIK + ++ + G IG+Q H S P +R
Sbjct: 871 TLFLNEYH-IEDGFDSRSSPEKYIKLVHKLQKKGAPVG---GIGIQGHITS--PVGHIVR 924
Query: 441 AGLDLLGATGFPIWLTETSIDP--QPHQEDYFDWILREGYAHPAVQGIIMF-MGPVQAGF 497
+ LD L G PIW TE + + + D + +L E +AHPAV+G++++ +
Sbjct: 925 SALDKLSTLGLPIWFTELDVSSTNEHIRGDDLEVMLWEAFAHPAVEGVMLWGFWELFMSR 984
Query: 498 KKAPLADENFKNTPIGDVVDMLIREWGTGPHEAKADSRGIVDISLHHGDYDV-IVTHPQK 556
+ + L + + + G + REW + + + G ++ +HG Y V +VT K
Sbjct: 985 EHSHLVNADGEVNEAGKRFLEIKREW-LSFVDGEIEDGGGLEFRGYHGSYTVEVVTSESK 1043
Query: 557 QISKTLNLSVRKGS 570
++ N V KG+
Sbjct: 1044 YVT---NFVVDKGN 1054
>At1g58370 unknown protein
Length = 917
Score = 197 bits (502), Expect = 1e-50
Identities = 132/509 (25%), Positives = 245/509 (47%), Gaps = 28/509 (5%)
Query: 85 FIVARNRTQPLDSFSQKI--QLKKGMLYTFSAWLQLSEGSDT---VSVVFKINGRELIRG 139
+I+ NRTQ +Q I +LK + Y S W+++ G ++ V+V I+ + + G
Sbjct: 420 YILVTNRTQTWMGPAQMITDKLKLFLTYQISVWVKVGSGINSPQNVNVALGIDS-QWVNG 478
Query: 140 GHVIAKHGCWTLMKGGL-VANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEE 198
G V W + G + S A + + + +++ + + P + H +
Sbjct: 479 GQVEINDDRWHEIGGSFRIEKNPSKALVYVQGPSSGIDLMVAGLQIFPVDRLARIKHLKR 538
Query: 199 SIERVRKSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRF 258
+++RK V + ++ + GA++ +Q + FP G I+ + + N ++ +FL F
Sbjct: 539 QCDKIRKRDVILKFAGVDSSKFSGASVRVRQIRNSFPVGTCISRSNIDNEDFVDFFLKNF 598
Query: 259 KYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSL 318
+ F N++KWY TE +G+ NY D M++L +N I RGH IFW+ QW++++
Sbjct: 599 NWAVFANELKWYWTEPEQGKLNYQDADDMLNLCSSNNIETRGHCIFWEVQATVQQWIQNM 658
Query: 319 SPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENASAAYYSTAHHLDP 378
+ +L A R+ + +RY G+ +DV NE LH +F++ LG++ + TAH LDP
Sbjct: 659 NQTDLNNAVQNRLTDLLNRYKGKFKHYDVNNEMLHGSFYQDKLGKDIRVNMFKTAHQLDP 718
Query: 379 NTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFASGQPNLAY 438
+ +F+N+++ IE D + P Y ++I + + G IG+Q H S P
Sbjct: 719 SATLFVNDYH-IEDGCDPKSCPEKYTEQILDLQEKGAPVG---GIGIQGHIDS--PVGPI 772
Query: 439 MRAGLDLLGATGFPIWLTETSIDP--QPHQEDYFDWILREGYAHPAVQGIIMFMGPVQAG 496
+ + LD LG G PIW TE + + + D + ++ E + HPAV+GI+++ G
Sbjct: 773 VCSALDKLGILGLPIWFTELDVSSVNEHIRADDLEVMMWEAFGHPAVEGIMLW------G 826
Query: 497 FKKAPLA-DENFKNTPIGDVVD-----MLIREWGTGPHEAKADSRGIVDISLHHGDYDV- 549
F + ++ D + GDV + + +++ D G + G+Y V
Sbjct: 827 FWELFMSRDNSHLVNAEGDVNEAGKRFLAVKKDWLSHANGHIDQNGAFPFRGYSGNYAVE 886
Query: 550 IVTHPQKQISKTLNLSVRKGSPQQTIQVK 578
++T ++ KT + S T+ ++
Sbjct: 887 VITTSSSKVLKTFGVDKEDSSQVITVDLQ 915
Score = 29.3 bits (64), Expect = 6.3
Identities = 40/185 (21%), Positives = 76/185 (40%), Gaps = 27/185 (14%)
Query: 18 ILISGLTVHSLSHSYDYSATTECLAEPHREHYGGGIIVNPGFDHNLEGWT-------VFG 70
+LI +T+H S T E I+VNP F+ L W+ +
Sbjct: 174 LLIQSVTIHRESEPELERVTAE----------DETIVVNPNFEDGLNNWSGRSCKIVLHD 223
Query: 71 NGAIEERRSNEGNAFIVARNRTQPLDSFSQKI--QLKKGMLYTFSAWLQLSEGSDTVSVV 128
+ A + G F A RTQ + Q+I ++++ +Y +A +++ + T + V
Sbjct: 224 SMADGKIVPESGKVFASATERTQNWNGIQQEITGKVQRKRVYEATAVVRIYGNNVTTATV 283
Query: 129 FKI------NGRELIRG-GHVIAKHGCWTLMKGGLVANFTSPAEILF-ESKNPTVEIWAD 180
N R+ G V A W +KG + N ++ +++ E P +I +
Sbjct: 284 QATLWVQNPNQRDQYIGISTVQATDKEWIHLKGKFLLNGSASRVVIYIEGPPPGTDILLN 343
Query: 181 SVSLQ 185
S++++
Sbjct: 344 SLTVK 348
>At4g08160 xylan endohydrolase like proteinchimeric
Length = 520
Score = 192 bits (489), Expect = 3e-49
Identities = 142/521 (27%), Positives = 244/521 (46%), Gaps = 39/521 (7%)
Query: 82 GNAFIVARNRTQPLDSFSQKI--QLKKGMLYTFSAWLQLSEG-------SDTVSVVFKIN 132
G +IV NRTQ +Q I ++K + Y SAW++L G V++ ++
Sbjct: 13 GGNYIVVTNRTQTWMGPAQMITDKIKLFLTYQISAWVKLGVGVSGSSMSPQNVNIALSVD 72
Query: 133 GRELIRGGHVIAKHG-CWTLMKGGLVANFTSPAEILFESKNPT--VEIWADSVSLQPFTK 189
+ + GG V G W + G P ++ + P +++ ++ + P +
Sbjct: 73 N-QWVNGGQVEVTVGDTWHEIAGSFRLE-KQPQNVMVYVQGPGAGIDLMIAALQIFPVDR 130
Query: 190 EQWRSHQEESIERVRKSKVRFQVTHINETA---LEGATITTKQTKLDFPFGCGINNNILT 246
+ + ++ VRK + + + +N+ L + KQT FP G IN +
Sbjct: 131 RERVRCLKRQVDEVRKRDIVLKFSGLNDDESFDLFPYIVKVKQTYNSFPVGTCINRTDID 190
Query: 247 NSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWD 306
N ++ +F F + F N++KWY+TE +G+ NY D M+ L N I+VRGH IFW+
Sbjct: 191 NEDFVDFFTKNFNWAVFGNELKWYATEAERGKVNYQDADDMLDLCIGNNINVRGHCIFWE 250
Query: 307 DPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENAS 366
WV+ L+ +L A KR+ + +RY G+ +DV NE LH +F++ LG+
Sbjct: 251 VESTVQPWVRQLNKTDLMNAVQKRLTDLLTRYKGKFKHYDVNNEMLHGSFYQDRLGKGVR 310
Query: 367 AAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQ 426
A ++ AH LDP+ +F+N+++ +E D +SP YIK + + + G IG+Q
Sbjct: 311 ALMFNIAHKLDPSPLLFVNDYH-VEDGDDPRSSPEKYIKLVLDLEAQGATVG---GIGIQ 366
Query: 427 CHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQPH--QEDYFDWILREGYAHPAVQ 484
H S P A + + LD+L G PIW TE + + + + +L E +AHP+V+
Sbjct: 367 GHIDS--PVGAIVCSALDMLSVLGRPIWFTELDVSSSNEYVRGEDLEVMLWEAFAHPSVE 424
Query: 485 GIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDMLIREWGTGPHEAKADSRGIVD----- 539
GI+++ GF + ++ EN V+ + + E + + GI++
Sbjct: 425 GIMLW------GFWELSMSRENANLVEGEGEVNEAGKRFLEVKQEWLSHAYGIINDESEF 478
Query: 540 -ISLHHGDYDVIVTHPQKQISKTLNLSVRKGSPQQTIQVKM 579
+HG Y V + P + KT V KG I + +
Sbjct: 479 TFRGYHGTYAVEICTPAGIVLKT--FVVEKGDTPLVISIDL 517
>At4g38300 unknown protein
Length = 181
Score = 175 bits (444), Expect = 5e-44
Identities = 80/163 (49%), Positives = 108/163 (66%)
Query: 202 RVRKSKVRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYT 261
+ RK V V+ N ++EGA +T +Q DFP G I+ IL N YQ+WF+ RF T
Sbjct: 10 QARKRAVTIHVSKENGESVEGAEVTVEQISKDFPIGSAISKTILGNIPYQEWFVKRFDAT 69
Query: 262 TFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPE 321
F N++KWY+TE +G+ NYT D MM+L + N I RGHNIFW+DPK P WV++L+ E
Sbjct: 70 VFENELKWYATESDQGKLNYTLADKMMNLVRANRIIARGHNIFWEDPKYNPDWVRNLTGE 129
Query: 322 ELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGEN 364
+LR A +RI S+ +RY GE + WDV NE LHF+F+E LG+N
Sbjct: 130 DLRSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYESRLGKN 172
>At1g64050 unknown protein
Length = 668
Score = 30.4 bits (67), Expect = 2.8
Identities = 22/101 (21%), Positives = 43/101 (41%)
Query: 306 DDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEGNLGENA 365
D+P +W K LS + ++ +K I R+ + + D+ + N + N A
Sbjct: 530 DNPNGILRWPKKLSQKSMKARKSKLIEKPLERHRTTVSSIDLNSSNNNNNKNHVRKDSAA 589
Query: 366 SAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKK 406
++ HH P+ R+ L+ + S ++SP +K
Sbjct: 590 EHNHHHHHHHPKPSKRLKLSTMENKKRSFPSSSSPIESDRK 630
>At1g18190 hypothetical protein
Length = 649
Score = 30.4 bits (67), Expect = 2.8
Identities = 29/86 (33%), Positives = 44/86 (50%), Gaps = 8/86 (9%)
Query: 95 LDSFSQKIQLKKGMLYTFSAWLQLSEGSDTVS-VVFKINGRELIRGGHVIAKHGCWTLMK 153
+++ S+ I+ KGM T ++ L G +S FK ++ IR G KH W +M+
Sbjct: 527 IEAVSRLIEENKGMSATEASSQDLEAGDWELSGSKFKPAFQDKIRSGK---KHLGWLVMQ 583
Query: 154 GGLVANFTSPAEILFESKNPTVEIWA 179
L A F S +F +NPT +IWA
Sbjct: 584 --LNAIFISGT--VFLRRNPTAKIWA 605
>At1g20270 unknown protein
Length = 287
Score = 29.6 bits (65), Expect = 4.8
Identities = 26/123 (21%), Positives = 52/123 (42%), Gaps = 16/123 (13%)
Query: 287 MMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWD 346
++SLAK + V+ + + K + V++ S LR+ K I ++ R +
Sbjct: 100 LISLAKPH--MVKSTVVDSETGKSKDSRVRTSSGTFLRRGRDKIIKTIEKRIAD------ 151
Query: 347 VVNENLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKK 406
+ F + GE +Y +P+ F++EFNT + G A+ Y+
Sbjct: 152 -------YTFIPADHGEGLQVLHYEAGQKYEPHYDYFVDEFNT-KNGGQRMATMLMYLSD 203
Query: 407 IQQ 409
+++
Sbjct: 204 VEE 206
>At1g26330 hypothetical protein
Length = 1196
Score = 29.3 bits (64), Expect = 6.3
Identities = 13/43 (30%), Positives = 23/43 (53%)
Query: 136 LIRGGHVIAKHGCWTLMKGGLVANFTSPAEILFESKNPTVEIW 178
L+ G + I + W + K LV+ F SP+ ++FE ++ W
Sbjct: 998 LVIGHNGIGEFTIWDISKRSLVSRFVSPSNLIFEFIPTSLFAW 1040
>At1g16370 putative transport protein
Length = 521
Score = 29.3 bits (64), Expect = 6.3
Identities = 15/57 (26%), Positives = 25/57 (43%), Gaps = 6/57 (10%)
Query: 472 WILREGYAHPAVQGIIMFMGPVQAGF-----KKAPLADENFKNTPIGDVVDMLIREW 523
W+ +G A+ ++ M P + + K PL ENF+ P + D R+W
Sbjct: 267 WLHMQGKDKEAID-VLTKMSPKEKAYLESVVSKLPLKQENFEQAPTYSIKDFFFRKW 322
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,622,729
Number of Sequences: 26719
Number of extensions: 609937
Number of successful extensions: 1380
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1326
Number of HSP's gapped (non-prelim): 19
length of query: 581
length of database: 11,318,596
effective HSP length: 105
effective length of query: 476
effective length of database: 8,513,101
effective search space: 4052236076
effective search space used: 4052236076
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0218.17


