

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.5
(316 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
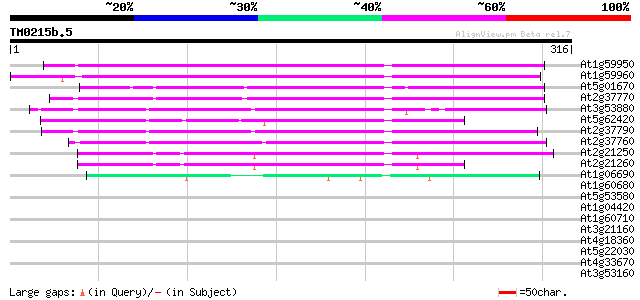
Score E
Sequences producing significant alignments: (bits) Value
At1g59950 hypothetical protein 202 1e-52
At1g59960 putative protein 196 1e-50
At5g01670 aldose reductase-like protein 143 1e-34
At2g37770 putative aldo/keto reductase 142 3e-34
At3g53880 putative alcohol dehydrogenase 138 4e-33
At5g62420 aldose reductase-like protein 132 2e-31
At2g37790 putative alcohol dehydrogenase 125 4e-29
At2g37760 putative alcohol dehydrogenase 124 9e-29
At2g21250 putative NADPH dependent mannose 6-phosphate reductase 107 6e-24
At2g21260 putative NADPH dependent mannose 6-phosphate reductase 101 5e-22
At1g06690 unknown protein 49 5e-06
At1g60680 unknown protein 39 0.003
At5g53580 aldo/keto reductase-like protein 37 0.014
At1g04420 unknown protein 35 0.053
At1g60710 unknown protein 32 0.35
At3g21160 putative alpha 1,2-mannosidase 30 1.7
At4g18360 glycolate oxidase - like protein 29 2.9
At5g22030 ubiquitin-specific protease 8 (UBP8) 28 5.0
At4g33670 putative protein 28 5.0
At3g53160 glucosyltransferase - like protein 28 6.5
>At1g59950 hypothetical protein
Length = 320
Score = 202 bits (515), Expect = 1e-52
Identities = 117/286 (40%), Positives = 172/286 (59%), Gaps = 9/286 (3%)
Query: 20 ILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIAS 79
+L +G + +P ++ VLEA+K G++HFD + Y E+ +G A+AE LI S
Sbjct: 17 VLALGTAASPPPEPIVLK-RTVLEAIKLGYRHFDTSPRYQTEEPLGEALAEAVSLGLIQS 75
Query: 80 KDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLE 139
+ LFVTSK+W D H L+VPA+Q+ L TL+L+YLDL+LIHWP ++K G K+PIE +
Sbjct: 76 RSELFVTSKLWCADAHGGLVVPAIQRSLETLKLDYLDLYLIHWPVSSKPGKYKFPIEEDD 135
Query: 140 IMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQV 199
+ + V S MEE QRLG+ K IGV+N S KKL + A I P ++QVE+ WQQ
Sbjct: 136 FLPMDYETVWSEMEECQRLGVAKCIGVSNFSCKKLQHILSIAKIPPSVNQVEMSPVWQQR 195
Query: 200 KLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQKEL-DTHGKTIAQICL*WI 257
KLR+ CK K + + A+S L G G + M++D+ KE+ + GKT+AQ+ + W
Sbjct: 196 KLRELCKSKGIV----VTAYSVLGSRGAFWGTHKIMESDVLKEIAEAKGKTVAQVSMRWA 251
Query: 258 YKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKIS-EIHQERLI 301
Y+ V+ + + L IF SLTE + ++IS EI Q R++
Sbjct: 252 YEEGVSMVVKSFRKDRLEENLKIFDWSLTEEEKQRISTEISQSRIV 297
>At1g59960 putative protein
Length = 326
Score = 196 bits (498), Expect = 1e-50
Identities = 120/308 (38%), Positives = 181/308 (57%), Gaps = 16/308 (5%)
Query: 1 MATTTVPKVVLQSS-TG*QRILVMGLSTA----PEAANMIST*DVVLEAVKQGFKHFDAA 55
M+ TTVP + ++S +G + V+G TA PE + T V+EA+K G++HFD +
Sbjct: 1 MSLTTVPTLAIRSGPSGHHSMPVLGFGTAASPLPEPTMLKET---VIEAIKLGYRHFDTS 57
Query: 56 AAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYL 115
Y E+ +G A+AE L+ S+ FVT+K+W D H L+VPA+++ L+ L+L+YL
Sbjct: 58 PRYQTEEPIGEALAEAVSLGLVRSRSEFFVTTKLWCADAHGGLVVPAIKRSLKNLKLDYL 117
Query: 116 DLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLD 175
DL++IHWP ++K G K+PI+ + M + V S MEE QRLGL K IGV+N S KKL
Sbjct: 118 DLYIIHWPVSSKPGKYKFPIDEDDFMPMDFEVVWSEMEECQRLGLAKCIGVSNFSCKKLQ 177
Query: 176 KLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAM 234
+ ATI P ++QVE+ WQQ KLR+ C+ + + A+S L G G M
Sbjct: 178 HILSIATIPPSVNQVEMSPIWQQRKLRELCRSNDIV----VTAYSVLGSRGAFWGTPKIM 233
Query: 235 DNDIQKEL-DTHGKTIAQICL*WIYKH-VNFCDEEL*QGEDATKLAIFGCSLTENDFEKI 292
++D+ KE+ + KT+AQ+ + W Y+ V+ + + L IF SLTE++ ++I
Sbjct: 234 ESDVLKEIAEAKEKTVAQVSMRWAYEQGVSMVVKSFTKERLEENLKIFDWSLTEDETQRI 293
Query: 293 S-EIHQER 299
S EI Q R
Sbjct: 294 STEIPQFR 301
>At5g01670 aldose reductase-like protein
Length = 322
Score = 143 bits (360), Expect = 1e-34
Identities = 87/263 (33%), Positives = 143/263 (54%), Gaps = 9/263 (3%)
Query: 40 VVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPELI 99
VV V+ G++H D A Y ++ VG I + H + +D LFVTSK+W + PE +
Sbjct: 42 VVTAIVEGGYRHIDTAWEYGDQREVGQGI-KRAMHAGLERRD-LFVTSKLWCTELSPERV 99
Query: 100 VPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLG 159
PALQ L+ LQLEYLDL+LIHWP ++GA K P + ++++F M+GV ME +
Sbjct: 100 RPALQNTLKELQLEYLDLYLIHWPIRLREGASK-PPKAGDVLDFDMEGVWREMENLSKDS 158
Query: 160 LTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQFGIIAF 219
L + IGV N ++ KL+KL FA + P + Q+E+ GW+ ++ +FCK+ ++ + A+
Sbjct: 159 LVRNIGVCNFTVTKLNKLLGFAELIPAVCQMEMHPGWRNDRILEFCKKNEIH----VTAY 214
Query: 220 SPLRMGVSRGANLAMDNDIQKELDTHGKTIAQICL*W-IYKHVNFCDEEL*QGEDATKLA 278
SPL G +L D + + KT QI + W + + + + L +
Sbjct: 215 SPLG-SQEGGRDLIHDQTVDRIAKKLNKTPGQILVKWGLQRGTSVIPKSLNPERIKENIK 273
Query: 279 IFGCSLTENDFEKISEIHQERLI 301
+F + E DF+ ++ I ++ +
Sbjct: 274 VFDWVIPEQDFQALNSITDQKRV 296
>At2g37770 putative aldo/keto reductase
Length = 315
Score = 142 bits (357), Expect = 3e-34
Identities = 93/282 (32%), Positives = 151/282 (52%), Gaps = 12/282 (4%)
Query: 23 MGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDG 82
+GL T + ++ D V AVK G++H D A Y E+ +GA + + + ++ +D
Sbjct: 19 VGLGTWQASPGLVG--DAVAAAVKIGYRHIDCAQIYGNEKEIGAVLKKLFEDRVVKRED- 75
Query: 83 LFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIME 142
LF+TSK+W D+ P+ + AL + L+ LQLEY+DL+LIHWP K+G+V I+ ++
Sbjct: 76 LFITSKLWCTDHDPQDVPEALNRTLKDLQLEYVDLYLIHWPARIKKGSV--GIKPENLLP 133
Query: 143 FGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLR 202
+ +ME G +AIGV+N S KKL L A + P ++QVE W+Q KL+
Sbjct: 134 VDIPSTWKAMEALYDSGKARAIGVSNFSTKKLADLLELARVPPAVNQVECHPSWRQTKLQ 193
Query: 203 DFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQKEL-DTHGKTIAQICL*W-IYK 259
+FCK K ++ + A+SPL G + + + N I + + GK+ AQ+ L W +
Sbjct: 194 EFCKSKGVH----LSAYSPLGSPGTTWLKSDVLKNPILNMVAEKLGKSPAQVALRWGLQM 249
Query: 260 HVNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLI 301
+ + +G +F S+ + F K +EI Q RL+
Sbjct: 250 GHSVLPKSTNEGRIKENFNVFDWSIPDYMFAKFAEIEQARLV 291
>At3g53880 putative alcohol dehydrogenase
Length = 315
Score = 138 bits (347), Expect = 4e-33
Identities = 98/300 (32%), Positives = 153/300 (50%), Gaps = 25/300 (8%)
Query: 12 QSSTG*QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEE 71
Q +TG +I +GL T A ++ D V AVK G++H D A+ Y E +G + +
Sbjct: 9 QLNTG-AKIPSVGLGTWQAAPGVVG--DAVAAAVKIGYQHIDCASRYGNEIEIGKVLKKL 65
Query: 72 HQH*LIASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAV 131
++ ++ LF+TSK+W+ D P + AL + L+ LQL+Y+DL+L+HWP K+G V
Sbjct: 66 FDDGVV-KREKLFITSKIWLTDLDPPDVQDALNRTLQDLQLDYVDLYLMHWPVRLKKGTV 124
Query: 132 KYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVE 191
+ E IM + +ME G +AIGV+N S KKL L A + P ++QVE
Sbjct: 125 DFKPE--NIMPIDIPSTWKAMEALVDSGKARAIGVSNFSTKKLSDLVEAARVPPAVNQVE 182
Query: 192 VDLGWQQVKLRDFCKEKALYWQFGIIAFSPL--------RMGVSRGANLAMDNDIQKELD 243
WQQ KL +FCK K ++ + +SPL + V + + M I KE+
Sbjct: 183 CHPSWQQHKLHEFCKSKGIH----LSGYSPLGSPGTTWVKADVLKSPVIEM---IAKEI- 234
Query: 244 THGKTIAQICL*W-IYKHVNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIK 302
GK+ AQ L W + + + +G + G S+ + F+K S+I Q RL++
Sbjct: 235 --GKSPAQTALRWGLQMGHSILPKSTNEGRIRENFDVLGWSIPKEMFDKFSKIEQARLVQ 292
>At5g62420 aldose reductase-like protein
Length = 316
Score = 132 bits (333), Expect = 2e-31
Identities = 85/243 (34%), Positives = 136/243 (54%), Gaps = 12/243 (4%)
Query: 18 QRILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LI 77
+ I ++G+ T + ST V +A+K G++HFD A Y E+++G A+ + + +
Sbjct: 12 ETIPLLGMGTYCPQKDRESTISAVHQAIKIGYRHFDTAKIYGSEEALGTALGQAISYGTV 71
Query: 78 ASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEV 137
+D LFVTSK+W D+H + AL + L+T+ L+YLD +L+HWP K G V PI
Sbjct: 72 -QRDDLFVTSKLWSSDHHDP--ISALIQTLKTMGLDYLDNYLVHWPIKLKPG-VSEPIPK 127
Query: 138 LEIME--FGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLG 195
+ E G++ ME +GL ++IGV+N S KK+ L FA++SP ++QVE+
Sbjct: 128 EDEFEKDLGIEETWQGMERCLEMGLCRSIGVSNFSSKKIFDLLDFASVSPSVNQVEMHPL 187
Query: 196 WQQVKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQKELD-THGKTIAQIC 253
W+Q KLR C+E ++ + +SPL G G+ +++ I K + H T AQ+
Sbjct: 188 WRQRKLRKVCEENNIH----VSGYSPLGGPGNCWGSTAVIEHPIIKSIALKHNATPAQVA 243
Query: 254 L*W 256
L W
Sbjct: 244 LRW 246
>At2g37790 putative alcohol dehydrogenase
Length = 350
Score = 125 bits (313), Expect = 4e-29
Identities = 88/282 (31%), Positives = 143/282 (50%), Gaps = 12/282 (4%)
Query: 19 RILVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIA 78
+I +GL T ++ + V AVK G++H D A Y E+ +G + + ++
Sbjct: 15 KIPSVGLGTWQADPGLVG--NAVDAAVKIGYRHIDCAQIYGNEKEIGLVLKKLFDGGVV- 71
Query: 79 SKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVL 138
++ +F+TSK+W + P+ + AL + L+ LQL+Y+DL+LIHWP + K+G+ + E
Sbjct: 72 KREEMFITSKLWCTYHDPQEVPEALNRTLQDLQLDYVDLYLIHWPVSLKKGSTGFKPE-- 129
Query: 139 EIMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQ 198
I+ + +ME G +AIGV+N S KKL L A + P ++QVE WQQ
Sbjct: 130 NILPTDIPSTWKAMESLFDSGKARAIGVSNFSSKKLADLLVVARVPPAVNQVECHPSWQQ 189
Query: 199 VKLRDFCKEKALYWQFGIIAFSPL-RMGVSRGANLAMDNDIQKEL-DTHGKTIAQICL*W 256
LRDFCK K ++ + +SPL G + + + N I + + GKT AQ+ L W
Sbjct: 190 NVLRDFCKSKGVH----LSGYSPLGSPGTTWLTSDVLKNPILGGVAEKLGKTPAQVALRW 245
Query: 257 -IYKHVNFCDEEL*QGEDATKLAIFGCSLTENDFEKISEIHQ 297
+ + + + +F S+ E+ K SEI Q
Sbjct: 246 GLQMGQSVLPKSTHEDRIKQNFDVFNWSIPEDMLSKFSEIGQ 287
>At2g37760 putative alcohol dehydrogenase
Length = 311
Score = 124 bits (310), Expect = 9e-29
Identities = 87/272 (31%), Positives = 144/272 (51%), Gaps = 12/272 (4%)
Query: 34 MIST*DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVID 93
M++T + +A+K G++H D A+ Y E+ +G + + + ++ LF+TSK+W D
Sbjct: 26 MVAT--AIEQAIKIGYRHIDCASIYGNEKEIGGVLKKLIGDGFV-KREELFITSKLWSND 82
Query: 94 NHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSME 153
+ PE + AL+K L+ LQ++Y+DL+LIHWP + K+ ++ E+L + + +ME
Sbjct: 83 HLPEDVPKALEKTLQDLQIDYVDLYLIHWPASLKKESLMPTPEMLTKPD--ITSTWKAME 140
Query: 154 ESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQ 213
G +AIGV+N S KKL L A ++P ++QVE WQQ L + CK K ++
Sbjct: 141 ALYDSGKARAIGVSNFSSKKLTDLLNVARVTPAVNQVECHPVWQQQGLHELCKSKGVH-- 198
Query: 214 FGIIAFSPLRMGVSRGANL-AMDNDIQKEL-DTHGKTIAQICL*WIYKHVNFCDEEL*QG 271
+ +SPL L + N I E+ + GKT AQ+ L W + + + G
Sbjct: 199 --LSGYSPLGSQSKGEVRLKVLQNPIVTEVAEKLGKTTAQVALRWGLQTGHSVLPKSSSG 256
Query: 272 EDATK-LAIFGCSLTENDFEKISEIHQERLIK 302
+ L +F S+ E+ F K S I QE+ +
Sbjct: 257 ARLKENLDVFDWSIPEDLFTKFSNIPQEKFCR 288
>At2g21250 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 107 bits (268), Expect = 6e-24
Identities = 78/281 (27%), Positives = 139/281 (48%), Gaps = 20/281 (7%)
Query: 39 DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL 98
D++L A+K G++H D AA Y E VG A+ E + L+ +D LF+T+K+W D+
Sbjct: 28 DLILNAIKIGYRHLDCAADYRNETEVGDALTEAFKTGLVKRED-LFITTKLWNSDHGH-- 84
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIE------VLEI-MEFGMQGV*SS 151
++ A + L+ LQL+YLDLFL+H+P ATK V VL+I ++
Sbjct: 85 VIEACKDSLKKLQLDYLDLFLVHFPVATKHTGVGTTDSALGDDGVLDIDTTISLETTWHD 144
Query: 152 MEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALY 211
ME+ +GL ++IG++N + ++ I P ++Q+E +Q+ L FC++ +
Sbjct: 145 MEKLVSMGLVRSIGISNYDVFLTRDCLAYSKIKPAVNQIETHPYFQRDSLVKFCQKHGIC 204
Query: 212 WQFGIIAFSPLRMGVSR----GANLAMDNDIQKEL-DTHGKTIAQICL*W-IYKHVNFCD 265
+ A +PL + G +D+ + K++ + + KT+AQ+ L W I +
Sbjct: 205 ----VTAHTPLGGATANAEWFGTVSCLDDPVLKDVAEKYKKTVAQVVLRWGIQRKTVVIP 260
Query: 266 EEL*QGEDATKLAIFGCSLTENDFEKISEIHQERLIKEPTK 306
+ +F L++ D E I + ++ +P K
Sbjct: 261 KTSKPARLEENFQVFDFELSKEDMEVIKSMERKYRTNQPAK 301
>At2g21260 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 101 bits (252), Expect = 5e-22
Identities = 68/230 (29%), Positives = 121/230 (52%), Gaps = 19/230 (8%)
Query: 39 DVVLEAVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL 98
D++++A+K G++H D AA Y E VG A+ E L+ +D LF+T+K+W D+
Sbjct: 28 DLIIDAIKIGYRHLDCAANYKNEAEVGEALTEAFTTGLVKRED-LFITTKLWSSDHGH-- 84
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIE------VLEI-MEFGMQGV*SS 151
++ A + L+ LQL+YLDLFL+H P ATK + VL+I ++
Sbjct: 85 VIEACKDSLKKLQLDYLDLFLVHIPIATKHTGIGTTDSALGDDGVLDIDTTISLETTWHD 144
Query: 152 MEESQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALY 211
ME+ +GL ++IG++N + ++ I P ++Q+E +Q+ L FC++ +
Sbjct: 145 MEKLVSMGLVRSIGISNYDVFLTRDCLAYSKIKPAVNQIETHPYFQRDSLVKFCQKHGIC 204
Query: 212 WQFGIIAFSPLRMGVSR----GANLAMDNDIQKEL-DTHGKTIAQICL*W 256
+ A +PL + G +D+ + K++ + + +T+AQI L W
Sbjct: 205 ----VTAHTPLGGATANAEWFGTVSCLDDPVLKDVAEKYKQTVAQIVLRW 250
>At1g06690 unknown protein
Length = 377
Score = 48.5 bits (114), Expect = 5e-06
Identities = 67/293 (22%), Positives = 112/293 (37%), Gaps = 59/293 (20%)
Query: 44 AVKQGFKHFDAAAAYAVEQSVGAAIAEEHQH*LIASKDGLFVTSKVWVIDNHPEL----- 98
++ G FD A Y + S+GA +E I + + ++V V L
Sbjct: 94 SLDNGIDFFDTAEVYGSKFSLGAISSETLLGRFIRERKERYPGAEVSVATKFAALPWRFG 153
Query: 99 ---IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEES 155
+V AL+ L L+L +DL+ +HWP +G +G + ++
Sbjct: 154 RESVVTALKDSLSRLELSSVDLYQLHWPGL-----------------WGNEGYLDGLGDA 196
Query: 156 QRLGLTKAIGVNNLSIKKLDKLF---CFATISPVIDQVEVDLGW---QQVKLRDFCKEKA 209
GL KA+GV+N S K+L + I +QV L + +Q ++ C E
Sbjct: 197 VEQGLVKAVGVSNYSEKRLRDAYERLKKRGIPLASNQVNYSLIYRAPEQTGVKAACDELG 256
Query: 210 LYWQFGIIAFSPLRMGVSRGANLAMD----------------------NDIQKELDTHGK 247
+ +IA+SP+ G G + N I++ + + K
Sbjct: 257 V----TLIAYSPIAQGALTGKYTPENPPSGPRGRIYTREFLTKLQPLLNRIKQIGENYSK 312
Query: 248 TIAQICL*WIYKHVNFCD-EEL*QGEDATKLA-IFGCSLTENDFEKISEIHQE 298
T QI L W+ N E A + A G SLT+N+ ++ + E
Sbjct: 313 TPTQIALNWLVAQGNVIPIPGAKNAEQAKEFAGAIGWSLTDNEVSELRSLASE 365
>At1g60680 unknown protein
Length = 346
Score = 39.3 bits (90), Expect = 0.003
Identities = 61/252 (24%), Positives = 106/252 (41%), Gaps = 63/252 (25%)
Query: 44 AVKQGFKHFDAAAAYAVEQS---VGAAIAEEHQH*L-IASKDGLFVTS-KVWVIDNHPEL 98
A+ G FD + Y E + +G A+ + + + +A+K G F+ ++ + PE
Sbjct: 49 AINSGVTFFDTSDMYGPETNELLLGKALKDGVKEKVELATKFGFFIVEGEISEVRGDPEY 108
Query: 99 IVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRL 158
+ A + L+ L + +DL+ H + + PIE+ +M E ++L
Sbjct: 109 VRAACEASLKRLDIACIDLYYQH------RIDTRVPIEI-------------TMRELKKL 149
Query: 159 ---GLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRD---FCKEKALYW 212
G K IG++ S + + I+ V Q+E L W + D C+E +
Sbjct: 150 VEEGKIKYIGLSEASASTIRRAHAVHPITAV--QIEWSL-WSRDAEEDIIPICRELGI-- 204
Query: 213 QFGIIAFSPLRMG-VSRGANLA--MDND--------IQKELDTHGK-------------- 247
GI+A+SPL G ++ G LA ++ND Q+E H K
Sbjct: 205 --GIVAYSPLGRGFLAAGPKLAENLENDDFRKTLPRFQQENVDHNKILFEKVSAMAEKKG 262
Query: 248 -TIAQICL*WIY 258
T AQ+ L W++
Sbjct: 263 CTPAQLALAWVH 274
>At5g53580 aldo/keto reductase-like protein
Length = 365
Score = 37.0 bits (84), Expect = 0.014
Identities = 46/203 (22%), Positives = 90/203 (43%), Gaps = 38/203 (18%)
Query: 44 AVKQGFKHFDAAAAYAV-------EQSVGAAIAEEHQH*LIASKDGLFVTSKV----WVI 92
A++ G FD A +Y E+ +G I E L ++ + V +K W +
Sbjct: 80 ALENGINLFDTADSYGTGRLNGQSERLLGKFIKESQG--LKGKQNEVVVATKFAAYPWRL 137
Query: 93 DNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSM 152
+ V A + L LQ++ L + +HW A+ P++ L + + +
Sbjct: 138 TSGQ--FVNACRASLDRLQIDQLGIGQLHWSTASYA-----PLQELVLWD--------GL 182
Query: 153 EESQRLGLTKAIGVNNLSIKKLDKLFCFATISPV---IDQVE---VDLGWQQVKLRDFCK 206
+ GL +A+GV+N ++L K+ + V QV+ + +G +Q++++ C
Sbjct: 183 VQMYEKGLVRAVGVSNYGPQQLVKIHDYLKTRGVPLCSAQVQFSLLSMGKEQLEIKSICD 242
Query: 207 EKALYWQFGIIAFSPLRMGVSRG 229
E + +I++SPL +G+ G
Sbjct: 243 ELGI----RLISYSPLGLGMLTG 261
>At1g04420 unknown protein
Length = 412
Score = 35.0 bits (79), Expect = 0.053
Identities = 54/247 (21%), Positives = 101/247 (40%), Gaps = 31/247 (12%)
Query: 21 LVMGLSTAPEAANMIST*DVVLEAVKQGFKHFDAAAAYAV---EQSVGAAI--------A 69
+ MG T E + +++ A+++G D A AY + +++ G +
Sbjct: 71 VTMGTMTFGEQNTEKESHEMLSYAIEEGINCIDTAEAYPIPMKKETQGKTDLYISSWLKS 130
Query: 70 EEHQH*LIASKDGLFVTSKVWVIDNHPELIVPA------LQKYLRTLQLEYLDLFLIHWP 123
++ ++A+K + ++ D+ L V A ++K L+ L +Y+DL IHWP
Sbjct: 131 QQRDKIVLATKVCGYSERSAYIRDSGEILRVDAANIKESVEKSLKRLGTDYIDLLQIHWP 190
Query: 124 --FATKQGAVKYPIEVLE-IMEFGMQGV*SSMEESQRLGLTKAIGVNNLSIKKLDKLFCF 180
+ G Y + F Q + ++ G + IGV+N + + +
Sbjct: 191 DRYVPLFGDFYYETSKWRPSVPFAEQ--LRAFQDLIVEGKVRYIGVSNETSYGVTEFVNT 248
Query: 181 ATISPVIDQVEVDLGWQ-------QVKLRDFCKEKALYWQFGIIAFSPLRMGVSRGANLA 233
A + + V + G+ +V L + C K G++A+SPL G G LA
Sbjct: 249 AKLEGLPKIVSIQNGYSLLVRCRYEVDLVEVCHPKNC--NVGLLAYSPLGGGSLSGKYLA 306
Query: 234 MDNDIQK 240
D + K
Sbjct: 307 TDQEATK 313
>At1g60710 unknown protein
Length = 345
Score = 32.3 bits (72), Expect = 0.35
Identities = 51/215 (23%), Positives = 91/215 (41%), Gaps = 39/215 (18%)
Query: 23 MGLST---APEAANMIST*DVVLEAVKQGFKHFDAAAAYAVEQS---VGAAIAEEHQH*L 76
MGLS AP+ N ++ A+ G D + Y E + +G A+ + + +
Sbjct: 27 MGLSAFYGAPKPENEAIA--LIHHAIHSGVTLLDTSDIYGPETNEVLLGKALKDGVREKV 84
Query: 77 -IASKDGLFVTSKVWVIDNHPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPI 135
+A+K G+ + PE + A + L+ L + +DL+ H + + PI
Sbjct: 85 ELATKFGISYAEGKREVRGDPEYVRAACEASLKRLDIACIDLYYQH------RVDTRVPI 138
Query: 136 EVLEIMEFGMQGV*SSMEESQRL---GLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEV 192
E+ +M E ++L G K IG++ S + + I+ V Q+E
Sbjct: 139 EI-------------TMGELKKLVEEGKIKYIGLSEASASTIRRAHAVHPITAV--QIEW 183
Query: 193 DLGWQQV--KLRDFCKEKALYWQFGIIAFSPLRMG 225
L + V ++ C+E + GI+A+SPL G
Sbjct: 184 SLWTRDVEEEIIPTCRELGI----GIVAYSPLGRG 214
>At3g21160 putative alpha 1,2-mannosidase
Length = 572
Score = 30.0 bits (66), Expect = 1.7
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 4/69 (5%)
Query: 114 YLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEESQRLGLTKAIGVN-NLSIK 172
+ + L W F K AVK+ ++ E M G+ S +++S L T + N I
Sbjct: 314 FYEYLLKVWVFGNKTSAVKH---YRDMWEKSMNGLLSLVKKSTPLSFTYICEKSGNSLID 370
Query: 173 KLDKLFCFA 181
K+D+L CFA
Sbjct: 371 KMDELACFA 379
>At4g18360 glycolate oxidase - like protein
Length = 368
Score = 29.3 bits (64), Expect = 2.9
Identities = 35/136 (25%), Positives = 55/136 (39%), Gaps = 12/136 (8%)
Query: 157 RLGLTKAIGVNN---LSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQ 213
R L + + + N L + K+DK S V QV+ L W+ +K + +
Sbjct: 171 RFALPRGLTLKNFEGLDLGKIDKTNDSGLASYVAGQVDQSLSWKDIKWLQSITSLPILVK 230
Query: 214 FGIIAFSPLRMGVSRG-ANLAMDNDIQKELDTHGKTIAQICL*WIYKHVN-----FCDEE 267
G+I R+ V G A + + N ++LD TI + L + K V F D
Sbjct: 231 -GVITAEDARIAVEYGAAGIIVSNHGARQLDYVPATI--VALEEVVKAVEGRIPVFLDGG 287
Query: 268 L*QGEDATKLAIFGCS 283
+ +G D K G S
Sbjct: 288 VRRGTDVFKALALGAS 303
>At5g22030 ubiquitin-specific protease 8 (UBP8)
Length = 901
Score = 28.5 bits (62), Expect = 5.0
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query: 272 EDATKLAIFGCSLTENDFEKISEIHQERLIK---EPTKPLLNIK 312
ED K + CSL E + ++E++ R+I+ EPT L I+
Sbjct: 522 EDLHKALVTACSLPEEETLLVTEVYNNRIIRFLEEPTDSLTLIR 565
>At4g33670 putative protein
Length = 319
Score = 28.5 bits (62), Expect = 5.0
Identities = 43/200 (21%), Positives = 82/200 (40%), Gaps = 39/200 (19%)
Query: 41 VLEAVKQGFKHFDAAAAYA---VEQSVGA---AIAEEHQH*LIASKDGLFVTSKVWVIDN 94
V EA + G FD + Y E+ +G A+ ++A+K G + D
Sbjct: 42 VREAFRLGINFFDTSPYYGGTLSEKMLGKGLKALQVPRSDYIVATKCGRYKEG----FDF 97
Query: 95 HPELIVPALQKYLRTLQLEYLDLFLIHWPFATKQGAVKYPIEVLEIMEFGMQGV*SSMEE 154
E + ++ + L LQL+Y+D+ H IE + + + + ++++
Sbjct: 98 SAERVRKSIDESLERLQLDYVDILHCH------------DIEFGSLDQIVSETI-PALQK 144
Query: 155 SQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRD--------FCK 206
++ G T+ IG+ L + +F + V+V L + + D + K
Sbjct: 145 LKQEGKTRFIGITGLPL----DIFTYVLDRVPPGTVDVILSYCHYGVNDSTLLDLLPYLK 200
Query: 207 EKALYWQFGIIAFSPLRMGV 226
K + G+I+ SPL MG+
Sbjct: 201 SKGV----GVISASPLAMGL 216
>At3g53160 glucosyltransferase - like protein
Length = 490
Score = 28.1 bits (61), Expect = 6.5
Identities = 21/86 (24%), Positives = 37/86 (42%), Gaps = 1/86 (1%)
Query: 155 SQRLGLTKAIGVNNLSIKKLDKLFCFATISPVIDQVEVDLGWQQVKLRDFCKEKALYWQF 214
SQR G+T I ++ K+ F+++ I+ VEV QQ L + C+ +
Sbjct: 31 SQRQGVTVCIITTTQNVAKIKTSLSFSSLFATINIVEVKFLSQQTGLPEGCESLDMLASM 90
Query: 215 G-IIAFSPLRMGVSRGANLAMDNDIQ 239
G ++ F + AM+ +Q
Sbjct: 91 GDMVKFFDAANSLEEQVEKAMEEMVQ 116
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.144 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,109,889
Number of Sequences: 26719
Number of extensions: 237926
Number of successful extensions: 838
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 801
Number of HSP's gapped (non-prelim): 20
length of query: 316
length of database: 11,318,596
effective HSP length: 99
effective length of query: 217
effective length of database: 8,673,415
effective search space: 1882131055
effective search space used: 1882131055
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0215b.5


