

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.16
(560 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
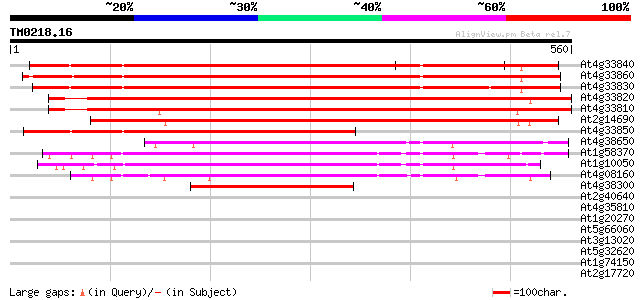
Sequences producing significant alignments: (bits) Value
At4g33840 putative protein 550 e-156
At4g33860 unknown protein 525 e-149
At4g33830 putative protein 523 e-148
At4g33820 putative glycosyl hydrolase family 10 protein 503 e-142
At4g33810 beta-xylan endohydrolase -like protein 486 e-137
At2g14690 1,4-beta-xylan endohydrolase 464 e-131
At4g33850 putative protein 344 7e-95
At4g38650 putative protein 306 3e-83
At1g58370 unknown protein 204 1e-52
At1g10050 putative xylan endohydrolase 201 7e-52
At4g08160 xylan endohydrolase like proteinchimeric 198 8e-51
At4g38300 unknown protein 165 5e-41
At2g40640 unknown protein 33 0.55
At4g35810 putative protein 32 0.72
At1g20270 unknown protein 31 1.6
At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein 30 2.7
At3g13020 hypothetical protein 30 2.7
At5g32620 putative protein 30 4.6
At1g74150 hypothetical protein 30 4.6
At2g17720 similar to prolyl 4-hydroxylase alpha subunit 29 6.1
>At4g33840 putative protein
Length = 669
Score = 550 bits (1416), Expect = e-156
Identities = 268/532 (50%), Positives = 358/532 (66%), Gaps = 7/532 (1%)
Query: 20 ISEPQREQYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKPLDSFSQ 79
+ P + QY GGIIVNP + +GW+ GN ++ R GN F+VA R + DS SQ
Sbjct: 127 LENPYKPQYNGGIIVNPDLQNGSQGWSQFGNAKVDFREFG-GNKFVVATQRNQSSDSISQ 185
Query: 80 KVQLKKGMIYTFSAWLQLSKGSETVSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANF 139
KV L+KG++YTFSAWLQ+S G VS VFK +G E G V+A+ CW++LKGGL +
Sbjct: 186 KVYLEKGILYTFSAWLQVSIGKSPVSAVFKKNG-EYKHAGSVVAESKCWSMLKGGLTVDE 244
Query: 140 TSPADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVTHINETAL 199
+ PA++ FES+N VEIW DSVSLQPFT+EEW SH E SI +VRK VR +V + +
Sbjct: 245 SGPAELFFESENTMVEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRVMNNKGETI 304
Query: 200 EGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQEN 259
ATISI+Q + +PFGC + NNIL N AY+ WF RF TTF N+MKWYSTE+++GQE+
Sbjct: 305 PNATISIEQKKLGYPFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYSTERIRGQED 364
Query: 260 YTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQDLREAARKRINSVVSRYKG 319
Y+ DAML+ +GI+VRGH +LWDD +QP WV SLS DL A ++R+ SVVSRYKG
Sbjct: 365 YSTADAMLSFFKSHGIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVYSVVSRYKG 424
Query: 320 DLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANP 379
L+ WDVVNENLH++F+E FG AS Y+ A+ +DP MF+NE+NT+E D+ ++P
Sbjct: 425 QLLGWDVVNENLHFSFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQPKDLTSSP 484
Query: 380 ANYIRKIREIQQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPIWLTETSI 439
A Y+ K+RE+Q IPL IGL+ HF++ PN+ YMRS LD GATG PIWLTE +
Sbjct: 485 ARYLGKLRELQSIRVAGKIPLAIGLESHFST--PNIPYMRSALDTFGATGLPIWLTEIDV 542
Query: 440 DPQPN-QEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPMGDVVDKL 498
D PN + YFE +LREG++HP V G++M+ G +G + L D NF N P GDVVDKL
Sbjct: 543 DAPPNVRANYFEQVLREGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLPTGDVVDKL 602
Query: 499 IREWGTGPHET--MADSRGIVDISLHHGDYDVIVSHPQTHFSKTLNLSVRKE 548
+REWG +T + D+ G+ + L HGDYD+ +SHP T+ + N ++ +
Sbjct: 603 LREWGGLRSQTTGVTDANGLFEAPLFHGDYDLRISHPLTNSKASYNFTLTSD 654
Score = 96.7 bits (239), Expect = 3e-20
Identities = 50/110 (45%), Positives = 72/110 (65%), Gaps = 3/110 (2%)
Query: 386 IREIQQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPIWLTETSIDPQPN- 444
++E+Q + I L IGL+ HF + PN+ YMRS LDIL ATG IWLTE ++ P+
Sbjct: 2 LKELQSIRISGYIRLAIGLESHFKT--PNIPYMRSALDILAATGLLIWLTEIDVEAPPSV 59
Query: 445 QEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPMGDV 494
Q +YFE +LR+G++HP V+G++++ G +G + L D NF N P GDV
Sbjct: 60 QAKYFEQVLRDGHAHPQVKGMVVWGGYSPSGCYRMCLTDGNFRNLPTGDV 109
>At4g33860 unknown protein
Length = 576
Score = 525 bits (1351), Expect = e-149
Identities = 265/541 (48%), Positives = 354/541 (64%), Gaps = 10/541 (1%)
Query: 13 TTSCLLLISEPQREQYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTK 72
T CL + P + QY GGIIV+P GWT GN ++ R+ N F VAR+R +
Sbjct: 30 TIECLEI---PLKPQYNGGIIVSPDVRDGTLGWTPFGNAKVDFRKIGNHN-FFVARDRKQ 85
Query: 73 PLDSFSQKVQLKKGMIYTFSAWLQLSKGSETVSVVFKIHGSELIRGGHVIAKHGCWTLLK 132
P DS SQKV L+KG++YTFSAWLQ+SKG V VFK +G E G V+A+ CW++LK
Sbjct: 86 PFDSVSQKVYLEKGLLYTFSAWLQVSKGKAPVKAVFKKNG-EYKLAGSVVAESKCWSMLK 144
Query: 133 GGLVANFTSPADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVT 192
GGL + + PA++ FES++ TVEIW DSVSLQPFT+EEW SH E SI++ RKR VR +
Sbjct: 145 GGLTVDESGPAELYFESEDTTVEIWVDSVSLQPFTQEEWNSHHEQSIQKERKRTVRIRAV 204
Query: 193 HINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTE 252
+ + ATISI+Q + FPFGC + NIL N AY+ WF RF TTF N+MKWYSTE
Sbjct: 205 NSKGEPIPKATISIEQRKLGFPFGCEVEKNILGNKAYQNWFTQRFTVTTFANEMKWYSTE 264
Query: 253 KVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQDLREAARKRINS 312
V+G+E+Y+ DAML +G++VRGH ILW+D +QP+WV +LS DL A ++R+ S
Sbjct: 265 VVRGKEDYSTADAMLRFFKQHGVAVRGHNILWNDPKYQPKWVNALSGNDLYNAVKRRVFS 324
Query: 313 VVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYS 372
VVSRYKG L WDVVNENLH++++E G AS + A DP MF+NE+NT+E S
Sbjct: 325 VVSRYKGQLAGWDVVNENLHFSYFEDKMGPKASYNIFKMAQAFDPTTTMFMNEYNTLEES 384
Query: 373 GDVVANPANYIRKIREIQQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPI 432
D ++ A Y++K+REI+ I LGIGL+ HF + PN+ YMRS LD L ATG PI
Sbjct: 385 SDSDSSLARYLQKLREIRSIRVCGNISLGIGLESHFKT--PNIPYMRSALDTLAATGLPI 442
Query: 433 WLTETSIDPQPN-QEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPM 491
WLTE ++ PN Q +YFE +LREG++HP V+GI+ + G +G + L D NF N P
Sbjct: 443 WLTEVDVEAPPNVQAKYFEQVLREGHAHPQVKGIVTWSGYSPSGCYRMCLTDGNFKNVPT 502
Query: 492 GDVVDKLIREWGTGPHET--MADSRGIVDISLHHGDYDVIVSHPQTHFSKTLNLSVRKEF 549
GDVVDKL+ EWG +T + D+ G + SL HGDYD+ ++HP T+ + + + +
Sbjct: 503 GDVVDKLLHEWGGFRRQTTGVTDADGYFEASLFHGDYDLKIAHPLTNSKASHSFKLTSDV 562
Query: 550 S 550
S
Sbjct: 563 S 563
>At4g33830 putative protein
Length = 544
Score = 523 bits (1347), Expect = e-148
Identities = 265/531 (49%), Positives = 352/531 (65%), Gaps = 8/531 (1%)
Query: 23 PQREQYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKPLDSFSQKVQ 82
P + QY GGIIVNP + +GW+ N + R GN F+VA R + DS SQKV
Sbjct: 6 PYKPQYNGGIIVNPDMQNGSQGWSQFENAKVNFREFG-GNKFVVATQRNQSSDSVSQKVY 64
Query: 83 LKKGMIYTFSAWLQLSKGSETVSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANFTSP 142
L+KG++YTFSAWLQ+S G VS VFK +G E G V+A+ CW++LKGGL + + P
Sbjct: 65 LEKGILYTFSAWLQVSTGKAPVSAVFKKNG-EYKHAGSVVAESKCWSMLKGGLTVDESGP 123
Query: 143 ADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVTHINETALEGA 202
A++ ES++ TVEIW DSVSLQPFT++EW +HQE SI+ RK VR +V + + A
Sbjct: 124 AELFVESEDTTVEIWVDSVSLQPFTQDEWNAHQEQSIDNSRKGPVRIRVVNNKGEKIPNA 183
Query: 203 TISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQENYTI 262
+I+I+Q R FPFG + NIL N AY+ WF RF TTF N+MKWYSTE V+G ENYT+
Sbjct: 184 SITIEQKRLGFPFGSAVAQNILGNQAYQNWFTQRFTVTTFENEMKWYSTESVRGIENYTV 243
Query: 263 PDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQDLREAARKRINSVVSRYKGDLI 322
DAML N +GI+VRGH ++WD +Q +WV SLS DL A ++R+ SVVSRYKG L
Sbjct: 244 ADAMLRFFNQHGIAVRGHNVVWDHPKYQSKWVTSLSRNDLYNAVKRRVFSVVSRYKGQLA 303
Query: 323 AWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANY 382
WDVVNENLH++F+E FG NAS ++ A+ +DP+ MF+NEF T+E D+ A+PA Y
Sbjct: 304 GWDVVNENLHHSFFESKFGPNASNNIFAMAHAIDPSTTMFMNEFYTLEDPTDLKASPAKY 363
Query: 383 IRKIREIQQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPIWLTETSID-P 441
+ K+RE+Q IPLGIGL+ HF++ PN+ YMRS LD LGATG PIWLTE + P
Sbjct: 364 LEKLRELQSIRVRGNIPLGIGLESHFST--PNIPYMRSALDTLGATGLPIWLTEIDVKAP 421
Query: 442 QPNQEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPMGDVVDKLIRE 501
+Q +YFE +LREG++HP V+G++ + + L D NF N P GDVVDKLIRE
Sbjct: 422 SSDQAKYFEQVLREGHAHPHVKGMVTWTAYAPNCYHMC-LTDGNFKNLPTGDVVDKLIRE 480
Query: 502 WGTGPHET--MADSRGIVDISLHHGDYDVIVSHPQTHFSKTLNLSVRKEFS 550
WG +T + D+ G + SL HGDYD+ +SHP T+ S + N ++ + S
Sbjct: 481 WGGLRSQTTEVTDADGFFEASLFHGDYDLNISHPLTNSSVSHNFTLTSDDS 531
>At4g33820 putative glycosyl hydrolase family 10 protein
Length = 570
Score = 503 bits (1294), Expect = e-142
Identities = 258/531 (48%), Positives = 342/531 (63%), Gaps = 30/531 (5%)
Query: 39 DHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKPLDSFSQKVQLKKGMIYTFSAWLQLS 98
D S E W + GNG I + +Q++QL +G IY+FSAW++L
Sbjct: 61 DDSDEEWKIDGNGFIRE---------------------MAQRIQLHQGNIYSFSAWVKLR 99
Query: 99 KGSET-VSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANFTSPADILFESKNPTVEIW 157
+G++ V VVF+ L+ GG V A CWTLLKGG+V +F+ P DI FES+N +I
Sbjct: 100 EGNDKKVGVVFRTENGRLVHGGEVRANQECWTLLKGGIVPDFSGPVDIFFESENRGAKIS 159
Query: 158 ADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVTHINETALEGATISIKQTRPHFPFGC 217
A +V L+ F+KEEW+ Q+ IE++RK KVRF+VT+ N+TA++G IS+KQT+ F GC
Sbjct: 160 AHNVLLKQFSKEEWKLKQDQLIEKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSFLLGC 219
Query: 218 GMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISV 277
GMN IL + YRKWF SRFK T+FTN+MKWY+TEK +GQENYT+ D+ML A DNGI V
Sbjct: 220 GMNFRILQSQGYRKWFASRFKITSFTNEMKWYATEKARGQENYTVADSMLKFAEDNGILV 279
Query: 278 RGHTILWDDRNFQPEWVKSL-SPQDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFY 336
RGHT+LWD+ QP WVK++ P D+ RINSV+ RYKG L WDVVNENLH++++
Sbjct: 280 RGHTVLWDNPKMQPSWVKNIKDPNDVMNVTLNRINSVMKRYKGKLTGWDVVNENLHWDYF 339
Query: 337 EQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIREIQQFPGTK 396
E+ G NAS +Y+ A+ +DP+ R+F+NE+NTIE + + A P + + EI +PG K
Sbjct: 340 EKMLGANASTSFYNLAFKIDPDVRLFVNEYNTIENTKEFTATPIKVKKMMEEILAYPGNK 399
Query: 397 GIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPIWLTETSIDPQPNQEEYFEWILREG 456
+ IG Q HF PNLAY+RS LD LG+ G PIWLTE + PNQ +Y E ILRE
Sbjct: 400 NMKGAIGAQGHFGPTQPNLAYIRSALDTLGSLGLPIWLTEVDMPKCPNQAQYVEDILREA 459
Query: 457 YSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPMGDVVDKLIREWGTGPHETMADSRGI 516
YSHPAV+GII+F GP +GF K LAD++F NT GDV+DKL++EW E +
Sbjct: 460 YSHPAVKGIIIFGGPEVSGFDKLTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQTNFTAD 519
Query: 517 VD-----ISLHHGDYDVIVSHPQ-THFSKTLNLSVRKEFSQ-QTIHVKMHA 560
D +SL HG Y+V VSHP + S + +L V KE Q Q I V + A
Sbjct: 520 SDNEEEEVSLLHGHYNVNVSHPWIANLSTSFSLEVTKEMDQDQVIRVVISA 570
>At4g33810 beta-xylan endohydrolase -like protein
Length = 536
Score = 486 bits (1251), Expect = e-137
Identities = 258/537 (48%), Positives = 338/537 (62%), Gaps = 36/537 (6%)
Query: 39 DHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKPLDSFSQKVQLKKGMIYTFSAWLQLS 98
D S E W + G+G+I + +Q++QL +G IY+FSAW++L
Sbjct: 21 DDSDEEWKIDGSGSIRE---------------------MTQRIQLHEGNIYSFSAWVKLR 59
Query: 99 KGS-ETVSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANFTSPADILFE-------SK 150
+G+ + V VVF+ + GG V AK CWTLLKGG+V + + DI FE S
Sbjct: 60 EGNNKKVGVVFRTENGRFVHGGEVRAKKRCWTLLKGGIVPDVSGSVDIFFEVQQLAIYSD 119
Query: 151 NPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVTHINETALEGATISIKQTR 210
+ +I A VSL+ F+K+EW+ Q+ IE++RK KVRF+VT+ N+TA++GA ISI+QT+
Sbjct: 120 DKEAKISASDVSLKQFSKQEWKLKQDQLIEKIRKSKVRFEVTYQNKTAVKGAVISIEQTK 179
Query: 211 PHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQENYTIPDAMLALA 270
P F GC MN IL + YR WF SRFK T+FTN+MKWY+TEK +G ENYT D+ML A
Sbjct: 180 PSFLLGCAMNFRILQSEGYRNWFASRFKITSFTNEMKWYTTEKERGHENYTAADSMLKFA 239
Query: 271 NDNGISVRGHTILWDDRNFQPEWVKSLS-PQDLREAARKRINSVVSRYKGDLIAWDVVNE 329
+NGI VRGHT+LWDD QP WV + P DL RINSV++RYKG L WDVVNE
Sbjct: 240 EENGILVRGHTVLWDDPLMQPTWVPKIEDPNDLMNVTLNRINSVMTRYKGKLTGWDVVNE 299
Query: 330 NLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIREI 389
N+H++++E+ G NAS+ +Y+ A+ LDP+ MF+NE+NTIE +V A P K+ EI
Sbjct: 300 NVHWDYFEKMLGANASSSFYNLAFKLDPDVTMFVNEYNTIENRVEVTATPVKVKEKMEEI 359
Query: 390 QQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGLDILGATGYPIWLTETSIDPQPNQEEYF 449
+PG I IG Q HF PNLAYMRS LD LG+ G PIWLTE + PNQE Y
Sbjct: 360 LAYPGNMNIKGAIGAQGHFRPTQPNLAYMRSALDTLGSLGLPIWLTEVDMPKCPNQEVYI 419
Query: 450 EWILREGYSHPAVQGIIMFMGPVQAGFQKAPLADENFTNTPMGDVVDKLIREWGTG---P 506
E ILRE YSHPAV+GII+F GP +GF K LAD+ F NT GDV+DKL++EW P
Sbjct: 420 EEILREAYSHPAVKGIIIFAGPEVSGFDKLTLADKYFNNTATGDVIDKLLKEWQQSSEIP 479
Query: 507 HETMADSRG-IVDISLHHGDYDVIVSHP-QTHFSKTLNLSVRKEFSQ-QTIHVKMHA 560
M DS ++SL HG Y+V VSHP + S + +L V KE Q Q + V ++A
Sbjct: 480 KIFMTDSENDEEEVSLLHGHYNVNVSHPWMKNMSTSFSLEVTKEMGQRQVVRVVINA 536
>At2g14690 1,4-beta-xylan endohydrolase
Length = 552
Score = 464 bits (1194), Expect = e-131
Identities = 239/507 (47%), Positives = 325/507 (63%), Gaps = 39/507 (7%)
Query: 81 VQLKKGMIYTFSAWLQLSKGSET-VSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANF 139
V L++G IY SAW++L S+ V + F + GG V+AK GCW+LLKGG+ A+F
Sbjct: 33 VDLREGNIYITSAWVKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGGITADF 92
Query: 140 TSPADILFESKNPTV------------------------EIWADSVSLQPFTKEEWRSHQ 175
+ P DI FE K V EI +V +Q F K +WR Q
Sbjct: 93 SGPIDIFFEVKELIVCSFVEISSTNVGIYTKQSDGLAGLEISVQNVRMQRFHKTQWRLQQ 152
Query: 176 EDSIERVRKRKVRFQVTHINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMS 235
+ IE++RK KVRFQ++ N++ALEG+ ISI+Q +P F GC MN IL + +YR+WF+S
Sbjct: 153 DQVIEKIRKNKVRFQMSFKNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYREWFVS 212
Query: 236 RFKHTTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVK 295
RF+ T+FTN+MKWY+TE V+GQENY I D+M+ LA +N I V+GHT+LWDD+ +QP WVK
Sbjct: 213 RFRLTSFTNEMKWYATEAVRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQPNWVK 272
Query: 296 SLS-PQDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYH 354
+++ P+DL+ R+NSV+ RYKG LI WDV+NEN+H+N++E G NASA+ YS A
Sbjct: 273 TITDPEDLKNVTLNRMNSVMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVYSLASK 332
Query: 355 LDPNARMFLNEFNTIEYSGDVVANPANYIRKIREIQQFPGTKGIPLGIGLQCHFASGNPN 414
LDP+ +FLNEFNT+EY D V +P N ++K++EI FPG I GIG Q HFA PN
Sbjct: 333 LDPDIPLFLNEFNTVEYDKDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFAPVQPN 392
Query: 415 LAYMRSGLDILGATGYPIWLTETSIDPQPNQEEYFEWILREGYSHPAVQGIIMFMGPVQA 474
LAYMR LD LG+ +P+WLTE + P+Q +Y E ILRE YSHPAV+ II++ GP +
Sbjct: 393 LAYMRYALDTLGSLSFPVWLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYGGPEVS 452
Query: 475 GFQKAPLADENFTNTPMGDVVDKLIREWGTGP-------HETMADSRGIV-----DISLH 522
GF K LAD++F NT GD++DKL++EW P HE + G + +ISL
Sbjct: 453 GFDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSPEISLL 512
Query: 523 HGDYDVIVSHP-QTHFSKTLNLSVRKE 548
HG Y V V++P + S ++ V KE
Sbjct: 513 HGHYRVTVTNPSMKNLSTRFSVEVTKE 539
>At4g33850 putative protein
Length = 371
Score = 344 bits (883), Expect = 7e-95
Identities = 167/332 (50%), Positives = 218/332 (65%), Gaps = 2/332 (0%)
Query: 14 TSCLLLISEPQREQYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVARNRTKP 73
++ L + P + QY GGIIVNP + +GW+ GN +E R + N ++VAR R +
Sbjct: 39 STSLECLENPYKPQYNGGIIVNPELQNGSQGWSKFGNAKVEFREFGD-NHYVVARQRNQS 97
Query: 74 LDSFSQKVQLKKGMIYTFSAWLQLSKGSETVSVVFKIHGSELIRGGHVIAKHGCWTLLKG 133
DS SQ V L+K ++YTFSAWLQ+S+G V +FK +G E G V+A+ CW++LKG
Sbjct: 98 FDSVSQTVYLEKELLYTFSAWLQVSEGKAPVRAIFKKNG-EYKNAGSVVAESKCWSMLKG 156
Query: 134 GLVANFTSPADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVRFQVTH 193
GL + + PA++ FES + VEIW DSVSLQPFT++EW HQE SI + RK VR +
Sbjct: 157 GLTVDESGPAELYFESDDTMVEIWVDSVSLQPFTQKEWNFHQEQSIYKARKGAVRIRAVD 216
Query: 194 INETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEK 253
+ ATISI+Q R FPFGC + NIL N AY WF RF TTF N+MKWYSTE
Sbjct: 217 SEGQPIPNATISIQQKRLGFPFGCEVEKNILGNQAYENWFTQRFTVTTFANEMKWYSTEV 276
Query: 254 VQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQDLREAARKRINSV 313
V+G+E+Y+ DAML +G++VRGH +LWDD +QP WV SLS DL A ++R+ SV
Sbjct: 277 VRGKEDYSTADAMLRFFKQHGVAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVFSV 336
Query: 314 VSRYKGDLIAWDVVNENLHYNFYEQNFGENAS 345
VSRYKG L WDVVNENLH++F+E G AS
Sbjct: 337 VSRYKGQLAGWDVVNENLHFSFFENKMGPKAS 368
>At4g38650 putative protein
Length = 433
Score = 306 bits (783), Expect = 3e-83
Identities = 165/439 (37%), Positives = 249/439 (56%), Gaps = 23/439 (5%)
Query: 135 LVANFTSPAD----ILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSIERV-------- 182
+VA +S D +L + +++ S SLQPFT+E+WR++Q+ I V
Sbjct: 1 MVAGLSSKEDSFLILLTSEDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTVIHTLISNA 60
Query: 183 RKRKVRFQVTHINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTF 242
RKR V V+ N ++EGA ++++Q F G ++ IL N Y++WF+ RF T F
Sbjct: 61 RKRAVTIHVSKENGESVEGAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVF 120
Query: 243 TNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQDL 302
N++KWY+TE QG+ NYT+ D M+ N I RGH I W+D + P+WV++L+ +DL
Sbjct: 121 ENELKWYATEPDQGKLNYTLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDL 180
Query: 303 REAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMF 362
R A +RI S+++RY+G+ + WDV NE LH++FYE G+NAS +++ A +D A +F
Sbjct: 181 RSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLF 240
Query: 363 LNEFNTIEYSGDVVANPANYIRKIREIQQFPGTKGIPLGIGLQCHFASGNPNLAYMRSGL 422
N+FN +E D + YI ++RE+Q++ G + GIGL+ HF + PN+A MR+ L
Sbjct: 241 FNDFNVVETCSDEKSTVDEYIARVRELQRYDGVR--MDGIGLEGHFTT--PNVALMRAIL 296
Query: 423 DILGATGYPIWLTETSID---PQPNQEEYFEWILREGYSHPAVQGIIMFMGPVQAGFQKA 479
D L PIWLTE I +Q Y E +LREG+SHP+V GI+++ G +
Sbjct: 297 DKLATLQLPIWLTEIDISSSLDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQM 356
Query: 480 PLADENFTNTPMGDVVDKLIREWGTGPHETMADSRGIVDISLHHGDYDVIVSHPQTHFSK 539
L D+ F N P GDVVD+ + EW TG + D G G+Y V + + K
Sbjct: 357 CLTDDKFRNLPAGDVVDQKLLEWKTGEVKATTDDHGSFSFFGFLGEYRVGIMYQ----GK 412
Query: 540 TLNLSVRKEFSQQTIHVKM 558
T+N S +T HV++
Sbjct: 413 TVNSSFSLSQGPETKHVRL 431
>At1g58370 unknown protein
Length = 917
Score = 204 bits (519), Expect = 1e-52
Identities = 156/571 (27%), Positives = 267/571 (46%), Gaps = 62/571 (10%)
Query: 33 IVNPAF-----------DHSLEGWTMVGNGAIEKRRSNE-----------------GNAF 64
I NPAF D + GW +GN + + +
Sbjct: 361 IENPAFGVNILTNSHLSDDTTNGWFSLGNCTLSVAEGSPRILPPMARDSLGAHERLSGRY 420
Query: 65 IVARNRTKPLDSFSQKV--QLKKGMIYTFSAWLQLSKG---SETVSVVFKIHGSELIRGG 119
I+ NRT+ +Q + +LK + Y S W+++ G + V+V I S+ + GG
Sbjct: 421 ILVTNRTQTWMGPAQMITDKLKLFLTYQISVWVKVGSGINSPQNVNVALGID-SQWVNGG 479
Query: 120 HVIAKHGCWTLLKGGL-VANFTSPADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDS 178
V W + G + S A + + + +++ + + P + H +
Sbjct: 480 QVEINDDRWHEIGGSFRIEKNPSKALVYVQGPSSGIDLMVAGLQIFPVDRLARIKHLKRQ 539
Query: 179 IERVRKRKVRFQVTHINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFK 238
+++RKR V + ++ + GA++ ++Q R FP G ++ + + N + +F+ F
Sbjct: 540 CDKIRKRDVILKFAGVDSSKFSGASVRVRQIRNSFPVGTCISRSNIDNEDFVDFFLKNFN 599
Query: 239 HTTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLS 298
F N++KWY TE QG+ NY D ML L + N I RGH I W+ + +W+++++
Sbjct: 600 WAVFANELKWYWTEPEQGKLNYQDADDMLNLCSSNNIETRGHCIFWEVQATVQQWIQNMN 659
Query: 299 PQDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPN 358
DL A + R+ +++RYKG +DV NE LH +FY+ G++ + TA+ LDP+
Sbjct: 660 QTDLNNAVQNRLTDLLNRYKGKFKHYDVNNEMLHGSFYQDKLGKDIRVNMFKTAHQLDPS 719
Query: 359 ARMFLNEFNTIEYSGDVVANPANYIRKIREIQQFPGTKGIPL-GIGLQCHFASGNPNLAY 417
A +F+N+++ IE D + P Y +I ++Q+ KG P+ GIG+Q H S P
Sbjct: 720 ATLFVNDYH-IEDGCDPKSCPEKYTEQILDLQE----KGAPVGGIGIQGHIDS--PVGPI 772
Query: 418 MRSGLDILGATGYPIWLTETSIDP--QPNQEEYFEWILREGYSHPAVQGIIMFMGPVQAG 475
+ S LD LG G PIW TE + + + + E ++ E + HPAV+GI+++ G
Sbjct: 773 VCSALDKLGILGLPIWFTELDVSSVNEHIRADDLEVMMWEAFGHPAVEGIMLW------G 826
Query: 476 FQKAPLA-DENFTNTPMGDVVD------KLIREWGTGPHETMADSRGIVDISLHHGDYDV 528
F + ++ D + GDV + + ++W + + + D G + G+Y V
Sbjct: 827 FWELFMSRDNSHLVNAEGDVNEAGKRFLAVKKDWLSHANGHI-DQNGAFPFRGYSGNYAV 885
Query: 529 IVSHPQTHFSKTL-NLSVRKEFSQQTIHVKM 558
V T SK L V KE S Q I V +
Sbjct: 886 EVI--TTSSSKVLKTFGVDKEDSSQVITVDL 914
Score = 28.9 bits (63), Expect = 7.9
Identities = 35/164 (21%), Positives = 70/164 (42%), Gaps = 20/164 (12%)
Query: 21 SEPQREQYGGG---IIVNPAFDHSLEGWT-------MVGNGAIEKRRSNEGNAFIVARNR 70
SEP+ E+ I+VNP F+ L W+ + + A K G F A R
Sbjct: 185 SEPELERVTAEDETIVVNPNFEDGLNNWSGRSCKIVLHDSMADGKIVPESGKVFASATER 244
Query: 71 TKPLDSFSQKV--QLKKGMIYTFSAWLQLSKGSETVSVVFKI-------HGSELIRGGHV 121
T+ + Q++ ++++ +Y +A +++ + T + V + I V
Sbjct: 245 TQNWNGIQQEITGKVQRKRVYEATAVVRIYGNNVTTATVQATLWVQNPNQRDQYIGISTV 304
Query: 122 IAKHGCWTLLKGGLVANFTSPADILF-ESKNPTVEIWADSVSLQ 164
A W LKG + N ++ +++ E P +I +S++++
Sbjct: 305 QATDKEWIHLKGKFLLNGSASRVVIYIEGPPPGTDILLNSLTVK 348
>At1g10050 putative xylan endohydrolase
Length = 1063
Score = 201 bits (512), Expect = 7e-52
Identities = 148/535 (27%), Positives = 254/535 (46%), Gaps = 43/535 (8%)
Query: 28 YGGGIIVNPAF-DHSLEGW-------TMVGNGA-----------IEKRRSNEGNAFIVAR 68
+G I+ N D ++EGW VG+G+ + K + +++A
Sbjct: 514 FGMNIVSNSHLSDGTIEGWFPLGDCHLKVGDGSPRILPPLARDSLRKTQGYLSGRYVLAT 573
Query: 69 NRTK----PLDSFSQKVQLKKGMIYTFSAWLQLSKGSET----VSVVFKIHGSELIRGGH 120
NR+ P + + KV+L + Y SAW+++ G T V++ + G+ + GG
Sbjct: 574 NRSGTWMGPAQTITDKVKLF--VTYQVSAWVKIGSGGRTSPQDVNIALSVDGN-WVNGGK 630
Query: 121 VIAKHGCWTLLKGGL-VANFTSPADILFESKNPTVEIWADSVSLQPFTKEEWRSHQEDSI 179
V G W + G + + + +P V++ + + ++ S+
Sbjct: 631 VEVDDGDWHEVVGSFRIEKEAKEVMLHVQGPSPGVDLMVAGLQIFAVDRKARLSYLRGQA 690
Query: 180 ERVRKRKVRFQVTHINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKH 239
+ VRKR V + + ++ + L GAT+ I+QTR FP G ++ + + N + +F++ F
Sbjct: 691 DVVRKRNVCLKFSGLDPSELSGATVKIRQTRNSFPLGSCISRSNIDNEDFVDFFLNNFDW 750
Query: 240 TTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSP 299
F ++KWY TE QG NY + M+ I RGH I W+ + WV+ L+
Sbjct: 751 AVFGYELKWYWTEPEQGNFNYRDANEMIEFCERYNIKTRGHCIFWEVESAIQPWVQQLTG 810
Query: 300 QDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNA 359
L A R+ +++RY G +DV NE LH +FY +A A + TA+ LDP A
Sbjct: 811 SKLEAAVENRVTDLLTRYNGKFRHYDVNNEMLHGSFYRDRLDSDARANMFKTAHELDPLA 870
Query: 360 RMFLNEFNTIEYSGDVVANPANYIRKIREIQQFPGTKGIPL-GIGLQCHFASGNPNLAYM 418
+FLNE++ IE D ++P YI+ + ++Q+ KG P+ GIG+Q H S P +
Sbjct: 871 TLFLNEYH-IEDGFDSRSSPEKYIKLVHKLQK----KGAPVGGIGIQGHITS--PVGHIV 923
Query: 419 RSGLDILGATGYPIWLTETSIDP--QPNQEEYFEWILREGYSHPAVQGIIMF-MGPVQAG 475
RS LD L G PIW TE + + + + E +L E ++HPAV+G++++ +
Sbjct: 924 RSALDKLSTLGLPIWFTELDVSSTNEHIRGDDLEVMLWEAFAHPAVEGVMLWGFWELFMS 983
Query: 476 FQKAPLADENFTNTPMGDVVDKLIREWGTGPHETMADSRGIVDISLHHGDYDVIV 530
+ + L + + G ++ REW + + D G+ + +HG Y V V
Sbjct: 984 REHSHLVNADGEVNEAGKRFLEIKREWLSFVDGEIEDGGGL-EFRGYHGSYTVEV 1037
>At4g08160 xylan endohydrolase like proteinchimeric
Length = 520
Score = 198 bits (503), Expect = 8e-51
Identities = 140/503 (27%), Positives = 246/503 (48%), Gaps = 37/503 (7%)
Query: 61 GNAFIVARNRTKPLDSFSQKV--QLKKGMIYTFSAWLQLSKG-------SETVSVVFKIH 111
G +IV NRT+ +Q + ++K + Y SAW++L G + V++ +
Sbjct: 13 GGNYIVVTNRTQTWMGPAQMITDKIKLFLTYQISAWVKLGVGVSGSSMSPQNVNIALSVD 72
Query: 112 GSELIRGGHVIAKHG-CWTLLKGGLVANFTSPADILFESKNPT--VEIWADSVSLQPFTK 168
++ + GG V G W + G P +++ + P +++ ++ + P +
Sbjct: 73 -NQWVNGGQVEVTVGDTWHEIAGSFRLE-KQPQNVMVYVQGPGAGIDLMIAALQIFPVDR 130
Query: 169 EEWRSHQEDSIERVRKRKVRFQVTHINETA---LEGATISIKQTRPHFPFGCGMNNNILT 225
E + ++ VRKR + + + +N+ L + +KQT FP G +N +
Sbjct: 131 RERVRCLKRQVDEVRKRDIVLKFSGLNDDESFDLFPYIVKVKQTYNSFPVGTCINRTDID 190
Query: 226 NSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWD 285
N + +F F F N++KWY+TE +G+ NY D ML L N I+VRGH I W+
Sbjct: 191 NEDFVDFFTKNFNWAVFGNELKWYATEAERGKVNYQDADDMLDLCIGNNINVRGHCIFWE 250
Query: 286 DRNFQPEWVKSLSPQDLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGENAS 345
+ WV+ L+ DL A +KR+ +++RYKG +DV NE LH +FY+ G+
Sbjct: 251 VESTVQPWVRQLNKTDLMNAVQKRLTDLLTRYKGKFKHYDVNNEMLHGSFYQDRLGKGVR 310
Query: 346 AMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIREIQQFPGTKGIPLGIGLQ 405
A+ ++ A+ LDP+ +F+N+++ +E D ++P YI+ + +++ T G GIG+Q
Sbjct: 311 ALMFNIAHKLDPSPLLFVNDYH-VEDGDDPRSSPEKYIKLVLDLEAQGATVG---GIGIQ 366
Query: 406 CHFASGNPNLAYMRSGLDILGATGYPIWLTETSIDPQPN--QEEYFEWILREGYSHPAVQ 463
H S P A + S LD+L G PIW TE + + E E +L E ++HP+V+
Sbjct: 367 GHIDS--PVGAIVCSALDMLSVLGRPIWFTELDVSSSNEYVRGEDLEVMLWEAFAHPSVE 424
Query: 464 GIIMFMGPVQAGFQKAPLADENFTNTPMGDVVDKLIREWGTGPHETMADSRGIVD----- 518
GI+++ GF + ++ EN V++ + + E ++ + GI++
Sbjct: 425 GIMLW------GFWELSMSRENANLVEGEGEVNEAGKRFLEVKQEWLSHAYGIINDESEF 478
Query: 519 -ISLHHGDYDVIVSHPQTHFSKT 540
+HG Y V + P KT
Sbjct: 479 TFRGYHGTYAVEICTPAGIVLKT 501
>At4g38300 unknown protein
Length = 181
Score = 165 bits (418), Expect = 5e-41
Identities = 71/163 (43%), Positives = 107/163 (65%)
Query: 181 RVRKRKVRFQVTHINETALEGATISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHT 240
+ RKR V V+ N ++EGA ++++Q FP G ++ IL N Y++WF+ RF T
Sbjct: 10 QARKRAVTIHVSKENGESVEGAEVTVEQISKDFPIGSAISKTILGNIPYQEWFVKRFDAT 69
Query: 241 TFTNQMKWYSTEKVQGQENYTIPDAMLALANDNGISVRGHTILWDDRNFQPEWVKSLSPQ 300
F N++KWY+TE QG+ NYT+ D M+ L N I RGH I W+D + P+WV++L+ +
Sbjct: 70 VFENELKWYATESDQGKLNYTLADKMMNLVRANRIIARGHNIFWEDPKYNPDWVRNLTGE 129
Query: 301 DLREAARKRINSVVSRYKGDLIAWDVVNENLHYNFYEQNFGEN 343
DLR A +RI S+++RY+G+ + WDV NE LH++FYE G+N
Sbjct: 130 DLRSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYESRLGKN 172
>At2g40640 unknown protein
Length = 383
Score = 32.7 bits (73), Expect = 0.55
Identities = 55/250 (22%), Positives = 89/250 (35%), Gaps = 38/250 (15%)
Query: 44 GWTMV------GNGAIEKRRSN--------EGNAFIVARNRTKPLDSFSQKVQLKKGMIY 89
GW +V GN I++ +GN RN + D S V+ ++
Sbjct: 137 GWELVVREDGEGNSTIDRHEMEFKVRITKPDGNVSNSHRNTQQKRDFAS--VEKERVTTT 194
Query: 90 TFSAWLQLSKGSETVSVVFKIHGSELIRGGHVIAKHGCWTLLKGGLVANFTSPADILFES 149
+ S+W L ++ + ++ GH G + K + A FT L S
Sbjct: 195 SVSSWESLKAILSDPVTGALMNDATILPCGHSFGAGGLKEVKK--MKACFTCSQPTLEGS 252
Query: 150 KNPTVEIWADSVSLQPFTKEEWRSHQEDSIERVRKRKVR------FQVTHINETALEGAT 203
+ P + + + + F +EE D I +++RK R F + +I ET
Sbjct: 253 EKPNLSL---RIVVHAFRQEE----DSDHIHTLKRRKERSDQKRSFCIPNITETPKSSRG 305
Query: 204 ISIKQTRPHFPFGCGMNNNILTNSAYRKWFMSRFKHTTFTNQMKWYSTEKVQGQENYTIP 263
I FPF G + I N F+ R WY + V E+ +
Sbjct: 306 IQ-------FPFSIGDHIIIEGNKRTPPRFVGRIAVIMTQCLNGWYVVKTVDNSESIKLQ 358
Query: 264 DAMLALANDN 273
LA +DN
Sbjct: 359 HCSLAKISDN 368
>At4g35810 putative protein
Length = 307
Score = 32.3 bits (72), Expect = 0.72
Identities = 20/80 (25%), Positives = 33/80 (41%), Gaps = 3/80 (3%)
Query: 320 DLIAWDVVNENLHYNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANP 379
D I ++ N + F GE ++Y +P+ F +EFN + G +A
Sbjct: 174 DEIVEEIENRISDFTFIPPENGEGLQVLHYEVGQRYEPHHDYFFDEFN-VRKGGQRIATV 232
Query: 380 ANYIRKIREIQQ--FPGTKG 397
Y+ + E + FP KG
Sbjct: 233 LMYLSDVDEGGETVFPAAKG 252
>At1g20270 unknown protein
Length = 287
Score = 31.2 bits (69), Expect = 1.6
Identities = 26/103 (25%), Positives = 43/103 (41%), Gaps = 5/103 (4%)
Query: 290 QPEWVKSL---SPQDLREAARKRINSVVSRYKG-DLIAWDVVNENLHYNFYEQNFGENAS 345
+P VKS S + +R R +S +G D I + Y F + GE
Sbjct: 105 KPHMVKSTVVDSETGKSKDSRVRTSSGTFLRRGRDKIIKTIEKRIADYTFIPADHGEGLQ 164
Query: 346 AMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIRE 388
++Y +P+ F++EFNT + G +A Y+ + E
Sbjct: 165 VLHYEAGQKYEPHYDYFVDEFNT-KNGGQRMATMLMYLSDVEE 206
>At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein
Length = 267
Score = 30.4 bits (67), Expect = 2.7
Identities = 17/67 (25%), Positives = 29/67 (42%), Gaps = 3/67 (4%)
Query: 333 YNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIREIQQ- 391
+ F GE ++Y +P+ F++E+NT G +A Y+ + E +
Sbjct: 154 FTFIPVEHGEGLQVLHYEIGQKYEPHYDYFMDEYNT-RNGGQRIATVLMYLSDVEEGGET 212
Query: 392 -FPGTKG 397
FP KG
Sbjct: 213 VFPAAKG 219
>At3g13020 hypothetical protein
Length = 605
Score = 30.4 bits (67), Expect = 2.7
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Query: 302 LREAARKRINSVVSRYKGDLIAWDVV----NENLHYNFYEQNFGENASAMYYSTAYHLDP 357
++ + +K N Y L WDV+ N++LH + + N ++ +YST +HLDP
Sbjct: 428 IKLSIKKEFNDEKKHY---LTLWDVIDDVWNKHLHNPLHAAGYYLNPTS-FYSTDFHLDP 483
>At5g32620 putative protein
Length = 301
Score = 29.6 bits (65), Expect = 4.6
Identities = 26/92 (28%), Positives = 37/92 (39%), Gaps = 18/92 (19%)
Query: 11 FFTTSCLLLISEPQRE---QYGGGIIVNPAFDHSLEGWTMVGNGAIEKRRSNEGNAFIVA 67
F+ +S + + + Q+E Q+ G PAF L S EG+ +
Sbjct: 7 FYQSSGFMDLLQSQQEDMSQFASGSTEVPAFSSQL---------------SEEGSELEGS 51
Query: 68 RNRTKPLDSFSQKVQLKKGMIYTFSAWLQLSK 99
+ KP S S+K K I SAWL SK
Sbjct: 52 EDEVKPKQSISRKKWTAKEDIVLVSAWLNPSK 83
>At1g74150 hypothetical protein
Length = 552
Score = 29.6 bits (65), Expect = 4.6
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 6/66 (9%)
Query: 481 LADENFTNTPMGDVVDKLIREWGTGPHETMADSRGIVDISLHHGDYDVI------VSHPQ 534
+AD N T+TPM D VD + GT ET A + D D ++ VSH Q
Sbjct: 468 VADSNDTDTPMIDTVDANVNMEGTHEAETAAVTIDAKDQDASQLDKGIVNTDPLPVSHDQ 527
Query: 535 THFSKT 540
+ T
Sbjct: 528 VNVIST 533
>At2g17720 similar to prolyl 4-hydroxylase alpha subunit
Length = 291
Score = 29.3 bits (64), Expect = 6.1
Identities = 17/67 (25%), Positives = 30/67 (44%), Gaps = 3/67 (4%)
Query: 333 YNFYEQNFGENASAMYYSTAYHLDPNARMFLNEFNTIEYSGDVVANPANYIRKIREIQQ- 391
+ F GE ++Y +P+ FL+EFNT + G +A Y+ + + +
Sbjct: 156 FTFIPVENGEGLQVLHYQVGQKYEPHYDYFLDEFNT-KNGGQRIATVLMYLSDVDDGGET 214
Query: 392 -FPGTKG 397
FP +G
Sbjct: 215 VFPAARG 221
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,348,723
Number of Sequences: 26719
Number of extensions: 598249
Number of successful extensions: 1270
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1212
Number of HSP's gapped (non-prelim): 27
length of query: 560
length of database: 11,318,596
effective HSP length: 104
effective length of query: 456
effective length of database: 8,539,820
effective search space: 3894157920
effective search space used: 3894157920
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0218.16


