

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.3
(66 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
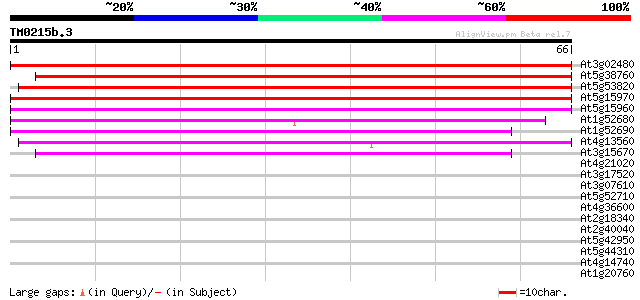
At3g02480 unknown protein 86 4e-18
At5g38760 pollen coat -like protein 84 2e-17
At5g53820 ABA-inducible protein-like 77 2e-15
At5g15970 cold-regulated protein COR6.6 (KIN2) 51 1e-07
At5g15960 cold and ABA inducible protein kin1 48 1e-06
At1g52680 unknown protein 46 3e-06
At1g52690 unknown protein 42 6e-05
At4g13560 unknown protein 39 6e-04
At3g15670 LEA76 homologue type2 39 6e-04
At4g21020 unknown protein (At4g21020) 33 0.020
At3g17520 unknown protein 30 0.22
At3g07610 hypothetical protein 28 0.65
At5g52710 unknown protein 28 0.85
At4g36600 putative protein 28 0.85
At2g18340 similar to late embryogenesis abundant proteins 27 1.9
At2g40040 unknown protein 27 2.5
At5g42950 putative protein 25 5.5
At5g44310 late embryogenesis abundant protein-like 25 9.4
At4g14740 unknown protein 25 9.4
At1g20760 unknown protein 25 9.4
>At3g02480 unknown protein
Length = 68
Score = 85.5 bits (210), Expect = 4e-18
Identities = 43/66 (65%), Positives = 48/66 (72%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
MD QN S+QAGQATGQ +EKA MM A++AA SAQ S Q+ GQQMK KAQGAAD VK
Sbjct: 1 MDNKQNASYQAGQATGQTKEKAGGMMDKAKDAAASAQDSLQQTGQQMKEKAQGAADVVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGMNK 66
>At5g38760 pollen coat -like protein
Length = 67
Score = 83.6 bits (205), Expect = 2e-17
Identities = 42/63 (66%), Positives = 50/63 (78%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
SQN+S QAGQA GQ QEKAS MM A NAAQSA++S +E GQQ+K KAQGA ++VK+A G
Sbjct: 5 SQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLKETGQQIKEKAQGATESVKNATG 64
Query: 64 ANK 66
NK
Sbjct: 65 MNK 67
>At5g53820 ABA-inducible protein-like
Length = 67
Score = 77.0 bits (188), Expect = 2e-15
Identities = 37/65 (56%), Positives = 49/65 (74%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
+ SQ++S AGQA GQ QEKASN++ A NAAQSA++S QE GQQ+K KAQGA++ +K
Sbjct: 3 NNSQSMSFNAGQAKGQTQEKASNLIDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEK 62
Query: 62 IGANK 66
G +K
Sbjct: 63 TGISK 67
>At5g15970 cold-regulated protein COR6.6 (KIN2)
Length = 66
Score = 50.8 bits (120), Expect = 1e-07
Identities = 25/66 (37%), Positives = 41/66 (61%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQA G+A+EK++ ++ A++AA +A S Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>At5g15960 cold and ABA inducible protein kin1
Length = 66
Score = 47.8 bits (112), Expect = 1e-06
Identities = 23/66 (34%), Positives = 38/66 (56%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQ G+A+EK++ ++ A++AA A Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>At1g52680 unknown protein
Length = 114
Score = 46.2 bits (108), Expect = 3e-06
Identities = 30/89 (33%), Positives = 37/89 (40%), Gaps = 26/89 (29%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNA--------------------------AQ 34
M SQ LSH AG+ TGQ Q K + N +A
Sbjct: 1 MSSSQQLSHSAGEVTGQVQLKKEEYLNNVSHAMNQNADHHTHSQSQLHSEHDQNNPSLIS 60
Query: 35 SAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
A Q+ G Q+K AQGAADAVK+ +G
Sbjct: 61 QASSVIQQTGGQVKNMAQGAADAVKNTLG 89
>At1g52690 unknown protein
Length = 169
Score = 42.0 bits (97), Expect = 6e-05
Identities = 21/59 (35%), Positives = 30/59 (50%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
M Q S++AG+ G+AQEK MG + Q+A+ QE Q + KA A + K
Sbjct: 1 MASHQEQSYKAGETRGKAQEKTGEAMGTMGDKTQAAKDKTQETAQSAQQKAHETAQSAK 59
Score = 30.0 bits (66), Expect = 0.22
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 18/63 (28%)
Query: 5 QNLSHQAGQATGQA----QEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
+N +H A + T + +EK S ++G + G+Q+K A GA DAVK
Sbjct: 92 KNKAHDAAEYTKETAEAGKEKTSGILG--------------QTGEQVKQMAMGATDAVKH 137
Query: 61 AIG 63
+G
Sbjct: 138 TLG 140
Score = 28.9 bits (63), Expect = 0.50
Identities = 22/76 (28%), Positives = 33/76 (42%), Gaps = 14/76 (18%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEA-----------GQQMKAK 50
D++Q + A Q +A E A + AAQ+ Q+ QE+ G+ +K K
Sbjct: 38 DKTQETAQSAQQ---KAHETAQSAKDKTSQAAQTTQERAQESKDKTGSYMSETGEAIKNK 94
Query: 51 AQGAADAVKSAIGANK 66
A AA+ K A K
Sbjct: 95 AHDAAEYTKETAEAGK 110
>At4g13560 unknown protein
Length = 109
Score = 38.5 bits (88), Expect = 6e-04
Identities = 21/69 (30%), Positives = 38/69 (54%), Gaps = 4/69 (5%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQ----EAGQQMKAKAQGAADA 57
D +Q+ +A T A++K ++ +A +AQ Q+ + G+ +K AQGA D
Sbjct: 39 DLTQSARDKAADLTQSARDKTADGSHSANKSAQHNQEQAAGLFGQTGESVKNMAQGALDG 98
Query: 58 VKSAIGANK 66
VK+++G N+
Sbjct: 99 VKNSLGMNE 107
Score = 35.0 bits (79), Expect = 0.007
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query: 11 AGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
AGQ GQAQEKA +A+ AQSA C + ++ AAD +SA
Sbjct: 8 AGQNRGQAQEKAEQWTESAKQTAQSA---CDKTADLTQSARDKAADLTQSA 55
>At3g15670 LEA76 homologue type2
Length = 225
Score = 38.5 bits (88), Expect = 6e-04
Identities = 18/56 (32%), Positives = 28/56 (49%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S S++AG+ G+AQEK MG R+ A+ + + Q + KA A + K
Sbjct: 3 SNQQSYKAGETRGKAQEKTGQAMGTMRDKAEEGRDKTSQTAQTAQQKAHETAQSAK 58
Score = 34.3 bits (77), Expect = 0.012
Identities = 20/56 (35%), Positives = 26/56 (45%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+Q +A + T A+EK S AR A A E G+ +K KAQ AA K
Sbjct: 87 AQTAQQKAHETTQAAKEKTSQAGDKAREAKDKAGSYLSETGEAIKNKAQDAAQYTK 142
Score = 32.7 bits (73), Expect = 0.034
Identities = 17/50 (34%), Positives = 22/50 (44%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+ GQA G ++KA AQ+AQQ E Q K K A A +
Sbjct: 20 KTGQAMGTMRDKAEEGRDKTSQTAQTAQQKAHETAQSAKDKTSQTAQAAQ 69
Score = 31.2 bits (69), Expect = 0.10
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 12/74 (16%)
Query: 2 DQSQNLSHQAGQAT-GQAQEKASNMMGNARNAAQSAQQSCQ-----------EAGQQMKA 49
D++ + + G+A +AQ+ A A+ AAQ +++ + + G+ +K
Sbjct: 117 DKAGSYLSETGEAIKNKAQDAAQYTKETAQGAAQYTKETAEAGRDKTGGFLSQTGEHVKQ 176
Query: 50 KAQGAADAVKSAIG 63
A GAADAVK G
Sbjct: 177 MAMGAADAVKHTFG 190
Score = 30.4 bits (67), Expect = 0.17
Identities = 18/57 (31%), Positives = 27/57 (46%), Gaps = 3/57 (5%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIGANK 66
QAG +A++KA + + A ++ Q+A Q K AQGAA K A +
Sbjct: 107 QAGDKAREAKDKAGSYLSETGEAIKN---KAQDAAQYTKETAQGAAQYTKETAEAGR 160
>At4g21020 unknown protein (At4g21020)
Length = 266
Score = 33.5 bits (75), Expect = 0.020
Identities = 15/43 (34%), Positives = 27/43 (61%)
Query: 17 QAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+A++ A + M NA+ A+ A++ +E G+ K KA+G + VK
Sbjct: 153 KAKDYAEDTMDNAKEKARHAKEKVKEYGEDTKEKAEGFKETVK 195
>At3g17520 unknown protein
Length = 298
Score = 30.0 bits (66), Expect = 0.22
Identities = 14/55 (25%), Positives = 28/55 (50%)
Query: 5 QNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+ +A T A+EKA + A ++SA++ +E+ + K+KA ++ K
Sbjct: 205 ETAKEKASDMTSAAKEKAEKLKEEAERESKSAKEKIKESYETAKSKADETLESAK 259
>At3g07610 hypothetical protein
Length = 1027
Score = 28.5 bits (62), Expect = 0.65
Identities = 11/52 (21%), Positives = 28/52 (53%)
Query: 11 AGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAI 62
+G+ + + +EK NM G + AA++ ++ ++ K ++ A + + + I
Sbjct: 891 SGEKSPEPEEKKQNMRGPKKGAAKAVAKALKDLSPSEKKSSEAAEEEISNGI 942
>At5g52710 unknown protein
Length = 451
Score = 28.1 bits (61), Expect = 0.85
Identities = 14/47 (29%), Positives = 24/47 (50%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMK 48
+ S+N SH +++ + SN GN+ ++QS S + GQ K
Sbjct: 401 NSSRNSSHSRSRSSENLSQFQSNSTGNSYTSSQSNTASGSKQGQSSK 447
>At4g36600 putative protein
Length = 347
Score = 28.1 bits (61), Expect = 0.85
Identities = 16/58 (27%), Positives = 28/58 (47%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
D++ + S +AGQA A +KA++ A N A+ + + + K A G + K
Sbjct: 193 DKAGSASEKAGQAKDFAYDKAAHAKDAAYNKAEDVIKMATDTSGEAKDSAYGTYERFK 250
Score = 26.9 bits (58), Expect = 1.9
Identities = 14/65 (21%), Positives = 32/65 (48%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
D++ ++ AG+A A++KA++ + AA ++ + G++ A ++ K A
Sbjct: 262 DKAHDVRETAGRAVDYAKDKANDAYESGSEAAGRFDEAMHKVGERYGAVKDSMSENTKEA 321
Query: 62 IGANK 66
+ K
Sbjct: 322 YESAK 326
>At2g18340 similar to late embryogenesis abundant proteins
Length = 456
Score = 26.9 bits (58), Expect = 1.9
Identities = 15/47 (31%), Positives = 21/47 (43%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAAD 56
+AG A A +K + G+A+ A S + EA KA A D
Sbjct: 288 KAGNAKDMAYDKVGSAYGSAQKAKDSGYEKAGEAKDYAYKKAGNAKD 334
>At2g40040 unknown protein
Length = 839
Score = 26.6 bits (57), Expect = 2.5
Identities = 15/56 (26%), Positives = 30/56 (52%), Gaps = 1/56 (1%)
Query: 3 QSQNLSHQAGQATGQAQ-EKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADA 57
QSQ+ S Q+ QAQ + S +++ +QS QS ++ Q ++++Q + +
Sbjct: 753 QSQSPSQTRAQSPSQAQAQSPSQTQSQSQSQSQSQSQSQSQSQSQSQSQSQSQSQS 808
Score = 26.6 bits (57), Expect = 2.5
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 3 QSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQ 52
QSQ+ S Q+ Q+Q + S +++ +QS Q+ ++ Q +A+AQ
Sbjct: 779 QSQSQSQSQSQSQSQSQSQ-SQSQSQSQSQSQSPSQTQTQSPSQTQAQAQ 827
>At5g42950 putative protein
Length = 1714
Score = 25.4 bits (54), Expect = 5.5
Identities = 10/42 (23%), Positives = 22/42 (51%)
Query: 21 KASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAI 62
+++N+M + N +++AQQ + + + QG D A+
Sbjct: 728 ESANLMPGSENVSENAQQPTRSPSSDLLSILQGVTDRSSPAV 769
>At5g44310 late embryogenesis abundant protein-like
Length = 331
Score = 24.6 bits (52), Expect = 9.4
Identities = 11/58 (18%), Positives = 24/58 (40%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
D ++ + + +A +KA ++ +N A+ + E + KA+ D K
Sbjct: 186 DFAEETKEKVNEGASRAADKAYDVKEKTKNYAEQTKDKVNEGASRAADKAEETKDKAK 243
>At4g14740 unknown protein
Length = 475
Score = 24.6 bits (52), Expect = 9.4
Identities = 17/52 (32%), Positives = 25/52 (47%), Gaps = 1/52 (1%)
Query: 11 AGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAI 62
A + G+ ++ A M A A A Q C EA + M A+ + A V SA+
Sbjct: 222 ASSSCGKDEQMAKTDMAVASAATLVAAQ-CVEAAEVMGAEREYLASVVSSAV 272
>At1g20760 unknown protein
Length = 1019
Score = 24.6 bits (52), Expect = 9.4
Identities = 10/42 (23%), Positives = 21/42 (49%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKA 51
Q G A G Q +A + + ++ + C++ G ++ +KA
Sbjct: 637 QGGSADGLLQVRADRIQSDLEELMKALTERCKKHGLEVNSKA 678
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.300 0.108 0.271
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 892,337
Number of Sequences: 26719
Number of extensions: 18353
Number of successful extensions: 109
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 66
Number of HSP's gapped (non-prelim): 43
length of query: 66
length of database: 11,318,596
effective HSP length: 42
effective length of query: 24
effective length of database: 10,196,398
effective search space: 244713552
effective search space used: 244713552
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0215b.3


