

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.7
(68 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
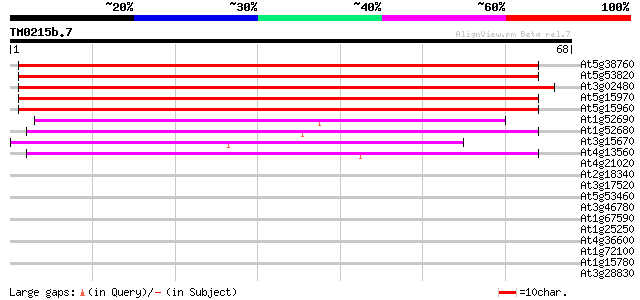
Score E
Sequences producing significant alignments: (bits) Value
At5g38760 pollen coat -like protein 87 2e-18
At5g53820 ABA-inducible protein-like 86 3e-18
At3g02480 unknown protein 73 2e-14
At5g15970 cold-regulated protein COR6.6 (KIN2) 55 6e-09
At5g15960 cold and ABA inducible protein kin1 52 5e-08
At1g52690 unknown protein 44 1e-05
At1g52680 unknown protein 42 4e-05
At3g15670 LEA76 homologue type2 40 2e-04
At4g13560 unknown protein 39 6e-04
At4g21020 unknown protein (At4g21020) 33 0.026
At2g18340 similar to late embryogenesis abundant proteins 30 0.22
At3g17520 unknown protein 28 1.1
At5g53460 NADH-dependent glutamate synthase 27 2.5
At3g46780 unknown protein 27 2.5
At1g67590 unknown protein 27 2.5
At1g25250 Indeterminate 1 like Zn finger containing protein 27 2.5
At4g36600 putative protein 26 3.2
At1g72100 unknown protein (At1g72100) 26 3.2
At1g15780 unknown protein 26 3.2
At3g28830 hypothetical protein 26 4.2
>At5g38760 pollen coat -like protein
Length = 67
Score = 87.0 bits (214), Expect = 2e-18
Identities = 42/63 (66%), Positives = 52/63 (81%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ +S+ AGQAKGQ +EK S +MD ASNAAQSAKE+++ GQQIK AQGA ++VKNAT
Sbjct: 4 NSQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLKETGQQIKEKAQGATESVKNAT 63
Query: 62 GMN 64
GMN
Sbjct: 64 GMN 66
>At5g53820 ABA-inducible protein-like
Length = 67
Score = 85.9 bits (211), Expect = 3e-18
Identities = 40/63 (63%), Positives = 52/63 (82%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ MS+NAGQAKGQ +EK SNL+D ASNAAQSAKE++Q GQQ+K AQGA++ +K T
Sbjct: 4 NSQSMSFNAGQAKGQTQEKASNLIDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEKT 63
Query: 62 GMN 64
G++
Sbjct: 64 GIS 66
>At3g02480 unknown protein
Length = 68
Score = 73.2 bits (178), Expect = 2e-14
Identities = 35/65 (53%), Positives = 46/65 (69%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+ Q SY AGQA GQ +EK +MD A +AA SA++++Q GQQ+K AQGAAD VK+ T
Sbjct: 3 NKQNASYQAGQATGQTKEKAGGMMDKAKDAAASAQDSLQQTGQQMKEKAQGAADVVKDKT 62
Query: 62 GMNNN 66
GMN +
Sbjct: 63 GMNKS 67
>At5g15970 cold-regulated protein COR6.6 (KIN2)
Length = 66
Score = 55.1 bits (131), Expect = 6e-09
Identities = 27/63 (42%), Positives = 43/63 (67%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQA G+AEEK++ L+D A +AA +A + Q AG+ I +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>At5g15960 cold and ABA inducible protein kin1
Length = 66
Score = 52.0 bits (123), Expect = 5e-08
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQ G+AEEK++ L+D A +AA A Q AG+ + +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>At1g52690 unknown protein
Length = 169
Score = 43.9 bits (102), Expect = 1e-05
Identities = 24/61 (39%), Positives = 33/61 (53%), Gaps = 4/61 (6%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAK----ETVQGAGQQIKASAQGAADAVKN 59
Q+ SY AG+ +G+A+EKT M + Q+AK ET Q A Q+ +AQ A D
Sbjct: 5 QEQSYKAGETRGKAQEKTGEAMGTMGDKTQAAKDKTQETAQSAQQKAHETAQSAKDKTSQ 64
Query: 60 A 60
A
Sbjct: 65 A 65
Score = 32.7 bits (73), Expect = 0.034
Identities = 16/53 (30%), Positives = 26/53 (48%), Gaps = 4/53 (7%)
Query: 18 EEKTSNLMDMASNAAQSAKETVQG----AGQQIKASAQGAADAVKNATGMNNN 66
+ K + + A++ KE G G+Q+K A GA DAVK+ G+ +
Sbjct: 92 KNKAHDAAEYTKETAEAGKEKTSGILGQTGEQVKQMAMGATDAVKHTLGLRTD 144
Score = 27.3 bits (59), Expect = 1.4
Identities = 15/54 (27%), Positives = 20/54 (36%)
Query: 5 KMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
K G+A G +KT D AQSA++ Q K AA +
Sbjct: 17 KAQEKTGEAMGTMGDKTQAAKDKTQETAQSAQQKAHETAQSAKDKTSQAAQTTQ 70
>At1g52680 unknown protein
Length = 114
Score = 42.4 bits (98), Expect = 4e-05
Identities = 27/88 (30%), Positives = 39/88 (43%), Gaps = 26/88 (29%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQS--------------------------AK 36
SQ++S++AG+ GQ + K ++ S+A A
Sbjct: 4 SQQLSHSAGEVTGQVQLKKEEYLNNVSHAMNQNADHHTHSQSQLHSEHDQNNPSLISQAS 63
Query: 37 ETVQGAGQQIKASAQGAADAVKNATGMN 64
+Q G Q+K AQGAADAVKN GM+
Sbjct: 64 SVIQQTGGQVKNMAQGAADAVKNTLGMS 91
>At3g15670 LEA76 homologue type2
Length = 225
Score = 40.0 bits (92), Expect = 2e-04
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLM----DMASNAAQSAKETVQGAGQQIKASAQGAAD 55
M S + SY AG+ +G+A+EKT M D A +T Q A Q+ +AQ A D
Sbjct: 1 MASNQQSYKAGETRGKAQEKTGQAMGTMRDKAEEGRDKTSQTAQTAQQKAHETAQSAKD 59
Score = 34.7 bits (78), Expect = 0.009
Identities = 26/91 (28%), Positives = 40/91 (43%), Gaps = 30/91 (32%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLM--------DMASNAAQSAKETVQGA------------ 42
+++ + AG +A++K + + + A +AAQ KET QGA
Sbjct: 101 AKEKTSQAGDKAREAKDKAGSYLSETGEAIKNKAQDAAQYTKETAQGAAQYTKETAEAGR 160
Query: 43 ----------GQQIKASAQGAADAVKNATGM 63
G+ +K A GAADAVK+ GM
Sbjct: 161 DKTGGFLSQTGEHVKQMAMGAADAVKHTFGM 191
Score = 27.3 bits (59), Expect = 1.4
Identities = 17/58 (29%), Positives = 20/58 (34%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
QK A AK + + A AQSAKE Q + A A K T
Sbjct: 48 QKAHETAQSAKDKTSQTAQAAQQKAHETAQSAKEKTSQTAQTAQQKAHETTQAAKEKT 105
>At4g13560 unknown protein
Length = 109
Score = 38.5 bits (88), Expect = 6e-04
Identities = 23/66 (34%), Positives = 33/66 (49%), Gaps = 4/66 (6%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQG----AGQQIKASAQGAADAVK 58
+Q A A +KT++ A+ +AQ +E G G+ +K AQGA D VK
Sbjct: 41 TQSARDKAADLTQSARDKTADGSHSANKSAQHNQEQAAGLFGQTGESVKNMAQGALDGVK 100
Query: 59 NATGMN 64
N+ GMN
Sbjct: 101 NSLGMN 106
>At4g21020 unknown protein (At4g21020)
Length = 266
Score = 33.1 bits (74), Expect = 0.026
Identities = 18/54 (33%), Positives = 28/54 (51%)
Query: 5 KMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
K + A K +A++ + MD A A+ AKE V+ G+ K A+G + VK
Sbjct: 142 KAADKAEDTKEKAKDYAEDTMDNAKEKARHAKEKVKEYGEDTKEKAEGFKETVK 195
Score = 30.8 bits (68), Expect = 0.13
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query: 9 NAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGA----GQQIKASAQGAADAVKNA 60
NA + A+EK + A+ KETV+G G++ K + +GA ++ KNA
Sbjct: 164 NAKEKARHAKEKVKEYGEDTKEKAEGFKETVKGKAEELGEKTKETVKGAWESTKNA 219
Score = 28.5 bits (62), Expect = 0.65
Identities = 16/54 (29%), Positives = 25/54 (45%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAV 57
+K+ K +AE + A + KETV+GA + K +AQ +AV
Sbjct: 174 EKVKEYGEDTKEKAEGFKETVKGKAEELGEKTKETVKGAWESTKNAAQTVTEAV 227
Score = 28.1 bits (61), Expect = 0.84
Identities = 15/48 (31%), Positives = 23/48 (47%)
Query: 12 QAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
QAK +A E D A NA + AK+ + ++ A AAD ++
Sbjct: 102 QAKDKAYETKEKAKDTAYNAKEKAKDYAERTKDKVNEGAYKAADKAED 149
>At2g18340 similar to late embryogenesis abundant proteins
Length = 456
Score = 30.0 bits (66), Expect = 0.22
Identities = 21/59 (35%), Positives = 27/59 (45%), Gaps = 1/59 (1%)
Query: 2 DSQKMSYN-AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
D++ M+Y AG AK A +K N DMA A AK+ G K A A K+
Sbjct: 141 DAKDMAYEKAGHAKDFAYDKAGNAKDMAYETAGYAKDFAYDKGGYAKDVAYDKAGNAKD 199
Score = 28.9 bits (63), Expect = 0.49
Identities = 20/59 (33%), Positives = 27/59 (44%), Gaps = 1/59 (1%)
Query: 2 DSQKMSY-NAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
D++ M+Y NAG AK A +K + DMA A AK+ K A A K+
Sbjct: 119 DAKNMAYENAGYAKDFAYDKAGDAKDMAYEKAGHAKDFAYDKAGNAKDMAYETAGYAKD 177
Score = 28.9 bits (63), Expect = 0.49
Identities = 19/67 (28%), Positives = 31/67 (45%), Gaps = 8/67 (11%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETV--------QGAGQQIKASAQGAADAVKNAT 61
AG+AK A +K N D+A AQ AK+ AG I+ + + +A + A
Sbjct: 318 AGEAKDYAYKKAGNAKDIAYEKAQDAKDFAYDKAGYGYDKAGDVIRMATDKSGEAYEGAK 377
Query: 62 GMNNNSK 68
+ ++K
Sbjct: 378 EKSKSAK 384
Score = 28.9 bits (63), Expect = 0.49
Identities = 18/53 (33%), Positives = 22/53 (40%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATG 62
AG AK A EK N DMA A+ K+ SAQ D+ + G
Sbjct: 194 AGNAKDMAYEKAGNAKDMAYEKAEHIKDFTYDKVGSAYGSAQSMMDSGYDKAG 246
Score = 28.1 bits (61), Expect = 0.84
Identities = 20/52 (38%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Query: 6 MSYN-AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADA 56
M+Y+ AG AK A EK DMA + A +AK+ SAQ A D+
Sbjct: 262 MAYDKAGDAKDVAYEKAGIAKDMAYDKAGNAKDMAYDKVGSAYGSAQKAKDS 313
Score = 27.3 bits (59), Expect = 1.4
Identities = 18/57 (31%), Positives = 27/57 (46%), Gaps = 7/57 (12%)
Query: 11 GQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIK-------ASAQGAADAVKNA 60
G AK A +K N DMA A +AK+ + IK SA G+A ++ ++
Sbjct: 184 GYAKDVAYDKAGNAKDMAYEKAGNAKDMAYEKAEHIKDFTYDKVGSAYGSAQSMMDS 240
Score = 25.4 bits (54), Expect = 5.5
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 3/54 (5%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATGM 63
AG AK A EK + DMA + A AK+ A ++ + A D NA M
Sbjct: 245 AGDAKDMAYEKAGIVKDMAYDKAGDAKDV---AYEKAGIAKDMAYDKAGNAKDM 295
Score = 25.0 bits (53), Expect = 7.1
Identities = 18/61 (29%), Positives = 30/61 (48%), Gaps = 1/61 (1%)
Query: 2 DSQKMSYN-AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
+++ M+Y AG AK A +K D+A + A +AK+ K A A+ +K+
Sbjct: 163 NAKDMAYETAGYAKDFAYDKGGYAKDVAYDKAGNAKDMAYEKAGNAKDMAYEKAEHIKDF 222
Query: 61 T 61
T
Sbjct: 223 T 223
>At3g17520 unknown protein
Length = 298
Score = 27.7 bits (60), Expect = 1.1
Identities = 14/58 (24%), Positives = 28/58 (48%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATGMNNNS 67
A A+EK L + A ++SAKE ++ + + K+ A ++ K+ + +S
Sbjct: 211 ASDMTSAAKEKAEKLKEEAERESKSAKEKIKESYETAKSKADETLESAKDKASQSYDS 268
Score = 25.0 bits (53), Expect = 7.1
Identities = 16/54 (29%), Positives = 25/54 (45%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADA 56
SQK AK +AEE + A +A ++AK V+ +K A + D+
Sbjct: 73 SQKAEDAKEAAKRKAEEAVGAAKEKAGSAYETAKSKVEEGLASVKDKASQSYDS 126
>At5g53460 NADH-dependent glutamate synthase
Length = 2216
Score = 26.6 bits (57), Expect = 2.5
Identities = 15/56 (26%), Positives = 28/56 (49%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
++ S A + + + E+ ++ + AA S+KE + G G +A +AVKN
Sbjct: 1655 ERASEEADETEEKELEEKDAFAELKNMAAASSKEEMSGNGVAAEARPSKVDNAVKN 1710
>At3g46780 unknown protein
Length = 510
Score = 26.6 bits (57), Expect = 2.5
Identities = 19/62 (30%), Positives = 31/62 (49%), Gaps = 3/62 (4%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKE---TVQGAGQQIKASAQGAADAVK 58
D ++ Y A+ +AEE+ D A AA++AKE +Q ++ +A A DA +
Sbjct: 343 DGRRKVYADAIARERAEEEAKVAADKAREAAEAAKEFEKQMQKLSEKEAEAASLAEDAQQ 402
Query: 59 NA 60
A
Sbjct: 403 KA 404
>At1g67590 unknown protein
Length = 347
Score = 26.6 bits (57), Expect = 2.5
Identities = 13/60 (21%), Positives = 26/60 (42%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M+ +KM A + K +AEEK +N + A+ + + + AD ++ +
Sbjct: 268 MEMKKMEVKAERMKARAEEKLANKLAATKRIAEERRANAEAKLNEKAVKTSEKADYIRRS 327
>At1g25250 Indeterminate 1 like Zn finger containing protein
Length = 385
Score = 26.6 bits (57), Expect = 2.5
Identities = 14/42 (33%), Positives = 25/42 (59%)
Query: 6 MSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIK 47
M+ + QA+ + +TS + A+ A+ A+ET Q A +QI+
Sbjct: 266 MARTSAQARHNEKRETSLTKERANEEARKAEETRQEAKRQIE 307
>At4g36600 putative protein
Length = 347
Score = 26.2 bits (56), Expect = 3.2
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 7/60 (11%)
Query: 10 AGQAKGQA-------EEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATG 62
+G+AK A +E + N D+AS+ A +ET A K A A ++ A G
Sbjct: 235 SGEAKDSAYGTYERFKEGSKNAKDIASDKAHDVRETAGRAVDYAKDKANDAYESGSEAAG 294
Score = 25.0 bits (53), Expect = 7.1
Identities = 21/73 (28%), Positives = 29/73 (38%), Gaps = 19/73 (26%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQI----------------- 46
QK Y G AE+ N + A NAA++AK+ A
Sbjct: 14 QKQCYGWGTET--AEDMVRNEAEHAKNAAETAKKMASDAAHDTKDKTASWAGWGFRQNFH 71
Query: 47 KASAQGAADAVKN 59
KA A+ AA++ KN
Sbjct: 72 KAEAEEAAESAKN 84
Score = 25.0 bits (53), Expect = 7.1
Identities = 17/44 (38%), Positives = 23/44 (51%), Gaps = 8/44 (18%)
Query: 2 DSQKMSYN-AGQAK-------GQAEEKTSNLMDMASNAAQSAKE 37
D++ M+Y+ GQAK G A EK DMA + A AK+
Sbjct: 128 DAKDMAYDKTGQAKYMAYDKAGSAYEKAGQAKDMAYDKAGQAKD 171
>At1g72100 unknown protein (At1g72100)
Length = 480
Score = 26.2 bits (56), Expect = 3.2
Identities = 18/48 (37%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKAS-AQGAADA 56
A AKG+ EK + + ++ A AKE+V K S AQ A DA
Sbjct: 190 AQYAKGRVTEKAHDPKEGVAHKAHDAKESVADKAHDAKESVAQKAHDA 237
>At1g15780 unknown protein
Length = 1335
Score = 26.2 bits (56), Expect = 3.2
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Query: 4 QKMSYNAGQ--AKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASA 50
Q + N GQ A G ++ TS L++ +S +AQS T+Q Q + S+
Sbjct: 774 QGNTLNNGQQVAMGSMQQNTSQLVNNSSASAQSGLSTLQSNVNQPQLSS 822
>At3g28830 hypothetical protein
Length = 536
Score = 25.8 bits (55), Expect = 4.2
Identities = 18/60 (30%), Positives = 25/60 (41%), Gaps = 1/60 (1%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQG-AGQQIKASAQGAADAVKNATGMNNNSK 68
+G K S +A+ + QG A Q ASAQG+A A + +G SK
Sbjct: 279 SGSPKASPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAMSK 338
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.295 0.108 0.262
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,039,554
Number of Sequences: 26719
Number of extensions: 24265
Number of successful extensions: 114
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 57
Number of HSP's gapped (non-prelim): 55
length of query: 68
length of database: 11,318,596
effective HSP length: 44
effective length of query: 24
effective length of database: 10,142,960
effective search space: 243431040
effective search space used: 243431040
T: 11
A: 40
X1: 17 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0215b.7


