

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.11
(360 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
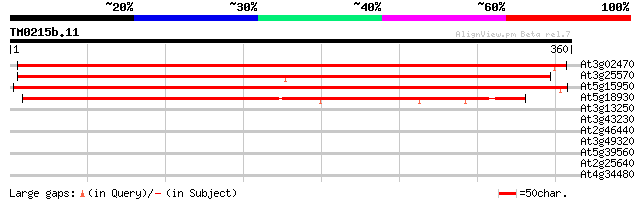
Score E
Sequences producing significant alignments: (bits) Value
At3g02470 S-adenosylmethionine decarboxylase (SAMdC) 442 e-124
At3g25570 S-adenosylmethionine decarboxylase, putative 437 e-123
At5g15950 S-adenosylmethionine decarboxylase (adoMetDC2) 436 e-123
At5g18930 S-adenosyl-L-methionine decarboxylase - like protein 259 2e-69
At3g13250 hypothetical protein 30 1.6
At3g43230 unknown protein 30 2.7
At2g46440 putative cyclic nucleotide-regulated ion channel protein 30 2.7
At3g49320 unknown protein 29 3.5
At5g39560 putative protein 29 4.5
At2g25640 putative transcription elongation factor S-II 29 4.5
At4g34480 putative protein (fragment) 28 5.9
>At3g02470 S-adenosylmethionine decarboxylase (SAMdC)
Length = 366
Score = 442 bits (1138), Expect = e-124
Identities = 224/363 (61%), Positives = 274/363 (74%), Gaps = 11/363 (3%)
Query: 6 SAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDS 65
SAIGFEGYEKRLE++F+E +F D +GLGLR L++ QLDEIL PA CTIV SLSND +DS
Sbjct: 4 SAIGFEGYEKRLEVTFFEPSIFQDSKGLGLRALTKSQLDEILTPAACTIVSSLSNDQLDS 63
Query: 66 YVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPEAQP 125
YVLSESS FVYPYKVIIKTCGTTKLLLSIP +LKLA L+++VKSV+YTRGSF+ P QP
Sbjct: 64 YVLSESSFFVYPYKVIIKTCGTTKLLLSIPPLLKLAGELSLSVKSVKYTRGSFLCPGGQP 123
Query: 126 FPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGS-SEAVYGLEM 184
FPHR+FSEEV+VL+ +F LG AY+MG+ D+++ WH+YAASA+ + + VY LEM
Sbjct: 124 FPHRSFSEEVSVLDGHFTQLGLNSVAYLMGNDDETKKWHVYAASAQDSSNCNNNVYTLEM 183
Query: 185 CMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSAIS 244
CMTGLD+E A+VF+KD MT NSGI KILP+S+ICDFEF+PCGYSMN IEG AIS
Sbjct: 184 CMTGLDREKAAVFYKDEADKTGSMTDNSGIRKILPKSEICDFEFEPCGYSMNSIEGDAIS 243
Query: 245 TIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKL-DKF 303
TIHVTPEDGFSYASFEAVGYD+ L L++LV RVL+CF P +FSVA+H + +
Sbjct: 244 TIHVTPEDGFSYASFEAVGYDFNTLDLSQLVTRVLSCFEPKQFSVAVHSSVGANSYKPEI 303
Query: 304 PLDIEGYYCDKRGTEELG-AGGAVMFHSFVRVDG-CASPKSILKCCW-------SEDEND 354
+D+E Y C +R E LG G VM+ +F ++ C SP+S LKC W SEDE D
Sbjct: 304 TVDLEDYGCRERTFESLGEESGTVMYQTFEKLGKYCGSPRSTLKCEWSSNNSCSSEDEKD 363
Query: 355 EEV 357
E +
Sbjct: 364 EGI 366
>At3g25570 S-adenosylmethionine decarboxylase, putative
Length = 349
Score = 437 bits (1125), Expect = e-123
Identities = 222/346 (64%), Positives = 267/346 (77%), Gaps = 4/346 (1%)
Query: 6 SAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDS 65
SA GFEG+EKRLEISF+E F DP+G LR L++ QLDEIL PA+CTIV SL+N +VDS
Sbjct: 4 SATGFEGFEKRLEISFFETTDFLDPQGKSLRSLTKSQLDEILTPAECTIVSSLTNSFVDS 63
Query: 66 YVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPEAQP 125
YVLSESSLFVYPYK+IIKTCGTTKLLLSIP IL+LADSL + VKSVRYTRGSFIFP AQ
Sbjct: 64 YVLSESSLFVYPYKIIIKTCGTTKLLLSIPHILRLADSLCLTVKSVRYTRGSFIFPGAQS 123
Query: 126 FPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKS-QIWHIYAASAEPKGS--SEAVYGL 182
+PHR+FSEEVA+L++YFG L +G +A+VMG D + Q WH+Y+AS+ + + + VY L
Sbjct: 124 YPHRSFSEEVALLDDYFGKLNAGSKAFVMGGSDNNPQRWHVYSASSTEESAVCDKPVYTL 183
Query: 183 EMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSA 242
EMCMTGLD ASVFFK N+ SA+ MT +SGI ILP S+ICDF F+PCGYSMN IEG A
Sbjct: 184 EMCMTGLDNIKASVFFKTNSVSASEMTISSGIRNILPGSEICDFNFEPCGYSMNSIEGDA 243
Query: 243 ISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKL-D 301
+STIHVTPEDGFSYASFE VGYD + L+ ELV+RVL CF P EFSVA+H ++ E L
Sbjct: 244 VSTIHVTPEDGFSYASFETVGYDLKALNFKELVDRVLVCFGPEEFSVAVHANLGTEVLAS 303
Query: 302 KFPLDIEGYYCDKRGTEELGAGGAVMFHSFVRVDGCASPKSILKCC 347
D+ GY+ +R EELG GG+V++ FV+ C SPKS L C
Sbjct: 304 DCVADVNGYFSQERELEELGLGGSVLYQRFVKTVECCSPKSTLGFC 349
>At5g15950 S-adenosylmethionine decarboxylase (adoMetDC2)
Length = 362
Score = 436 bits (1121), Expect = e-123
Identities = 216/361 (59%), Positives = 272/361 (74%), Gaps = 5/361 (1%)
Query: 3 LTSSAIGFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDY 62
+ SAIGFEGYEKRLE++F+E G+F D +G GLR L++ Q+DEIL PA+CTIV SLSND
Sbjct: 1 MAMSAIGFEGYEKRLEVTFFEPGLFLDTQGKGLRALAKSQIDEILQPAECTIVSSLSNDQ 60
Query: 63 VDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPE 122
+DSYVLSESSLF++PYK++IKTCGTTKLLLSI +L+LA L++ VK+VRYTRGSF+ P
Sbjct: 61 LDSYVLSESSLFIFPYKIVIKTCGTTKLLLSIEPLLRLAGELSLDVKAVRYTRGSFLCPG 120
Query: 123 AQPFPHRNFSEEVAVLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGL 182
QPFPHRNFSEEV+VL+ +F LG AY+MG+ D+++ WH+Y+AS+ + VY L
Sbjct: 121 GQPFPHRNFSEEVSVLDGHFAKLGLSSVAYLMGNDDETKKWHVYSASSANSNNKNNVYTL 180
Query: 183 EMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPCGYSMNGIEGSA 242
EMCMTGLDK+ ASVF+K+ +SSA MT NSGI KILPQS ICDFEF+PCGYSMN IEG A
Sbjct: 181 EMCMTGLDKDKASVFYKNESSSAGSMTDNSGIRKILPQSQICDFEFEPCGYSMNSIEGDA 240
Query: 243 ISTIHVTPEDGFSYASFEAVGYDYEDLSLTELVERVLACFHPAEFSVALHMDMHGEKLDK 302
ISTIHVTPEDGFSYASFEAVGYD+ + L+ LV +VL CF P +FSVA+H + + D
Sbjct: 241 ISTIHVTPEDGFSYASFEAVGYDFTTMDLSHLVSKVLTCFKPKQFSVAVHSTVAQKSYDS 300
Query: 303 -FPLDIEGYYCDKRGTEELG-AGGAVMFHSFVRVDG-CASPKSILKCCWSEDE--NDEEV 357
+D++ Y C + E LG G VM+ F ++ C SP+S LKC WS + N E+
Sbjct: 301 GLSVDLDDYGCKESTMESLGEERGTVMYQRFEKLGRYCGSPRSTLKCEWSSNSSCNSEDE 360
Query: 358 K 358
K
Sbjct: 361 K 361
>At5g18930 S-adenosyl-L-methionine decarboxylase - like protein
Length = 347
Score = 259 bits (662), Expect = 2e-69
Identities = 150/342 (43%), Positives = 212/342 (61%), Gaps = 23/342 (6%)
Query: 9 GFEGYEKRLEISFYERGVFADPEGLGLRELSRDQLDEILNPAQCTIVGSLSNDYVDSYVL 68
GFEG+EKRLE+ F++ +GLR + + LD++LN QCT+V +++N D+YVL
Sbjct: 5 GFEGFEKRLELRFFDDDKPITKNPMGLRLIDFESLDQVLNEVQCTVVSAVANRSFDAYVL 64
Query: 69 SESSLFVYPYKVIIKTCGTTKLLLSIPVILKLADSLNIAVKSVRYTRGSFIFPEAQPFPH 128
SESSLFVYP K+IIKTCGTT+LL SI ++ LA +L + +++ RY+RGSFIFP+AQPFP+
Sbjct: 65 SESSLFVYPTKIIIKTCGTTQLLKSIRPLIHLARNLGLTLRACRYSRGSFIFPKAQPFPY 124
Query: 129 RNFSEEVAVLNNYFGNLGSGGQAYVMGDPDK-SQIWHIYAASAEPKGSSEAVYGLEMCMT 187
+F +EV V+ +A VM + S+ WH++ ASA+ + S E V +E+CMT
Sbjct: 125 TSFKDEVIVVEESLPKSLCYRKASVMTPSNNPSRAWHVFTASADVE-SDEHVVVVEVCMT 183
Query: 188 GLDKESASVFF------KDNTSSAAL-MTTNSGINKILPQSDICDFEFDPCGYSMNGIEG 240
LD+ +A FF K+N+ SA MT SGI+ I + ICDF FDPCGYSMNG++G
Sbjct: 184 ELDRVNARSFFKRKGDEKNNSDSAGKEMTRLSGIDNINANAYICDFAFDPCGYSMNGVDG 243
Query: 241 SAISTIHVTPEDGFSYASFEA--VGYDYEDLSLTELVERVLACFHPAEFSVAL------- 291
STIHVTPEDGFSYASFE YD ++E++ R + F P++ S+A
Sbjct: 244 DRYSTIHVTPEDGFSYASFECGLSLYDNGHEDISEVLSRAIDVFRPSDVSIATTYGGEDY 303
Query: 292 --HMDMHGEKLDKFPLDIEGYYCDKRGTEELGAGGAVMFHSF 331
+ E++ LD++ C R +E G V++ SF
Sbjct: 304 NHEVTKRVERVLAKKLDLK---CRSRLMDEFPGSGTVVYQSF 342
>At3g13250 hypothetical protein
Length = 1419
Score = 30.4 bits (67), Expect = 1.6
Identities = 24/94 (25%), Positives = 37/94 (38%), Gaps = 13/94 (13%)
Query: 137 VLNNYFGNLGSGGQAYVMGDPDKSQIWHIYAASAEPKGSSEAVYGLEMCMTGLDKESASV 196
+ N F +LG Y G K+ IW AA+ KG ++C+ AS+
Sbjct: 951 ITNAVFNDLGGVFFVYGFGGTGKTFIWKTLAAAVRSKG--------QICLNVASSGIASL 1002
Query: 197 FFKDNTS-----SAALMTTNSGINKILPQSDICD 225
+ + S L + KI P+SD+ D
Sbjct: 1003 LLEGGRTAHSRFSIPLNPDEFSVCKIKPKSDLAD 1036
>At3g43230 unknown protein
Length = 485
Score = 29.6 bits (65), Expect = 2.7
Identities = 25/89 (28%), Positives = 45/89 (50%), Gaps = 15/89 (16%)
Query: 41 DQLDEILNPAQCTIVGSLSNDYVDSYVLSESSLFVYPYKVIIKTCGTTKLLLSIPVILKL 100
D E L+P QC ++ S+SN ++ V + V+ TC ++ L++PV L +
Sbjct: 230 DSCYERLDPLQCVLINSISN-----------AVQVAKHDVVDWTC--SRGWLNLPVGLSM 276
Query: 101 ADSLNIAVKSVR-YTRGSFIFPEAQPFPH 128
D + A ++R Y + + + PE + PH
Sbjct: 277 EDEIYKAANTLRGYCQVARLDPE-KSIPH 304
>At2g46440 putative cyclic nucleotide-regulated ion channel protein
Length = 588
Score = 29.6 bits (65), Expect = 2.7
Identities = 26/102 (25%), Positives = 45/102 (43%), Gaps = 8/102 (7%)
Query: 89 KLLLSIPVIL----KLADSLNIAVKSVRYTRGSFIFPEAQPFPHRNFSEEVAVLN--NYF 142
KLL +P++ +L D+L +K+V YT S+I E +P F +++ Y
Sbjct: 395 KLLKKVPLLQAMDDQLLDALCARLKTVHYTEKSYIVREGEPVEDMLFIMRGNLISTTTYG 454
Query: 143 GNLGSGGQAYVMGDPDKSQI--WHIYAASAEPKGSSEAVYGL 182
G G ++ + W +Y+ S++ SS V L
Sbjct: 455 GRTGFFNSVDLIAGDSCGDLLTWALYSLSSQFPISSRTVQAL 496
>At3g49320 unknown protein
Length = 354
Score = 29.3 bits (64), Expect = 3.5
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 1/44 (2%)
Query: 152 YVMGDPDKSQIWHIYAASAEPKGSSEAVYGLEMCMTGLDKESAS 195
YV+ D+S+ W I A S P+ E+ L + GL+KE S
Sbjct: 276 YVLYQDDRSENWRIQAVSVSPE-RFESRKALPLAWRGLEKEKLS 318
>At5g39560 putative protein
Length = 395
Score = 28.9 bits (63), Expect = 4.5
Identities = 27/107 (25%), Positives = 46/107 (42%), Gaps = 6/107 (5%)
Query: 200 DNTSSAALMTTNSGI--NKILPQSDICDFEFDPCGYSMNGIEGSAISTIHV-TPEDGFSY 256
D+ ++ L+ N K+ + I D+ ++P + + S+I + V + Y
Sbjct: 229 DSQTAVRLINVNPNALEGKLYVAAQINDYTYEPKDDTWKVVSKSSIRRVKVWCVIENVMY 288
Query: 257 ASFEAV--GYDYEDLSLTELVERVLACFHPAE-FSVALHMDMHGEKL 300
A + YDYED E+ C+HP FS A + +G KL
Sbjct: 289 ACHDVFLRWYDYEDRRWREIQGLEELCYHPTRGFSGAERIVNYGGKL 335
>At2g25640 putative transcription elongation factor S-II
Length = 643
Score = 28.9 bits (63), Expect = 4.5
Identities = 15/58 (25%), Positives = 26/58 (43%)
Query: 174 GSSEAVYGLEMCMTGLDKESASVFFKDNTSSAALMTTNSGINKILPQSDICDFEFDPC 231
GS + L+ M+ +D ES S F +T ++ N+ + ++L S D C
Sbjct: 439 GSLPPIVSLDEFMSSIDSESPSGFLSSDTEKKPSVSDNNDVEEVLVSSPKESANIDLC 496
>At4g34480 putative protein (fragment)
Length = 356
Score = 28.5 bits (62), Expect = 5.9
Identities = 29/108 (26%), Positives = 51/108 (46%), Gaps = 14/108 (12%)
Query: 102 DSLNIAVKSVRYTRGSFIFPEAQPFPHRNFSEEVAVLNN---YFGNL-----GSGGQAYV 153
D+++ A+KS+ + + + E + +E A ++N Y GNL G +
Sbjct: 244 DAVHSALKSMGFEKVEIVVAETGWASRGDANEVGASVDNAKAYNGNLIAHLRSMVGTPLM 303
Query: 154 MGDPDKSQIWHIYAASAEPKGSSEAVYGL---EMCM---TGLDKESAS 195
G P + I+ +Y + +P SSE +GL ++ M GL K S+S
Sbjct: 304 PGKPVDTYIFALYDENLKPGPSSERAFGLFKTDLSMVYDVGLAKSSSS 351
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,411,445
Number of Sequences: 26719
Number of extensions: 366572
Number of successful extensions: 796
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 780
Number of HSP's gapped (non-prelim): 11
length of query: 360
length of database: 11,318,596
effective HSP length: 101
effective length of query: 259
effective length of database: 8,619,977
effective search space: 2232574043
effective search space used: 2232574043
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0215b.11


