

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215a.1
(623 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
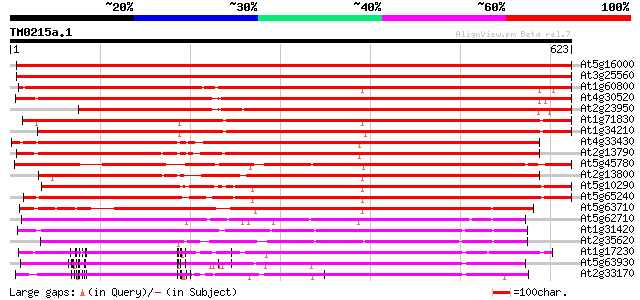
At5g16000 receptor protein kinase-like protein 967 0.0
At3g25560 unknown protein 899 0.0
At1g60800 Putative Serine/Threonine protein kinase 765 0.0
At4g30520 receptor-like kinase homolog 744 0.0
At2g23950 putative LRR receptor protein kinase 679 0.0
At1g71830 protein kinase, putative 626 e-179
At1g34210 putative protein 620 e-178
At4g33430 somatic embryogenesis receptor-like kinase 3 (SERK3) 605 e-173
At2g13790 putative receptor protein kinase (F13J11.14/At2g13790) 582 e-166
At5g45780 receptor-like protein kinase 561 e-160
At2g13800 receptor-like protein kinase 556 e-159
At5g10290 protein serine/threonine kinase-like protein 539 e-153
At5g65240 receptor-like protein kinase 535 e-152
At5g63710 receptor-like protein kinase 450 e-126
At5g62710 receptor - like protein kinase - like 330 2e-90
At1g31420 unknown protein 312 4e-85
At2g35620 putative receptor-like protein kinase 299 4e-81
At1g17230 putative leucine-rich receptor protein kinase 297 1e-80
At5g63930 receptor-like protein kinase 293 2e-79
At2g33170 putative receptor-like protein kinase 290 1e-78
>At5g16000 receptor protein kinase-like protein
Length = 638
Score = 967 bits (2500), Expect = 0.0
Identities = 491/625 (78%), Positives = 543/625 (86%), Gaps = 9/625 (1%)
Query: 8 SVLCFAIFL--VWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWA 65
S CF FL + SS LLSPKGVNFEVQALM IK LHDP GVLDNWD DAVDPCSW
Sbjct: 14 SFFCFLGFLCLLCSSVHGLLSPKGVNFEVQALMDIKASLHDPHGVLDNWDRDAVDPCSWT 73
Query: 66 MVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQT 125
MVTCSS+N VI LGTPSQNLSGTLS +I NLTNL+ VLLQNNNI G IP+ +G+L +L+T
Sbjct: 74 MVTCSSENFVIGLGTPSQNLSGTLSPSITNLTNLRIVLLQNNNIKGKIPAEIGRLTRLET 133
Query: 126 LDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPV 185
LDLS+N F GE+P S+G+L+SLQYLRLNNNSL G P SL+NMTQL+FLDLSYNNLSGPV
Sbjct: 134 LDLSDNFFHGEIPFSVGYLQSLQYLRLNNNSLSGVFPLSLSNMTQLAFLDLSYNNLSGPV 193
Query: 186 PRILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGL 245
PR AK+FSI+GNPL+C TG EP+C+G TL+PMSMNLN T L +G + HKMAIA G
Sbjct: 194 PRFAAKTFSIVGNPLICPTGTEPDCNGTTLIPMSMNLNQTGVPLYAGGSRNHKMAIAVGS 253
Query: 246 SLGCLCLILLGFGAFLWWRHKHNQQAFFDVKD-RHHEEVYLGNLKRFPFRELQIATHNFS 304
S+G + LI + G FLWWR +HNQ FFDVKD HHEEV LGNL+RF FRELQIAT+NFS
Sbjct: 254 SVGTVSLIFIAVGLFLWWRQRHNQNTFFDVKDGNHHEEVSLGNLRRFGFRELQIATNNFS 313
Query: 305 NKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLY 364
+KN+LGKGG+GNVYKG+L D T+VAVKRLKDG A+GGEIQFQTEVEMISLAVHRNLL+LY
Sbjct: 314 SKNLLGKGGYGNVYKGILGDSTVVAVKRLKDGGALGGEIQFQTEVEMISLAVHRNLLRLY 373
Query: 365 GFCMTPTERLLVYPYMSNGSVALRLKGKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIH 424
GFC+T TE+LLVYPYMSNGSVA R+K KPVLDW RK IA+GAARGL+YLHEQCDPKIIH
Sbjct: 374 GFCITQTEKLLVYPYMSNGSVASRMKAKPVLDWSIRKRIAIGAARGLVYLHEQCDPKIIH 433
Query: 425 RDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTD 484
RDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTD
Sbjct: 434 RDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTD 493
Query: 485 VFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDL--KSNYDQI 542
VFGFGILLLEL+TGQRA EFGKAANQKG MLDWVKKIH EKKLELLVDK+L K +YD+I
Sbjct: 494 VFGFGILLLELVTGQRAFEFGKAANQKGVMLDWVKKIHQEKKLELLVDKELLKKKSYDEI 553
Query: 543 ELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRADT-SKC--RPHE-SS 598
EL+EMV+VALLCTQYLPGHRPKMSEVVRMLEGDGLAE+WEASQR+D+ SKC R +E S
Sbjct: 554 ELDEMVRVALLCTQYLPGHRPKMSEVVRMLEGDGLAEKWEASQRSDSVSKCSNRINELMS 613
Query: 599 SSDRYSDLTDDSLLLVQAMELSGPR 623
SSDRYSDLTDDS LLVQAMELSGPR
Sbjct: 614 SSDRYSDLTDDSSLLVQAMELSGPR 638
>At3g25560 unknown protein
Length = 635
Score = 899 bits (2323), Expect = 0.0
Identities = 452/620 (72%), Positives = 525/620 (83%), Gaps = 5/620 (0%)
Query: 8 SVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMV 67
S F SS+SA L+ KGVNFEV AL+GIK L DP GVL NWD AVDPCSW M+
Sbjct: 17 STFFFFFICFLSSSSAELTDKGVNFEVVALIGIKSSLTDPHGVLMNWDDTAVDPCSWNMI 76
Query: 68 TCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLD 127
TCS D VI L PSQNLSGTLS++IGNLTNLQTVLLQNN I+G IP +GKL KL+TLD
Sbjct: 77 TCS-DGFVIRLEAPSQNLSGTLSSSIGNLTNLQTVLLQNNYITGNIPHEIGKLMKLKTLD 135
Query: 128 LSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
LS N+F+G++P +L + ++LQYLR+NNNSL G P SLANMTQL+FLDLSYNNLSGPVPR
Sbjct: 136 LSTNNFTGQIPFTLSYSKNLQYLRVNNNSLTGTIPSSLANMTQLTFLDLSYNNLSGPVPR 195
Query: 188 ILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSL 247
LAK+F+++GN +C TG E +C+G PMS+ LN++++ G K K+A+ FG+SL
Sbjct: 196 SLAKTFNVMGNSQICPTGTEKDCNGTQPKPMSITLNSSQNKSSDGGTKNRKIAVVFGVSL 255
Query: 248 GCLCLILLGFGAFLWWRHKHNQQA-FFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNK 306
C+CL+++GFG LWWR +HN+Q FFD+ +++ EE+ LGNL+RF F+ELQ AT NFS+K
Sbjct: 256 TCVCLLIIGFGFLLWWRRRHNKQVLFFDINEQNKEEMCLGNLRRFNFKELQSATSNFSSK 315
Query: 307 NILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGF 366
N++GKGGFGNVYKG L DG+++AVKRLKD N GGE+QFQTE+EMISLAVHRNLL+LYGF
Sbjct: 316 NLVGKGGFGNVYKGCLHDGSIIAVKRLKDINNGGGEVQFQTELEMISLAVHRNLLRLYGF 375
Query: 367 CMTPTERLLVYPYMSNGSVALRLKGKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRD 426
C T +ERLLVYPYMSNGSVA RLK KPVLDWGTRK IALGA RGLLYLHEQCDPKIIHRD
Sbjct: 376 CTTSSERLLVYPYMSNGSVASRLKAKPVLDWGTRKRIALGAGRGLLYLHEQCDPKIIHRD 435
Query: 427 VKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVF 486
VKAANILLDDY EAVVGDFGLAKLLDH++SHVTTAVRGTVGHIAPEYLSTGQSSEKTDVF
Sbjct: 436 VKAANILLDDYFEAVVGDFGLAKLLDHEESHVTTAVRGTVGHIAPEYLSTGQSSEKTDVF 495
Query: 487 GFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQIELEE 546
GFGILLLELITG RALEFGKAANQ+GA+LDWVKK+ EKKLE +VDKDLKSNYD+IE+EE
Sbjct: 496 GFGILLLELITGLRALEFGKAANQRGAILDWVKKLQQEKKLEQIVDKDLKSNYDRIEVEE 555
Query: 547 MVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEA-SQRADTSK--CRPHESSSSDRY 603
MVQVALLCTQYLP HRPKMSEVVRMLEGDGL E+WEA SQRA+T++ +P+E SSS+RY
Sbjct: 556 MVQVALLCTQYLPIHRPKMSEVVRMLEGDGLVEKWEASSQRAETNRSYSKPNEFSSSERY 615
Query: 604 SDLTDDSLLLVQAMELSGPR 623
SDLTDDS +LVQAMELSGPR
Sbjct: 616 SDLTDDSSVLVQAMELSGPR 635
>At1g60800 Putative Serine/Threonine protein kinase
Length = 632
Score = 765 bits (1975), Expect = 0.0
Identities = 391/627 (62%), Positives = 486/627 (77%), Gaps = 18/627 (2%)
Query: 10 LCFAIFLVW--SSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMV 67
L F +F VW +SA LSP GVN+EV AL+ +K L+DP VL+NWD ++VDPCSW MV
Sbjct: 11 LGFLVF-VWFFDISSATLSPTGVNYEVTALVAVKNELNDPYKVLENWDVNSVDPCSWRMV 69
Query: 68 TCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLD 127
+C+ D V SL PSQ+LSGTLS IGNLT LQ+V+LQNN I+GPIP +G+L KLQ+LD
Sbjct: 70 SCT-DGYVSSLDLPSQSLSGTLSPRIGNLTYLQSVVLQNNAITGPIPETIGRLEKLQSLD 128
Query: 128 LSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
LSNNSF+GE+P SLG L++L YLRLNNNSL+G CPESL+ + L+ +D+SYNNLSG +P+
Sbjct: 129 LSNNSFTGEIPASLGELKNLNYLRLNNNSLIGTCPESLSKIEGLTLVDISYNNLSGSLPK 188
Query: 188 ILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSL 247
+ A++F ++GN L+C NC + P+++ + +++ + H +A+AF S
Sbjct: 189 VSARTFKVIGNALICGPKAVSNCSAVP-EPLTLPQDGPDES--GTRTNGHHVALAFAASF 245
Query: 248 GCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKN 307
+ G FLWWR++ N+Q FFDV +++ EV LG+LKR+ F+EL+ AT++F++KN
Sbjct: 246 SAAFFVFFTSGMFLWWRYRRNKQIFFDVNEQYDPEVSLGHLKRYTFKELRSATNHFNSKN 305
Query: 308 ILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFC 367
ILG+GG+G VYKG L+DGTLVAVKRLKD N GGE+QFQTEVE ISLA+HRNLL+L GFC
Sbjct: 306 ILGRGGYGIVYKGHLNDGTLVAVKRLKDCNIAGGEVQFQTEVETISLALHRNLLRLRGFC 365
Query: 368 MTPTERLLVYPYMSNGSVALRLK----GKPVLDWGTRKHIALGAARGLLYLHEQCDPKII 423
+ ER+LVYPYM NGSVA RLK G+P LDW RK IA+G ARGL+YLHEQCDPKII
Sbjct: 366 SSNQERILVYPYMPNGSVASRLKDNIRGEPALDWSRRKKIAVGTARGLVYLHEQCDPKII 425
Query: 424 HRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKT 483
HRDVKAANILLD+ EAVVGDFGLAKLLDH+DSHVTTAVRGTVGHIAPEYLSTGQSSEKT
Sbjct: 426 HRDVKAANILLDEDFEAVVGDFGLAKLLDHRDSHVTTAVRGTVGHIAPEYLSTGQSSEKT 485
Query: 484 DVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQIE 543
DVFGFGILLLELITGQ+AL+FG++A+QKG MLDWVKK+H E KL+ L+DKDL +D++E
Sbjct: 486 DVFGFGILLLELITGQKALDFGRSAHQKGVMLDWVKKLHQEGKLKQLIDKDLNDKFDRVE 545
Query: 544 LEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRA----DTSKCRPHESSS 599
LEE+VQVALLCTQ+ P HRPKMSEV++MLEGDGLAERWEA+Q P SS
Sbjct: 546 LEEIVQVALLCTQFNPSHRPKMSEVMKMLEGDGLAERWEATQNGTGEHQPPPLPPGMVSS 605
Query: 600 SDR---YSDLTDDSLLLVQAMELSGPR 623
S R YSD +S L+V+A+ELSGPR
Sbjct: 606 SPRVRYYSDYIQESSLVVEAIELSGPR 632
>At4g30520 receptor-like kinase homolog
Length = 648
Score = 744 bits (1920), Expect = 0.0
Identities = 393/644 (61%), Positives = 476/644 (73%), Gaps = 35/644 (5%)
Query: 7 VSVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAM 66
+ +L +FL +S+ + P+ N EV+AL+ I+ LHDP G L+NWD +VDPCSWAM
Sbjct: 13 IHLLYSFLFLCFSTLTLSSEPR--NPEVEALISIRNNLHDPHGALNNWDEFSVDPCSWAM 70
Query: 67 VTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTL 126
+TCS DNLVI LG PSQ+LSG LS +IGNLTNL+ V LQNNNISG IP LG LPKLQTL
Sbjct: 71 ITCSPDNLVIGLGAPSQSLSGGLSESIGNLTNLRQVSLQNNNISGKIPPELGFLPKLQTL 130
Query: 127 DLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
DLSNN FSG++P S+ L SLQYLRLNNNSL G P SL+ + LSFLDLSYNNLSGPVP
Sbjct: 131 DLSNNRFSGDIPVSIDQLSSLQYLRLNNNSLSGPFPASLSQIPHLSFLDLSYNNLSGPVP 190
Query: 187 RILAKSFSILGNPLVCATGKEPNCHG-ITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGL 245
+ A++F++ GNPL+C + C G I P+S++L+++ SG+ +++++AIA +
Sbjct: 191 KFPARTFNVAGNPLICRSNPPEICSGSINASPLSVSLSSS-----SGR-RSNRLAIALSV 244
Query: 246 SLGCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVY-LGNLKRFPFRELQIATHNFS 304
SLG + +++L G+F W+R K + ++ D+ E + LGNL+ F FREL + T FS
Sbjct: 245 SLGSVVILVLALGSFCWYRKKQRRLLILNLNDKQEEGLQGLGNLRSFTFRELHVYTDGFS 304
Query: 305 NKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLY 364
+KNILG GGFGNVY+G L DGT+VAVKRLKD N G+ QF+ E+EMISLAVH+NLL+L
Sbjct: 305 SKNILGAGGFGNVYRGKLGDGTMVAVKRLKDINGTSGDSQFRMELEMISLAVHKNLLRLI 364
Query: 365 GFCMTPTERLLVYPYMSNGSVALRLKGKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIH 424
G+C T ERLLVYPYM NGSVA +LK KP LDW RK IA+GAARGLLYLHEQCDPKIIH
Sbjct: 365 GYCATSGERLLVYPYMPNGSVASKLKSKPALDWNMRKRIAIGAARGLLYLHEQCDPKIIH 424
Query: 425 RDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTD 484
RDVKAANILLD+ EAVVGDFGLAKLL+H DSHVTTAVRGTVGHIAPEYLSTGQSSEKTD
Sbjct: 425 RDVKAANILLDECFEAVVGDFGLAKLLNHADSHVTTAVRGTVGHIAPEYLSTGQSSEKTD 484
Query: 485 VFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQIEL 544
VFGFGILLLELITG RALEFGK +QKGAML+WV+K+H E K+E L+D++L +NYD+IE+
Sbjct: 485 VFGFGILLLELITGLRALEFGKTVSQKGAMLEWVRKLHEEMKVEELLDRELGTNYDKIEV 544
Query: 545 EEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQR-----------------A 587
EM+QVALLCTQYLP HRPKMSEVV MLEGDGLAERW AS +
Sbjct: 545 GEMLQVALLCTQYLPAHRPKMSEVVLMLEGDGLAERWAASHNHSHFYHANISFKTISSLS 604
Query: 588 DTSKCR--------PHESSSSDRYSDLTDDSLLLVQAMELSGPR 623
TS R ++ S + D D L AMELSGPR
Sbjct: 605 TTSVSRLDAHCNDPTYQMFGSSAFDDDDDHQPLDSFAMELSGPR 648
>At2g23950 putative LRR receptor protein kinase
Length = 607
Score = 679 bits (1751), Expect = 0.0
Identities = 358/564 (63%), Positives = 428/564 (75%), Gaps = 24/564 (4%)
Query: 77 SLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGE 136
S G PSQ+LSGTLS +IGNLTNL+ V LQNNNISG IP + LPKLQTLDLSNN FSGE
Sbjct: 51 SRGAPSQSLSGTLSGSIGNLTNLRQVSLQNNNISGKIPPEICSLPKLQTLDLSNNRFSGE 110
Query: 137 VPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSIL 196
+P S+ L +LQYLRLNNNSL G P SL+ + LSFLDLSYNNL GPVP+ A++F++
Sbjct: 111 IPGSVNQLSNLQYLRLNNNSLSGPFPASLSQIPHLSFLDLSYNNLRGPVPKFPARTFNVA 170
Query: 197 GNPLVCATGKEPNCHG-ITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILL 255
GNPL+C C G I+ P+S++L ++ SG+ +T+ +A+A G+SLG ++L
Sbjct: 171 GNPLICKNSLPEICSGSISASPLSVSLRSS-----SGR-RTNILAVALGVSLGFAVSVIL 224
Query: 256 GFGAFLWWRHKHNQQAFFDVKDRHHEEVY-LGNLKRFPFRELQIATHNFSNKNILGKGGF 314
G F+W+R K + + D+ E + LGNL+ F FREL +AT FS+K+ILG GGF
Sbjct: 225 SLG-FIWYRKKQRRLTMLRISDKQEEGLLGLGNLRSFTFRELHVATDGFSSKSILGAGGF 283
Query: 315 GNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERL 374
GNVY+G DGT+VAVKRLKD N G QF+TE+EMISLAVHRNLL+L G+C + +ERL
Sbjct: 284 GNVYRGKFGDGTVVAVKRLKDVNGTSGNSQFRTELEMISLAVHRNLLRLIGYCASSSERL 343
Query: 375 LVYPYMSNGSVALRLKGKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILL 434
LVYPYMSNGSVA RLK KP LDW TRK IA+GAARGL YLHEQCDPKIIHRDVKAANILL
Sbjct: 344 LVYPYMSNGSVASRLKAKPALDWNTRKKIAIGAARGLFYLHEQCDPKIIHRDVKAANILL 403
Query: 435 DDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLE 494
D+Y EAVVGDFGLAKLL+H+DSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLE
Sbjct: 404 DEYFEAVVGDFGLAKLLNHEDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLE 463
Query: 495 LITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLC 554
LITG RALEFGK+ +QKGAML+WV+K+H E K+E LVD++L + YD+IE+ EM+QVALLC
Sbjct: 464 LITGMRALEFGKSVSQKGAMLEWVRKLHKEMKVEELVDRELGTTYDRIEVGEMLQVALLC 523
Query: 555 TQYLPGHRPKMSEVVRMLEGDGLAERWEASQ------------RADTSKCRPHESS---S 599
TQ+LP HRPKMSEVV+MLEGDGLAERW AS R TS +++
Sbjct: 524 TQFLPAHRPKMSEVVQMLEGDGLAERWAASHDHSHFYHANMSYRTITSTDGNNQTKHLFG 583
Query: 600 SDRYSDLTDDSLLLVQAMELSGPR 623
S + D D+ L AMELSGPR
Sbjct: 584 SSGFEDEDDNQALDSFAMELSGPR 607
>At1g71830 protein kinase, putative
Length = 625
Score = 626 bits (1614), Expect = e-179
Identities = 352/625 (56%), Positives = 422/625 (67%), Gaps = 20/625 (3%)
Query: 15 FLVWSSASALLSPK------GVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVT 68
++V+ S +L P N E AL ++ L DP VL +WD V+PC+W VT
Sbjct: 5 YVVFILLSLILLPNHSLWLASANLEGDALHTLRVTLVDPNNVLQSWDPTLVNPCTWFHVT 64
Query: 69 CSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDL 128
C+++N VI + + LSG L +G L NLQ + L +NNI+GPIPS LG L L +LDL
Sbjct: 65 CNNENSVIRVDLGNAELSGHLVPELGVLKNLQYLELYSNNITGPIPSNLGNLTNLVSLDL 124
Query: 129 SNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR- 187
NSFSG +P SLG L L++LRLNNNSL G P SL N+T L LDLS N LSG VP
Sbjct: 125 YLNSFSGPIPESLGKLSKLRFLRLNNNSLTGSIPMSLTNITTLQVLDLSNNRLSGSVPDN 184
Query: 188 ---ILAKSFSILGNPLVCATGKEPNCHGIT-LMPMSMNLNNTEDALPSGKPKTHKMAIAF 243
L S N +C C G P + + PSG T AIA
Sbjct: 185 GSFSLFTPISFANNLDLCGPVTSHPCPGSPPFSPPPPFIQPPPVSTPSGYGITG--AIAG 242
Query: 244 GLSLGCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNF 303
G++ G L AF WWR + FFDV EV+LG LKRF RELQ+A+ F
Sbjct: 243 GVAAGAALLFAAPAIAFAWWRRRKPLDIFFDVPAEEDPEVHLGQLKRFSLRELQVASDGF 302
Query: 304 SNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKL 363
SNKNILG+GGFG VYKG L+DGTLVAVKRLK+ GGE+QFQTEVEMIS+AVHRNLL+L
Sbjct: 303 SNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLLRL 362
Query: 364 YGFCMTPTERLLVYPYMSNGSVALRLK----GKPVLDWGTRKHIALGAARGLLYLHEQCD 419
GFCMTPTERLLVYPYM+NGSVA L+ +P LDW TRK IALG+ARGL YLH+ CD
Sbjct: 363 RGFCMTPTERLLVYPYMANGSVASCLRERPPSQPPLDWPTRKRIALGSARGLSYLHDHCD 422
Query: 420 PKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQS 479
PKIIHRDVKAANILLD+ EAVVGDFGLAKL+D++D+HVTTAVRGT+GHIAPEYLSTG+S
Sbjct: 423 PKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKS 482
Query: 480 SEKTDVFGFGILLLELITGQRALEFGKAANQKGAM-LDWVKKIHLEKKLELLVDKDLKSN 538
SEKTDVFG+GI+LLELITGQRA + + AN M LDWVK + EKKLE+LVD DL++N
Sbjct: 483 SEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEMLVDPDLQTN 542
Query: 539 YDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRADTSKCRPHESS 598
Y++ ELE+++QVALLCTQ P RPKMSEVVRMLEGDGLAE+W+ Q+ + R
Sbjct: 543 YEERELEQVIQVALLCTQGSPMERPKMSEVVRMLEGDGLAEKWDEWQKVEI--LREEIDL 600
Query: 599 SSDRYSDLTDDSLLLVQAMELSGPR 623
S + SD DS + A+ELSGPR
Sbjct: 601 SPNPNSDWILDSTYNLHAVELSGPR 625
>At1g34210 putative protein
Length = 628
Score = 620 bits (1599), Expect = e-178
Identities = 344/603 (57%), Positives = 412/603 (68%), Gaps = 14/603 (2%)
Query: 31 NFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCSSDNLVISLGTPSQNLSGTLS 90
N E AL ++ L DP VL +WD V+PC+W VTC+++N VI + + +LSG L
Sbjct: 30 NMEGDALHSLRANLVDPNNVLQSWDPTLVNPCTWFHVTCNNENSVIRVDLGNADLSGQLV 89
Query: 91 ATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQYL 150
+G L NLQ + L +NNI+GP+PS LG L L +LDL NSF+G +P SLG L L++L
Sbjct: 90 PQLGQLKNLQYLELYSNNITGPVPSDLGNLTNLVSLDLYLNSFTGPIPDSLGKLFKLRFL 149
Query: 151 RLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR----ILAKSFSILGNPLVCATGK 206
RLNNNSL G P SL N+ L LDLS N LSG VP L S N +C
Sbjct: 150 RLNNNSLTGPIPMSLTNIMTLQVLDLSNNRLSGSVPDNGSFSLFTPISFANNLDLCGPVT 209
Query: 207 EPNCHGIT-LMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGAFLWWRH 265
C G P + P G T AIA G++ G L AF WWR
Sbjct: 210 SRPCPGSPPFSPPPPFIPPPIVPTPGGYSATG--AIAGGVAAGAALLFAAPALAFAWWRR 267
Query: 266 KHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDG 325
+ Q+ FFDV EV+LG LKRF RELQ+AT +FSNKNILG+GGFG VYKG L+DG
Sbjct: 268 RKPQEFFFDVPAEEDPEVHLGQLKRFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADG 327
Query: 326 TLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSV 385
TLVAVKRLK+ GGE+QFQTEVEMIS+AVHRNLL+L GFCMTPTERLLVYPYM+NGSV
Sbjct: 328 TLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSV 387
Query: 386 ALRLKGKP----VLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAV 441
A L+ +P L W R+ IALG+ARGL YLH+ CDPKIIHRDVKAANILLD+ EAV
Sbjct: 388 ASCLRERPPSQLPLAWSIRQQIALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAV 447
Query: 442 VGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRA 501
VGDFGLA+L+D++D+HVTTAVRGT+GHIAPEYLSTG+SSEKTDVFG+GI+LLELITGQRA
Sbjct: 448 VGDFGLARLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRA 507
Query: 502 LEFGKAANQKGAM-LDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPG 560
+ + AN M LDWVK + EKKLE+LVD DL+SNY + E+E+++QVALLCTQ P
Sbjct: 508 FDLARLANDDDVMLLDWVKGLLKEKKLEMLVDPDLQSNYTEAEVEQLIQVALLCTQSSPM 567
Query: 561 HRPKMSEVVRMLEGDGLAERWEASQRADTSKCRPHESSSSDRYSDLTDDSLLLVQAMELS 620
RPKMSEVVRMLEGDGLAE+W+ Q+ + R SS SD DS + AMELS
Sbjct: 568 ERPKMSEVVRMLEGDGLAEKWDEWQKVEV--LRQEVELSSHPTSDWILDSTDNLHAMELS 625
Query: 621 GPR 623
GPR
Sbjct: 626 GPR 628
>At4g33430 somatic embryogenesis receptor-like kinase 3 (SERK3)
Length = 615
Score = 605 bits (1561), Expect = e-173
Identities = 335/593 (56%), Positives = 407/593 (68%), Gaps = 21/593 (3%)
Query: 3 MPRGVSVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPC 62
M R + + CF F + +L G N E AL +K L DP VL +WD V PC
Sbjct: 1 MERRLMIPCF--FWLILVLDLVLRVSG-NAEGDALSALKNSLADPNKVLQSWDATLVTPC 57
Query: 63 SWAMVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPK 122
+W VTC+SDN V + + NLSG L +G L NLQ + L +NNI+G IP LG L +
Sbjct: 58 TWFHVTCNSDNSVTRVDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTE 117
Query: 123 LQTLDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLS 182
L +LDL N+ SG +P +LG L+ L++LRLNNNSL GE P SL + L LDLS N L+
Sbjct: 118 LVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLT 177
Query: 183 GPVPRILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKM--A 240
G +P + SFS+ P+ A K L P+ + P ++++ A
Sbjct: 178 GDIP--VNGSFSLF-TPISFANTK--------LTPLPASPPPPISPTPPSPAGSNRITGA 226
Query: 241 IAFGLSLGCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIAT 300
IA G++ G L + A WWR K Q FFDV EV+LG LKRF RELQ+A+
Sbjct: 227 IAGGVAAGAALLFAVPAIALAWWRRKKPQDHFFDVPAEEDPEVHLGQLKRFSLRELQVAS 286
Query: 301 HNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNL 360
NFSNKNILG+GGFG VYKG L+DGTLVAVKRLK+ GGE+QFQTEVEMIS+AVHRNL
Sbjct: 287 DNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRNL 346
Query: 361 LKLYGFCMTPTERLLVYPYMSNGSVAL----RLKGKPVLDWGTRKHIALGAARGLLYLHE 416
L+L GFCMTPTERLLVYPYM+NGSVA R + +P LDW R+ IALG+ARGL YLH+
Sbjct: 347 LRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLAYLHD 406
Query: 417 QCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLST 476
CDPKIIHRDVKAANILLD+ EAVVGDFGLAKL+D++D+HVTTAVRGT+GHIAPEYLST
Sbjct: 407 HCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLST 466
Query: 477 GQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAM-LDWVKKIHLEKKLELLVDKDL 535
G+SSEKTDVFG+G++LLELITGQRA + + AN M LDWVK + EKKLE LVD DL
Sbjct: 467 GKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDVDL 526
Query: 536 KSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRAD 588
+ NY E+E+++QVALLCTQ P RPKMSEVVRMLEGDGLAERWE Q+ +
Sbjct: 527 QGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEGDGLAERWEEWQKEE 579
>At2g13790 putative receptor protein kinase (F13J11.14/At2g13790)
Length = 615
Score = 582 bits (1499), Expect = e-166
Identities = 328/590 (55%), Positives = 404/590 (67%), Gaps = 24/590 (4%)
Query: 8 SVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLH--DP-RGVLDNWDGDAVDPCSW 64
S+LCF L+ + + ++ N E AL +K L DP VL +WD V PC+W
Sbjct: 5 SLLCFLYLLLLFNFTLRVAG---NAEGDALTQLKNSLSSGDPANNVLQSWDATLVTPCTW 61
Query: 65 AMVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQ 124
VTC+ +N V + + LSG L +G L NLQ + L +NNI+G IP LG L +L
Sbjct: 62 FHVTCNPENKVTRVDLGNAKLSGKLVPELGQLLNLQYLELYSNNITGEIPEELGDLVELV 121
Query: 125 TLDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGP 184
+LDL NS SG +P SLG L L++LRLNNNSL GE P +L ++ QL LD+S N LSG
Sbjct: 122 SLDLYANSISGPIPSSLGKLGKLRFLRLNNNSLSGEIPMTLTSV-QLQVLDISNNRLSGD 180
Query: 185 VPRILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALP-SGKPKTHKMAIAF 243
+P + SFS+ P+ A + +T +P + + P SG T AIA
Sbjct: 181 IP--VNGSFSLF-TPISFAN------NSLTDLPEPPPTSTSPTPPPPSGGQMT--AAIAG 229
Query: 244 GLSLGCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNF 303
G++ G L + AF WW + Q FFDV EV+LG LKRF REL +AT NF
Sbjct: 230 GVAAGAALLFAVPAIAFAWWLRRKPQDHFFDVPAEEDPEVHLGQLKRFTLRELLVATDNF 289
Query: 304 SNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKL 363
SNKN+LG+GGFG VYKG L+DG LVAVKRLK+ GGE+QFQTEVEMIS+AVHRNLL+L
Sbjct: 290 SNKNVLGRGGFGKVYKGRLADGNLVAVKRLKEERTKGGELQFQTEVEMISMAVHRNLLRL 349
Query: 364 YGFCMTPTERLLVYPYMSNGSVAL----RLKGKPVLDWGTRKHIALGAARGLLYLHEQCD 419
GFCMTPTERLLVYPYM+NGSVA R +G P LDW RKHIALG+ARGL YLH+ CD
Sbjct: 350 RGFCMTPTERLLVYPYMANGSVASCLRERPEGNPALDWPKRKHIALGSARGLAYLHDHCD 409
Query: 420 PKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQS 479
KIIHRDVKAANILLD+ EAVVGDFGLAKL+++ DSHVTTAVRGT+GHIAPEYLSTG+S
Sbjct: 410 QKIIHRDVKAANILLDEEFEAVVGDFGLAKLMNYNDSHVTTAVRGTIGHIAPEYLSTGKS 469
Query: 480 SEKTDVFGFGILLLELITGQRALEFGKAANQKGAM-LDWVKKIHLEKKLELLVDKDLKSN 538
SEKTDVFG+G++LLELITGQ+A + + AN M LDWVK++ EKKLE LVD +L+
Sbjct: 470 SEKTDVFGYGVMLLELITGQKAFDLARLANDDDIMLLDWVKEVLKEKKLESLVDAELEGK 529
Query: 539 YDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRAD 588
Y + E+E+++Q+ALLCTQ RPKMSEVVRMLEGDGLAERWE Q+ +
Sbjct: 530 YVETEVEQLIQMALLCTQSSAMERPKMSEVVRMLEGDGLAERWEEWQKEE 579
>At5g45780 receptor-like protein kinase
Length = 570
Score = 561 bits (1447), Expect = e-160
Identities = 315/626 (50%), Positives = 407/626 (64%), Gaps = 75/626 (11%)
Query: 6 GVSVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWA 65
G+ V +++ S+ +LLSPKGVN+EV ALM +K + D + VL WD ++VDPC+W
Sbjct: 12 GIWVYYYSVLDSVSAMDSLLSPKGVNYEVAALMSVKNKMKDEKEVLSGWDINSVDPCTWN 71
Query: 66 MVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQT 125
MV CSS+ V+SL LLQNN ++GPIPS LG+L +L+T
Sbjct: 72 MVGCSSEGFVVSL------------------------LLQNNQLTGPIPSELGQLSELET 107
Query: 126 LDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPV 185
LDLS N FSGE+P SLG L L YLRL+ N L G+ P +A ++ LSFL
Sbjct: 108 LDLSGNRFSGEIPASLGFLTHLNYLRLSRNLLSGQVPHLVAGLSGLSFL----------- 156
Query: 186 PRILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGL 245
I+GN +C + C T + + L+ +++ K + ++ AFG+
Sbjct: 157 ---------IVGNAFLCGPASQELCSDATPVRNATGLSEKDNS----KHHSLVLSFAFGI 203
Query: 246 SLGCLCLILLGFGAFLWWRHK----HNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATH 301
+ + ++ F LW R + H QQ + E +G+LKRF FRE+Q AT
Sbjct: 204 VVAFIISLMFLFFWVLWHRSRLSRSHVQQDY---------EFEIGHLKRFSFREIQTATS 254
Query: 302 NFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLL 361
NFS KNILG+GGFG VYKG L +GT+VAVKRLKD GE+QFQTEVEMI LAVHRNLL
Sbjct: 255 NFSPKNILGQGGFGMVYKGYLPNGTVVAVKRLKD-PIYTGEVQFQTEVEMIGLAVHRNLL 313
Query: 362 KLYGFCMTPTERLLVYPYMSNGSVALRLKG----KPVLDWGTRKHIALGAARGLLYLHEQ 417
+L+GFCMTP ER+LVYPYM NGSVA RL+ KP LDW R IALGAARGL+YLHEQ
Sbjct: 314 RLFGFCMTPEERMLVYPYMPNGSVADRLRDNYGEKPSLDWNRRISIALGAARGLVYLHEQ 373
Query: 418 CDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTG 477
C+PKIIHRDVKAANILLD+ EA+VGDFGLAKLLD +DSHVTTAVRGT+GHIAPEYLSTG
Sbjct: 374 CNPKIIHRDVKAANILLDESFEAIVGDFGLAKLLDQRDSHVTTAVRGTIGHIAPEYLSTG 433
Query: 478 QSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKKIHLEKKLELLVDKDLKS 537
QSSEKTDVFGFG+L+LELITG + ++ G +KG +L WV+ + EK+ +VD+DLK
Sbjct: 434 QSSEKTDVFGFGVLILELITGHKMIDQGNGQVRKGMILSWVRTLKAEKRFAEMVDRDLKG 493
Query: 538 NYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRADTSKCRPHES 597
+D + LEE+V++ALLCTQ P RP+MS+V+++LE GL E+ E A
Sbjct: 494 EFDDLVLEEVVELALLCTQPHPNLRPRMSQVLKVLE--GLVEQCEGGYEA-------RAP 544
Query: 598 SSSDRYSDLTDDSLLLVQAMELSGPR 623
S S YS+ ++ +++A+ELSGPR
Sbjct: 545 SVSRNYSNGHEEQSFIIEAIELSGPR 570
>At2g13800 receptor-like protein kinase
Length = 601
Score = 556 bits (1434), Expect = e-159
Identities = 309/564 (54%), Positives = 373/564 (65%), Gaps = 33/564 (5%)
Query: 33 EVQALMGIKELLHD---PRGVLDNWDGDAVDPCSWAMVTCSSDNLVISLGTPSQNLSGTL 89
+V AL+ ++ L +L +W+ V PCSW VTC+++N V L S NLSG L
Sbjct: 27 QVDALIALRSSLSSGDHTNNILQSWNATHVTPCSWFHVTCNTENSVTRLDLGSANLSGEL 86
Query: 90 SATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQY 149
+ L NLQ + L NNNI+G IP LG L +L +LDL N+ SG +P SLG L L++
Sbjct: 87 VPQLAQLPNLQYLELFNNNITGEIPEELGDLMELVSLDLFANNISGPIPSSLGKLGKLRF 146
Query: 150 LRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSILGNPLVCATGKEPN 209
LRL NNSL GE P SL + L LD+S N LSG +P + SFS
Sbjct: 147 LRLYNNSLSGEIPRSLTALP-LDVLDISNNRLSGDIP--VNGSFS--------------- 188
Query: 210 CHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGAFLWWRHKHNQ 269
T M + N A PS P AI G++ G L L WW + Q
Sbjct: 189 --QFTSMSFANNKLRPRPASPSPSPSGTSAAIVVGVAAGAALLFALA-----WWLRRKLQ 241
Query: 270 QAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVA 329
F DV EVYLG KRF REL +AT FS +N+LGKG FG +YKG L+D TLVA
Sbjct: 242 GHFLDVPAEEDPEVYLGQFKRFSLRELLVATEKFSKRNVLGKGRFGILYKGRLADDTLVA 301
Query: 330 VKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRL 389
VKRL + GGE+QFQTEVEMIS+AVHRNLL+L GFCMTPTERLLVYPYM+NGSVA L
Sbjct: 302 VKRLNEERTKGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCL 361
Query: 390 K----GKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDF 445
+ G P LDW RKHIALG+ARGL YLH+ CD KIIH DVKAANILLD+ EAVVGDF
Sbjct: 362 RERPEGNPALDWPKRKHIALGSARGLAYLHDHCDQKIIHLDVKAANILLDEEFEAVVGDF 421
Query: 446 GLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFG 505
GLAKL+++ DSHVTTAVRGT+GHIAPEYLSTG+SSEKTDVFG+G++LLELITGQ+A +
Sbjct: 422 GLAKLMNYNDSHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQKAFDLA 481
Query: 506 KAANQKGAM-LDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPK 564
+ AN M LDWVK++ EKKLE LVD +L+ Y + E+E+++Q+ALLCTQ RPK
Sbjct: 482 RLANDDDIMLLDWVKEVLKEKKLESLVDAELEGKYVETEVEQLIQMALLCTQSSAMERPK 541
Query: 565 MSEVVRMLEGDGLAERWEASQRAD 588
MSEVVRMLEGDGLAERWE Q+ +
Sbjct: 542 MSEVVRMLEGDGLAERWEEWQKEE 565
>At5g10290 protein serine/threonine kinase-like protein
Length = 613
Score = 539 bits (1389), Expect = e-153
Identities = 307/596 (51%), Positives = 389/596 (64%), Gaps = 23/596 (3%)
Query: 36 ALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCSSDNLVISLGTPSQNLSGTLSATIGN 95
AL ++ L L +W+ + V+PC+W+ V C N V SL N SGTLS+ +G
Sbjct: 33 ALFALRISLRALPNQLSDWNQNQVNPCTWSQVICDDKNFVTSLTLSDMNFSGTLSSRVGI 92
Query: 96 LTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQYLRLNNN 155
L NL+T+ L+ N I+G IP G L L +LDL +N +G +P ++G+L+ LQ+L L+ N
Sbjct: 93 LENLKTLTLKGNGITGEIPEDFGNLTSLTSLDLEDNQLTGRIPSTIGNLKKLQFLTLSRN 152
Query: 156 SLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSILGNPLVCATGKEPNCHGITL 215
L G PESL + L L L N+LSG +P+ L F I P T NC G
Sbjct: 153 KLNGTIPESLTGLPNLLNLLLDSNSLSGQIPQSL---FEI---PKYNFTSNNLNCGGRQP 206
Query: 216 MPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGAFLWWRHKHN---QQAF 272
P + ++ D S KPKT I G+ G + ++L G FL+ + +H + F
Sbjct: 207 HPCVSAVAHSGD---SSKPKT---GIIAGVVAG-VTVVLFGILLFLFCKDRHKGYRRDVF 259
Query: 273 FDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGVLSDGTLVAVKR 332
DV + G LKRF +RELQ+AT NFS KN+LG+GGFG VYKGVL D T VAVKR
Sbjct: 260 VDVAGEVDRRIAFGQLKRFAWRELQLATDNFSEKNVLGQGGFGKVYKGVLPDNTKVAVKR 319
Query: 333 LKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLK-- 390
L D + GG+ FQ EVEMIS+AVHRNLL+L GFC T TERLLVYP+M N S+A RL+
Sbjct: 320 LTDFESPGGDAAFQREVEMISVAVHRNLLRLIGFCTTQTERLLVYPFMQNLSLAHRLREI 379
Query: 391 --GKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLA 448
G PVLDW TRK IALGAARG YLHE C+PKIIHRDVKAAN+LLD+ EAVVGDFGLA
Sbjct: 380 KAGDPVLDWETRKRIALGAARGFEYLHEHCNPKIIHRDVKAANVLLDEDFEAVVGDFGLA 439
Query: 449 KLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQRALEFGKAA 508
KL+D + ++VTT VRGT+GHIAPEYLSTG+SSE+TDVFG+GI+LLEL+TGQRA++F +
Sbjct: 440 KLVDVRRTNVTTQVRGTMGHIAPEYLSTGKSSERTDVFGYGIMLLELVTGQRAIDFSRLE 499
Query: 509 NQKGA-MLDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSE 567
+ +LD VKK+ EK+L +VDK+L Y + E+E M+QVALLCTQ P RP MSE
Sbjct: 500 EEDDVLLLDHVKKLEREKRLGAIVDKNLDGEYIKEEVEMMIQVALLCTQGSPEDRPVMSE 559
Query: 568 VVRMLEGDGLAERWEASQRADTSKCRPHESSSSDRYSDLTDDSLLLVQAMELSGPR 623
VVRMLEG+GLAERWE Q + + R HE R D +DS+ A+ELSG R
Sbjct: 560 VVRMLEGEGLAERWEEWQNVEVT--RRHEFERLQRRFDWGEDSMHNQDAIELSGGR 613
>At5g65240 receptor-like protein kinase
Length = 607
Score = 535 bits (1377), Expect = e-152
Identities = 309/617 (50%), Positives = 398/617 (64%), Gaps = 27/617 (4%)
Query: 16 LVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCSSDNLV 75
LV+SS + +SP + AL ++ L L +W+ + VDPC+W+ V C V
Sbjct: 9 LVFSSLWSSVSPDA---QGDALFALRSSLRASPEQLSDWNQNQVDPCTWSQVICDDKKHV 65
Query: 76 ISLGTPSQNLS-GTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFS 134
S+ N S GTLS+ IG LT L+T+ L+ N I G IP ++G L L +LDL +N +
Sbjct: 66 TSVTLSYMNFSSGTLSSGIGILTTLKTLTLKGNGIMGGIPESIGNLSSLTSLDLEDNHLT 125
Query: 135 GEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFS 194
+P +LG+L++LQ+L L+ N+L G P+SL +++L + L NNLSG +P+ L K
Sbjct: 126 DRIPSTLGNLKNLQFLTLSRNNLNGSIPDSLTGLSKLINILLDSNNLSGEIPQSLFKI-- 183
Query: 195 ILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLIL 254
P T +C G P ++ PSG + K I G+ G + +IL
Sbjct: 184 ----PKYNFTANNLSCGGTFPQPC------VTESSPSGDSSSRKTGIIAGVVSG-IAVIL 232
Query: 255 LGFGAFLWWRHKHN---QQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGK 311
LGF F + + KH + F DV + G L+RF +RELQ+AT FS KN+LG+
Sbjct: 233 LGFFFFFFCKDKHKGYKRDVFVDVAGEVDRRIAFGQLRRFAWRELQLATDEFSEKNVLGQ 292
Query: 312 GGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPT 371
GGFG VYKG+LSDGT VAVKRL D GG+ FQ EVEMIS+AVHRNLL+L GFC T T
Sbjct: 293 GGFGKVYKGLLSDGTKVAVKRLTDFERPGGDEAFQREVEMISVAVHRNLLRLIGFCTTQT 352
Query: 372 ERLLVYPYMSNGSVALRLK----GKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDV 427
ERLLVYP+M N SVA L+ G PVLDW RK IALGAARGL YLHE C+PKIIHRDV
Sbjct: 353 ERLLVYPFMQNLSVAYCLREIKPGDPVLDWFRRKQIALGAARGLEYLHEHCNPKIIHRDV 412
Query: 428 KAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFG 487
KAAN+LLD+ EAVVGDFGLAKL+D + ++VTT VRGT+GHIAPE +STG+SSEKTDVFG
Sbjct: 413 KAANVLLDEDFEAVVGDFGLAKLVDVRRTNVTTQVRGTMGHIAPECISTGKSSEKTDVFG 472
Query: 488 FGILLLELITGQRALEFGKAANQKGA-MLDWVKKIHLEKKLELLVDKDLKSNYDQIELEE 546
+GI+LLEL+TGQRA++F + + +LD VKK+ EK+LE +VDK L +Y + E+E
Sbjct: 473 YGIMLLELVTGQRAIDFSRLEEEDDVLLLDHVKKLEREKRLEDIVDKKLDEDYIKEEVEM 532
Query: 547 MVQVALLCTQYLPGHRPKMSEVVRMLEGDGLAERWEASQRADTSKCRPHESSSSDRYSDL 606
M+QVALLCTQ P RP MSEVVRMLEG+GLAERWE Q + + R E R D
Sbjct: 533 MIQVALLCTQAAPEERPAMSEVVRMLEGEGLAERWEEWQNLEVT--RQEEFQRLQRRFDW 590
Query: 607 TDDSLLLVQAMELSGPR 623
+DS+ A+ELSG R
Sbjct: 591 GEDSINNQDAIELSGGR 607
>At5g63710 receptor-like protein kinase
Length = 614
Score = 450 bits (1157), Expect = e-126
Identities = 263/584 (45%), Positives = 353/584 (60%), Gaps = 58/584 (9%)
Query: 11 CFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPC-SWAMVTC 69
CF S+ P + E AL+ +++ L+D L W D V PC SW+ VTC
Sbjct: 34 CFMALAFVGITSSTTQP---DIEGGALLQLRDSLNDSSNRL-KWTRDFVSPCYSWSYVTC 89
Query: 70 SSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLS 129
+ V++L S +GTLS A+ KL L TL+L
Sbjct: 90 RGQS-VVALNLASSGFTGTLSP------------------------AITKLKFLVTLELQ 124
Query: 130 NNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR-- 187
NNS SG +P SLG++ +LQ L L+ NS G P S + ++ L LDLS NNL+G +P
Sbjct: 125 NNSLSGALPDSLGNMVNLQTLNLSVNSFSGSIPASWSQLSNLKHLDLSSNNLTGSIPTQF 184
Query: 188 ILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSL 247
+F G L+C C + +P++ + D L+
Sbjct: 185 FSIPTFDFSGTQLICGKSLNQPCSSSSRLPVTSSKKKLRDIT---------------LTA 229
Query: 248 GCLCLILLGFGAFLWWRHKHNQQA----FFDVKDRHHEEVYLGNLKRFPFRELQIATHNF 303
C+ I+L GA + + H ++ FFDV ++ G LKRF RE+Q+AT +F
Sbjct: 230 SCVASIILFLGAMVMYHHHRVRRTKYDIFFDVAGEDDRKISFGQLKRFSLREIQLATDSF 289
Query: 304 SNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKL 363
+ N++G+GGFG VY+G+L D T VAVKRL D + GGE FQ E+++IS+AVH+NLL+L
Sbjct: 290 NESNLIGQGGFGKVYRGLLPDKTKVAVKRLADYFSPGGEAAFQREIQLISVAVHKNLLRL 349
Query: 364 YGFCMTPTERLLVYPYMSNGSVALRLK----GKPVLDWGTRKHIALGAARGLLYLHEQCD 419
GFC T +ER+LVYPYM N SVA RL+ G+ LDW TRK +A G+A GL YLHE C+
Sbjct: 350 IGFCTTSSERILVYPYMENLSVAYRLRDLKAGEEGLDWPTRKRVAFGSAHGLEYLHEHCN 409
Query: 420 PKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQS 479
PKIIHRD+KAANILLD+ E V+GDFGLAKL+D +HVTT VRGT+GHIAPEYL TG+S
Sbjct: 410 PKIIHRDLKAANILLDNNFEPVLGDFGLAKLVDTSLTHVTTQVRGTMGHIAPEYLCTGKS 469
Query: 480 SEKTDVFGFGILLLELITGQRALEFGKAANQKG-AMLDWVKKIHLEKKLELLVDKDLKSN 538
SEKTDVFG+GI LLEL+TGQRA++F + ++ +LD +KK+ E++L +VD +L +
Sbjct: 470 SEKTDVFGYGITLLELVTGQRAIDFSRLEEEENILLLDHIKKLLREQRLRDIVDSNL-TT 528
Query: 539 YDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEG-DGLAERW 581
YD E+E +VQVALLCTQ P RP MSEVV+ML+G GLAE+W
Sbjct: 529 YDSKEVETIVQVALLCTQGSPEDRPAMSEVVKMLQGTGGLAEKW 572
>At5g62710 receptor - like protein kinase - like
Length = 604
Score = 330 bits (845), Expect = 2e-90
Identities = 209/585 (35%), Positives = 325/585 (54%), Gaps = 33/585 (5%)
Query: 14 IFLVWSSASALLSPK-GVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCS-S 71
+F V S A+ +S + + AL+ +K +D R L+NW PCSW V+C+
Sbjct: 7 VFSVISVATLFVSCSFALTLDGFALLELKSGFNDTRNSLENWKDSDESPCSWTGVSCNPQ 66
Query: 72 DNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNN 131
D V+S+ P L G +S +IG L+ LQ + L N++ G IP+ + +L+ + L N
Sbjct: 67 DQRVVSINLPYMQLGGIISPSIGKLSRLQRLALHQNSLHGNIPNEITNCTELRAMYLRAN 126
Query: 132 SFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRI-LA 190
G +PP LG+L L L L++N+L G P S++ +T+L L+LS N SG +P I +
Sbjct: 127 FLQGGIPPDLGNLTFLTILDLSSNTLKGAIPSSISRLTRLRSLNLSTNFFSGEIPDIGVL 186
Query: 191 KSFSI---LGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSL 247
F + GN +C C P+ L + E A S PK I G+ +
Sbjct: 187 SRFGVETFTGNLDLCGRQIRKPCRSSMGFPVV--LPHAESADESDSPKRSSRLIK-GILI 243
Query: 248 GCLCLILLGF---GAFLW-W---RHKHNQQAFFDVKDRHHEEVYLGNLKRF----PFREL 296
G + + L F FLW W + + + + +VK + L F P+
Sbjct: 244 GAMSTMALAFIVIFVFLWIWMLSKKERKVKKYTEVKKQKDPSETSKKLITFHGDLPYSST 303
Query: 297 QIAT--HNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISL 354
++ + ++I+G GGFG VY+ V++D AVK++ D + G + F+ EVE++
Sbjct: 304 ELIEKLESLDEEDIVGSGGFGTVYRMVMNDLGTFAVKKI-DRSRQGSDRVFEREVEILGS 362
Query: 355 AVHRNLLKLYGFCMTPTERLLVYPYMSNGSVA----LRLKGKPVLDWGTRKHIALGAARG 410
H NL+ L G+C P+ RLL+Y Y++ GS+ R + +L+W R IALG+ARG
Sbjct: 363 VKHINLVNLRGYCRLPSSRLLIYDYLTLGSLDDLLHERAQEDGLLNWNARLKIALGSARG 422
Query: 411 LLYLHEQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIA 470
L YLH C PKI+HRD+K++NILL+D E V DFGLAKLL +D+HVTT V GT G++A
Sbjct: 423 LAYLHHDCSPKIVHRDIKSSNILLNDKLEPRVSDFGLAKLLVDEDAHVTTVVAGTFGYLA 482
Query: 471 PEYLSTGQSSEKTDVFGFGILLLELITGQRALE--FGKAANQKGAMLDWVKKIHLEKKLE 528
PEYL G+++EK+DV+ FG+LLLEL+TG+R + F K ++ W+ + E +LE
Sbjct: 483 PEYLQNGRATEKSDVYSFGVLLLELVTGKRPTDPIFVKRGLN---VVGWMNTVLKENRLE 539
Query: 529 LLVDKDLKSNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLE 573
++DK ++ D+ +E ++++A CT P +RP M++V ++LE
Sbjct: 540 DVIDKRC-TDVDEESVEALLEIAERCTDANPENRPAMNQVAQLLE 583
>At1g31420 unknown protein
Length = 592
Score = 312 bits (799), Expect = 4e-85
Identities = 197/579 (34%), Positives = 323/579 (55%), Gaps = 27/579 (4%)
Query: 9 VLCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVT 68
+L ++ S+ S +SP G +AL+ + + + W + DPC+W VT
Sbjct: 14 LLLISLLCSLSNESQAISPDG-----EALLSFRNAVTRSDSFIHQWRPEDPDPCNWNGVT 68
Query: 69 CSSDNL-VISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLD 127
C + VI+L + G L IG L +L+ ++L NN + G IP+ALG L+ +
Sbjct: 69 CDAKTKRVITLNLTYHKIMGPLPPDIGKLDHLRLLMLHNNALYGAIPTALGNCTALEEIH 128
Query: 128 LSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
L +N F+G +P +G L LQ L +++N+L G P SL + +LS ++S N L G +P
Sbjct: 129 LQSNYFTGPIPAEMGDLPGLQKLDMSSNTLSGPIPASLGQLKKLSNFNVSNNFLVGQIPS 188
Query: 188 --ILA--KSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAF 243
+L+ S +GN +C + C + P S + + SGK + I+
Sbjct: 189 DGVLSGFSKNSFIGNLNLCGKHVDVVCQDDSGNPSSHSQSGQNQKKNSGK-----LLISA 243
Query: 244 GLSLGCLCLILLG--FGAFLWWR--HKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIA 299
++G L L+ L +G FL+ + + DV ++ G+L + +++
Sbjct: 244 SATVGALLLVALMCFWGCFLYKKLGKVEIKSLAKDVGGGASIVMFHGDLP-YSSKDIIKK 302
Query: 300 THNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRN 359
+ ++I+G GGFG VYK + DG + A+KR+ N G + F+ E+E++ HR
Sbjct: 303 LEMLNEEHIIGCGGFGTVYKLAMDDGKVFALKRILKLNE-GFDRFFERELEILGSIKHRY 361
Query: 360 LLKLYGFCMTPTERLLVYPYMSNGSV--ALRLKGKPVLDWGTRKHIALGAARGLLYLHEQ 417
L+ L G+C +PT +LL+Y Y+ GS+ AL ++ LDW +R +I +GAA+GL YLH
Sbjct: 362 LVNLRGYCNSPTSKLLLYDYLPGGSLDEALHVERGEQLDWDSRVNIIIGAAKGLSYLHHD 421
Query: 418 CDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTG 477
C P+IIHRD+K++NILLD EA V DFGLAKLL+ ++SH+TT V GT G++APEY+ +G
Sbjct: 422 CSPRIIHRDIKSSNILLDGNLEARVSDFGLAKLLEDEESHITTIVAGTFGYLAPEYMQSG 481
Query: 478 QSSEKTDVFGFGILLLELITGQRALEFGKAANQKGA-MLDWVKKIHLEKKLELLVDKDLK 536
+++EKTDV+ FG+L+LE+++G+R + + +KG ++ W+K + EK+ +VD + +
Sbjct: 482 RATEKTDVYSFGVLVLEVLSGKRPTD--ASFIEKGLNVVGWLKFLISEKRPRDIVDPNCE 539
Query: 537 SNYDQIELEEMVQVALLCTQYLPGHRPKMSEVVRMLEGD 575
L+ ++ +A C P RP M VV++LE +
Sbjct: 540 -GMQMESLDALLSIATQCVSPSPEERPTMHRVVQLLESE 577
>At2g35620 putative receptor-like protein kinase
Length = 567
Score = 299 bits (765), Expect = 4e-81
Identities = 189/557 (33%), Positives = 313/557 (55%), Gaps = 41/557 (7%)
Query: 35 QALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCSSDNL-VISLGTPSQNLSGTLSATI 93
+AL+ + + GV+ W + DPC+W VTC + VI+L L G L +
Sbjct: 21 EALLSFRNGVLASDGVIGLWRPEDPDPCNWKGVTCDAKTKRVIALSLTYHKLRGPLPPEL 80
Query: 94 GNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQYLRLN 153
G L L+ ++L NN + IP++LG L+ + L NN +G +P +G+L L+ L L+
Sbjct: 81 GKLDQLRLLMLHNNALYQSIPASLGNCTALEGIYLQNNYITGTIPSEIGNLSGLKNLDLS 140
Query: 154 NNSLVGECPESLANMTQLSFLDLSYNNLSGPVP------RILAKSFSILGNPLVCATGKE 207
NN+L G P SL + +L+ ++S N L G +P R+ SF+ GN +C +
Sbjct: 141 NNNLNGAIPASLGQLKRLTKFNVSNNFLVGKIPSDGLLARLSRDSFN--GNRNLCGKQID 198
Query: 208 PNCHGITLMPMSMNLNNTEDALPSGKPKTH--KMAIAFGLSLGCLCLILLG--FGAFLWW 263
C+ + N+T P+G+ + ++ I+ ++G L L+ L +G FL+
Sbjct: 199 IVCND--------SGNSTASGSPTGQGGNNPKRLLISASATVGGLLLVALMCFWGCFLYK 250
Query: 264 R--HKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGGFGNVYKGV 321
+ ++ DV G + +++ + + ++I+G GGFG VYK
Sbjct: 251 KLGRVESKSLVIDV----------GGDLPYASKDIIKKLESLNEEHIIGCGGFGTVYKLS 300
Query: 322 LSDGTLVAVKRLKDGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLLVYPYMS 381
+ DG + A+KR+ N G + F+ E+E++ HR L+ L G+C +PT +LL+Y Y+
Sbjct: 301 MDDGNVFALKRIVKLNE-GFDRFFERELEILGSIKHRYLVNLRGYCNSPTSKLLLYDYLP 359
Query: 382 NGSV--ALRLKGKPVLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANILLDDYCE 439
GS+ AL +G+ LDW +R +I +GAA+GL YLH C P+IIHRD+K++NILLD E
Sbjct: 360 GGSLDEALHKRGEQ-LDWDSRVNIIIGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLE 418
Query: 440 AVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQ 499
A V DFGLAKLL+ ++SH+TT V GT G++APEY+ +G+++EKTDV+ FG+L+LE+++G+
Sbjct: 419 ARVSDFGLAKLLEDEESHITTIVAGTFGYLAPEYMQSGRATEKTDVYSFGVLVLEVLSGK 478
Query: 500 RALEFGKAANQKG-AMLDWVKKIHLEKKLELLVDKDLKSNYDQIELEEMVQVALLCTQYL 558
L + +KG ++ W+ + E + + +VD + ++ L+ ++ +A C
Sbjct: 479 --LPTDASFIEKGFNIVGWLNFLISENRAKEIVDLSCE-GVERESLDALLSIATKCVSSS 535
Query: 559 PGHRPKMSEVVRMLEGD 575
P RP M VV++LE +
Sbjct: 536 PDERPTMHRVVQLLESE 552
>At1g17230 putative leucine-rich receptor protein kinase
Length = 1133
Score = 297 bits (761), Expect = 1e-80
Identities = 200/535 (37%), Positives = 299/535 (55%), Gaps = 35/535 (6%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQ-TLDLSNNSFSGEVPPSLGH 143
L+G + + G+LT L + L N +S IP LGKL LQ +L++S+N+ SG +P SLG+
Sbjct: 583 LTGEIPHSFGDLTRLMELQLGGNLLSENIPVELGKLTSLQISLNISHNNLSGTIPDSLGN 642
Query: 144 LRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILA----KSFSILGNP 199
L+ L+ L LN+N L GE P S+ N+ L ++S NNL G VP S + GN
Sbjct: 643 LQMLEILYLNDNKLSGEIPASIGNLMSLLICNISNNNLVGTVPDTAVFQRMDSSNFAGNH 702
Query: 200 LVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGA 259
+C + + H L+P S + N L +G + + I + + LG
Sbjct: 703 GLCNSQRS---HCQPLVPHSDSKLNW---LINGSQRQKILTITCIVIGSVFLITFLG--- 753
Query: 260 FLWWRHKHNQQAFFDVKDRHHEEV---YLGNLKRFPFRELQIATHNFSNKNILGKGGFGN 316
L W K + AF ++D+ +V Y K F ++ L AT NFS +LG+G G
Sbjct: 754 -LCWTIKRREPAFVALEDQTKPDVMDSYYFPKKGFTYQGLVDATRNFSEDVVLGRGACGT 812
Query: 317 VYKGVLSDGTLVAVKRLKD-GNAIGGEIQFQTEVEMISLAVHRNLLKLYGFCMTPTERLL 375
VYK +S G ++AVK+L G + F+ E+ + HRN++KLYGFC LL
Sbjct: 813 VYKAEMSGGEVIAVKKLNSRGEGASSDNSFRAEISTLGKIRHRNIVKLYGFCYHQNSNLL 872
Query: 376 VYPYMSNGSVALRL-KGKP--VLDWGTRKHIALGAARGLLYLHEQCDPKIIHRDVKAANI 432
+Y YMS GS+ +L +G+ +LDW R IALGAA GL YLH C P+I+HRD+K+ NI
Sbjct: 873 LYEYMSKGSLGEQLQRGEKNCLLDWNARYRIALGAAEGLCYLHHDCRPQIVHRDIKSNNI 932
Query: 433 LLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILL 492
LLD+ +A VGDFGLAKL+D S +AV G+ G+IAPEY T + +EK D++ FG++L
Sbjct: 933 LLDERFQAHVGDFGLAKLIDLSYSKSMSAVAGSYGYIAPEYAYTMKVTEKCDIYSFGVVL 992
Query: 493 LELITGQRALEFGKAANQKGAMLDWVKKI--HLEKKLELLVDKDLKSNYDQI--ELEEMV 548
LELITG+ ++ Q G +++WV++ ++ +E+ D L +N + E+ ++
Sbjct: 993 LELITGKPPVQ---PLEQGGDLVNWVRRSIRNMIPTIEMF-DARLDTNDKRTVHEMSLVL 1048
Query: 549 QVALLCTQYLPGHRPKMSEVVRML-EGDGLAERWEASQRADTSKCRPHESSSSDR 602
++AL CT P RP M EVV M+ E G + +S ++T P E ++S +
Sbjct: 1049 KIALFCTSNSPASRPTMREVVAMITEARGSSSLSSSSITSET----PLEEANSSK 1099
Score = 108 bits (269), Expect = 1e-23
Identities = 64/182 (35%), Positives = 95/182 (52%), Gaps = 1/182 (0%)
Query: 10 LCFAIFLVWSSASALLSPKGVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTC 69
+CF ++ S S +L + +N E + L+ K L+D G L +W+ +PC+W + C
Sbjct: 5 ICFLAIVILCSFSFILV-RSLNEEGRVLLEFKAFLNDSNGYLASWNQLDSNPCNWTGIAC 63
Query: 70 SSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLS 129
+ V S+ NLSGTLS I L L+ + + N ISGPIP L L+ LDL
Sbjct: 64 THLRTVTSVDLNGMNLSGTLSPLICKLHGLRKLNVSTNFISGPIPQDLSLCRSLEVLDLC 123
Query: 130 NNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
N F G +P L + +L+ L L N L G P + N++ L L + NNL+G +P +
Sbjct: 124 TNRFHGVIPIQLTMIITLKKLYLCENYLFGSIPRQIGNLSSLQELVIYSNNLTGVIPPSM 183
Query: 190 AK 191
AK
Sbjct: 184 AK 185
Score = 83.2 bits (204), Expect = 4e-16
Identities = 58/183 (31%), Positives = 91/183 (49%), Gaps = 7/183 (3%)
Query: 68 TCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLD 127
TC S + L L+G+L + NL NL + L N +SG I + LGKL L+ L
Sbjct: 449 TCKS---LTKLMLGDNQLTGSLPIELFNLQNLTALELHQNWLSGNISADLGKLKNLERLR 505
Query: 128 LSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
L+NN+F+GE+PP +G+L + +++N L G P+ L + + LDLS N SG + +
Sbjct: 506 LANNNFTGEIPPEIGNLTKIVGFNISSNQLTGHIPKELGSCVTIQRLDLSGNKFSGYIAQ 565
Query: 188 ILAK--SFSILGNPLVCATGKEPNCHG--ITLMPMSMNLNNTEDALPSGKPKTHKMAIAF 243
L + IL TG+ P+ G LM + + N + +P K + I+
Sbjct: 566 ELGQLVYLEILRLSDNRLTGEIPHSFGDLTRLMELQLGGNLLSENIPVELGKLTSLQISL 625
Query: 244 GLS 246
+S
Sbjct: 626 NIS 628
Score = 77.8 bits (190), Expect = 2e-14
Identities = 38/110 (34%), Positives = 60/110 (54%)
Query: 78 LGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEV 137
LG L G+L + L NL ++L N +SG IP ++G + +L+ L L N F+G +
Sbjct: 216 LGLAENLLEGSLPKQLEKLQNLTDLILWQNRLSGEIPPSVGNISRLEVLALHENYFTGSI 275
Query: 138 PPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
P +G L ++ L L N L GE P + N+ + +D S N L+G +P+
Sbjct: 276 PREIGKLTKMKRLYLYTNQLTGEIPREIGNLIDAAEIDFSENQLTGFIPK 325
Score = 77.4 bits (189), Expect = 2e-14
Identities = 41/122 (33%), Positives = 69/122 (55%), Gaps = 2/122 (1%)
Query: 74 LVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSF 133
+++SLG S LSG + + +L ++L +N ++G +P L L L L+L N
Sbjct: 430 ILLSLG--SNKLSGNIPRDLKTCKSLTKLMLGDNQLTGSLPIELFNLQNLTALELHQNWL 487
Query: 134 SGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSF 193
SG + LG L++L+ LRL NN+ GE P + N+T++ ++S N L+G +P+ L
Sbjct: 488 SGNISADLGKLKNLERLRLANNNFTGEIPPEIGNLTKIVGFNISSNQLTGHIPKELGSCV 547
Query: 194 SI 195
+I
Sbjct: 548 TI 549
Score = 77.4 bits (189), Expect = 2e-14
Identities = 38/104 (36%), Positives = 58/104 (55%)
Query: 86 SGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLR 145
+G++ IG LT ++ + L N ++G IP +G L +D S N +G +P GH+
Sbjct: 272 TGSIPREIGKLTKMKRLYLYTNQLTGEIPREIGNLIDAAEIDFSENQLTGFIPKEFGHIL 331
Query: 146 SLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
+L+ L L N L+G P L +T L LDLS N L+G +P+ L
Sbjct: 332 NLKLLHLFENILLGPIPRELGELTLLEKLDLSINRLNGTIPQEL 375
Score = 75.1 bits (183), Expect = 1e-13
Identities = 37/107 (34%), Positives = 59/107 (54%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
LSG + ++GN++ L+ + L N +G IP +GKL K++ L L N +GE+P +G+L
Sbjct: 247 LSGEIPPSVGNISRLEVLALHENYFTGSIPREIGKLTKMKRLYLYTNQLTGEIPREIGNL 306
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAK 191
+ + N L G P+ ++ L L L N L GP+PR L +
Sbjct: 307 IDAAEIDFSENQLTGFIPKEFGHILNLKLLHLFENILLGPIPRELGE 353
Score = 74.3 bits (181), Expect = 2e-13
Identities = 41/102 (40%), Positives = 60/102 (58%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L+G + G++ NL+ + L N + GPIP LG+L L+ LDLS N +G +P L L
Sbjct: 319 LTGFIPKEFGHILNLKLLHLFENILLGPIPRELGELTLLEKLDLSINRLNGTIPQELQFL 378
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
L L+L +N L G+ P + + S LD+S N+LSGP+P
Sbjct: 379 PYLVDLQLFDNQLEGKIPPLIGFYSNFSVLDMSANSLSGPIP 420
Score = 73.9 bits (180), Expect = 2e-13
Identities = 38/102 (37%), Positives = 60/102 (58%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L G++ IGNL++LQ +++ +NN++G IP ++ KL +L+ + N FSG +P +
Sbjct: 151 LFGSIPRQIGNLSSLQELVIYSNNLTGVIPPSMAKLRQLRIIRAGRNGFSGVIPSEISGC 210
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
SL+ L L N L G P+ L + L+ L L N LSG +P
Sbjct: 211 ESLKVLGLAENLLEGSLPKQLEKLQNLTDLILWQNRLSGEIP 252
Score = 70.1 bits (170), Expect = 4e-12
Identities = 36/102 (35%), Positives = 55/102 (53%)
Query: 86 SGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHLR 145
SG + + I +L+ + L N + G +P L KL L L L N SGE+PPS+G++
Sbjct: 200 SGVIPSEISGCESLKVLGLAENLLEGSLPKQLEKLQNLTDLILWQNRLSGEIPPSVGNIS 259
Query: 146 SLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
L+ L L+ N G P + +T++ L L N L+G +PR
Sbjct: 260 RLEVLALHENYFTGSIPREIGKLTKMKRLYLYTNQLTGEIPR 301
Score = 67.8 bits (164), Expect = 2e-11
Identities = 38/110 (34%), Positives = 57/110 (51%)
Query: 82 SQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSL 141
S NL+G + ++ L L+ + N SG IPS + L+ L L+ N G +P L
Sbjct: 172 SNNLTGVIPPSMAKLRQLRIIRAGRNGFSGVIPSEISGCESLKVLGLAENLLEGSLPKQL 231
Query: 142 GHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAK 191
L++L L L N L GE P S+ N+++L L L N +G +PR + K
Sbjct: 232 EKLQNLTDLILWQNRLSGEIPPSVGNISRLEVLALHENYFTGSIPREIGK 281
Score = 66.2 bits (160), Expect = 5e-11
Identities = 38/105 (36%), Positives = 52/105 (49%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L G + +G LT L+ + L N ++G IP L LP L L L +N G++PP +G
Sbjct: 343 LLGPIPRELGELTLLEKLDLSINRLNGTIPQELQFLPYLVDLQLFDNQLEGKIPPLIGFY 402
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
+ L ++ NSL G P L L L N LSG +PR L
Sbjct: 403 SNFSVLDMSANSLSGPIPAHFCRFQTLILLSLGSNKLSGNIPRDL 447
Score = 65.9 bits (159), Expect = 7e-11
Identities = 39/117 (33%), Positives = 57/117 (48%)
Query: 75 VISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFS 134
++ L L G + IG +N + + N++SGPIP+ + L L L +N S
Sbjct: 381 LVDLQLFDNQLEGKIPPLIGFYSNFSVLDMSANSLSGPIPAHFCRFQTLILLSLGSNKLS 440
Query: 135 GEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAK 191
G +P L +SL L L +N L G P L N+ L+ L+L N LSG + L K
Sbjct: 441 GNIPRDLKTCKSLTKLMLGDNQLTGSLPIELFNLQNLTALELHQNWLSGNISADLGK 497
>At5g63930 receptor-like protein kinase
Length = 1102
Score = 293 bits (750), Expect = 2e-79
Identities = 196/520 (37%), Positives = 282/520 (53%), Gaps = 36/520 (6%)
Query: 84 NLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGH 143
N SGTL + +G+L L+ + L NNN+SG IP ALG L +L L + N F+G +P LG
Sbjct: 564 NFSGTLPSEVGSLYQLELLKLSNNNLSGTIPVALGNLSRLTELQMGGNLFNGSIPRELGS 623
Query: 144 LRSLQY-LRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSILGNPLV- 201
L LQ L L+ N L GE P L+N+ L FL L+ NNLSG +P A S+LG
Sbjct: 624 LTGLQIALNLSYNKLTGEIPPELSNLVMLEFLLLNNNNLSGEIPSSFANLSSLLGYNFSY 683
Query: 202 -CATGKEPNCHGITLMPMSMN-------LNN---TEDALPS---GKP---KTHKMAIAFG 244
TG P I++ N LN T+ PS GKP ++ K+
Sbjct: 684 NSLTGPIPLLRNISMSSFIGNEGLCGPPLNQCIQTQPFAPSQSTGKPGGMRSSKIIAITA 743
Query: 245 LSLGCLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEE----VYLGNLKRFPFRELQIAT 300
+G + L+L+ +L R + +D E +Y + F F++L AT
Sbjct: 744 AVIGGVSLMLIALIVYLMRRPVRTVAS--SAQDGQPSEMSLDIYFPPKEGFTFQDLVAAT 801
Query: 301 HNFSNKNILGKGGFGNVYKGVLSDGTLVAVKRL----KDGNAIGGEIQFQTEVEMISLAV 356
NF ++G+G G VYK VL G +AVK+L + GN + F+ E+ +
Sbjct: 802 DNFDESFVVGRGACGTVYKAVLPAGYTLAVKKLASNHEGGNNNNVDNSFRAEILTLGNIR 861
Query: 357 HRNLLKLYGFCMTPTERLLVYPYMSNGSVALRLKGKPV-LDWGTRKHIALGAARGLLYLH 415
HRN++KL+GFC LL+Y YM GS+ L LDW R IALGAA+GL YLH
Sbjct: 862 HRNIVKLHGFCNHQGSNLLLYEYMPKGSLGEILHDPSCNLDWSKRFKIALGAAQGLAYLH 921
Query: 416 EQCDPKIIHRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLS 475
C P+I HRD+K+ NILLDD EA VGDFGLAK++D S +A+ G+ G+IAPEY
Sbjct: 922 HDCKPRIFHRDIKSNNILLDDKFEAHVGDFGLAKVIDMPHSKSMSAIAGSYGYIAPEYAY 981
Query: 476 TGQSSEKTDVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKK-IHLEKKLELLVDKD 534
T + +EK+D++ +G++LLEL+TG+ ++ +Q G +++WV+ I + ++D
Sbjct: 982 TMKVTEKSDIYSYGVVLLELLTGKAPVQ---PIDQGGDVVNWVRSYIRRDALSSGVLDAR 1038
Query: 535 LKSNYDQI--ELEEMVQVALLCTQYLPGHRPKMSEVVRML 572
L ++I + ++++ALLCT P RP M +VV ML
Sbjct: 1039 LTLEDERIVSHMLTVLKIALLCTSVSPVARPSMRQVVLML 1078
Score = 117 bits (293), Expect = 2e-26
Identities = 71/179 (39%), Positives = 97/179 (53%), Gaps = 4/179 (2%)
Query: 13 AIFLVWSSASALLSPK-GVNFEVQALMGIKELLHDPRGVLDNWDGDAVDPCSWAMVTCS- 70
A+F + L+S G+N E Q L+ IK D + L NW+ + PC W V CS
Sbjct: 9 AVFFISLLLILLISETTGLNLEGQYLLEIKSKFVDAKQNLRNWNSNDSVPCGWTGVMCSN 68
Query: 71 --SDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDL 128
SD V+SL S LSG LS +IG L +L+ + L N +SG IP +G L+ L L
Sbjct: 69 YSSDPEVLSLNLSSMVLSGKLSPSIGGLVHLKQLDLSYNGLSGKIPKEIGNCSSLEILKL 128
Query: 129 SNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
+NN F GE+P +G L SL+ L + NN + G P + N+ LS L NN+SG +PR
Sbjct: 129 NNNQFDGEIPVEIGKLVSLENLIIYNNRISGSLPVEIGNLLSLSQLVTYSNNISGQLPR 187
Score = 78.6 bits (192), Expect = 1e-14
Identities = 40/110 (36%), Positives = 61/110 (55%)
Query: 77 SLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGE 136
+L L G + +G+L +L+ + L N ++G IP +G L +D S N+ +GE
Sbjct: 269 TLALYKNQLVGPIPKELGDLQSLEFLYLYRNGLNGTIPREIGNLSYAIEIDFSENALTGE 328
Query: 137 VPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
+P LG++ L+ L L N L G P L+ + LS LDLS N L+GP+P
Sbjct: 329 IPLELGNIEGLELLYLFENQLTGTIPVELSTLKNLSKLDLSINALTGPIP 378
Score = 78.2 bits (191), Expect = 1e-14
Identities = 45/112 (40%), Positives = 59/112 (52%)
Query: 78 LGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEV 137
L T S N+SG L +IGNL L + N ISG +PS +G L L L+ N SGE+
Sbjct: 174 LVTYSNNISGQLPRSIGNLKRLTSFRAGQNMISGSLPSEIGGCESLVMLGLAQNQLSGEL 233
Query: 138 PPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
P +G L+ L + L N G P ++N T L L L N L GP+P+ L
Sbjct: 234 PKEIGMLKKLSQVILWENEFSGFIPREISNCTSLETLALYKNQLVGPIPKEL 285
Score = 77.0 bits (188), Expect = 3e-14
Identities = 44/126 (34%), Positives = 69/126 (53%), Gaps = 4/126 (3%)
Query: 70 SSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLS 129
S +NL+I + +SG+L IGNL +L ++ +NNISG +P ++G L +L +
Sbjct: 146 SLENLIIY----NNRISGSLPVEIGNLLSLSQLVTYSNNISGQLPRSIGNLKRLTSFRAG 201
Query: 130 NNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
N SG +P +G SL L L N L GE P+ + + +LS + L N SG +PR +
Sbjct: 202 QNMISGSLPSEIGGCESLVMLGLAQNQLSGELPKEIGMLKKLSQVILWENEFSGFIPREI 261
Query: 190 AKSFSI 195
+ S+
Sbjct: 262 SNCTSL 267
Score = 77.0 bits (188), Expect = 3e-14
Identities = 42/112 (37%), Positives = 57/112 (50%)
Query: 75 VISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFS 134
++ LG LSG L IG L L V+L N SG IP + L+TL L N
Sbjct: 219 LVMLGLAQNQLSGELPKEIGMLKKLSQVILWENEFSGFIPREISNCTSLETLALYKNQLV 278
Query: 135 GEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
G +P LG L+SL++L L N L G P + N++ +D S N L+G +P
Sbjct: 279 GPIPKELGDLQSLEFLYLYRNGLNGTIPREIGNLSYAIEIDFSENALTGEIP 330
Score = 75.9 bits (185), Expect = 6e-14
Identities = 41/116 (35%), Positives = 64/116 (54%), Gaps = 2/116 (1%)
Query: 71 SDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSN 130
S+ ++++LGT NLSG + I L + L NN+ G PS L K + ++L
Sbjct: 433 SNMIILNLGT--NNLSGNIPTGITTCKTLVQLRLARNNLVGRFPSNLCKQVNVTAIELGQ 490
Query: 131 NSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
N F G +P +G+ +LQ L+L +N GE P + ++QL L++S N L+G VP
Sbjct: 491 NRFRGSIPREVGNCSALQRLQLADNGFTGELPREIGMLSQLGTLNISSNKLTGEVP 546
Score = 75.5 bits (184), Expect = 8e-14
Identities = 42/119 (35%), Positives = 66/119 (55%), Gaps = 3/119 (2%)
Query: 69 CSSDNLV-ISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLD 127
C N+ I LG G++ +GN + LQ + L +N +G +P +G L +L TL+
Sbjct: 478 CKQVNVTAIELG--QNRFRGSIPREVGNCSALQRLQLADNGFTGELPREIGMLSQLGTLN 535
Query: 128 LSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
+S+N +GEVP + + + LQ L + N+ G P + ++ QL L LS NNLSG +P
Sbjct: 536 ISSNKLTGEVPSEIFNCKMLQRLDMCCNNFSGTLPSEVGSLYQLELLKLSNNNLSGTIP 594
Score = 75.1 bits (183), Expect = 1e-13
Identities = 40/103 (38%), Positives = 58/103 (55%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
+SG+L + IG +L + L N +SG +P +G L KL + L N FSG +P + +
Sbjct: 205 ISGSLPSEIGGCESLVMLGLAQNQLSGELPKEIGMLKKLSQVILWENEFSGFIPREISNC 264
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
SL+ L L N LVG P+ L ++ L FL L N L+G +PR
Sbjct: 265 TSLETLALYKNQLVGPIPKELGDLQSLEFLYLYRNGLNGTIPR 307
Score = 75.1 bits (183), Expect = 1e-13
Identities = 56/190 (29%), Positives = 84/190 (43%), Gaps = 17/190 (8%)
Query: 66 MVTCSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQT 125
+ TC + ++ L NL G + + N+ + L N G IP +G LQ
Sbjct: 453 ITTCKT---LVQLRLARNNLVGRFPSNLCKQVNVTAIELGQNRFRGSIPREVGNCSALQR 509
Query: 126 LDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPV 185
L L++N F+GE+P +G L L L +++N L GE P + N L LD+ NN SG +
Sbjct: 510 LQLADNGFTGELPREIGMLSQLGTLNISSNKLTGEVPSEIFNCKMLQRLDMCCNNFSGTL 569
Query: 186 P---------RILAKSFSILGNPLVCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKT 236
P +L S + L + A G N +T + M NL N ++P
Sbjct: 570 PSEVGSLYQLELLKLSNNNLSGTIPVALG---NLSRLTELQMGGNLFN--GSIPRELGSL 624
Query: 237 HKMAIAFGLS 246
+ IA LS
Sbjct: 625 TGLQIALNLS 634
Score = 72.8 bits (177), Expect = 5e-13
Identities = 41/105 (39%), Positives = 58/105 (55%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L+G + +GN+ L+ + L N ++G IP L L L LDLS N+ +G +P +L
Sbjct: 325 LTGEIPLELGNIEGLELLYLFENQLTGTIPVELSTLKNLSKLDLSINALTGPIPLGFQYL 384
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRIL 189
R L L+L NSL G P L + L LD+S N+LSG +P L
Sbjct: 385 RGLFMLQLFQNSLSGTIPPKLGWYSDLWVLDMSDNHLSGRIPSYL 429
Score = 68.6 bits (166), Expect = 1e-11
Identities = 39/102 (38%), Positives = 53/102 (51%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L+GT+ + L NL + L N ++GPIP L L L L NS SG +PP LG
Sbjct: 349 LTGTIPVELSTLKNLSKLDLSINALTGPIPLGFQYLRGLFMLQLFQNSLSGTIPPKLGWY 408
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
L L +++N L G P L + + L+L NNLSG +P
Sbjct: 409 SDLWVLDMSDNHLSGRIPSYLCLHSNMIILNLGTNNLSGNIP 450
Score = 65.9 bits (159), Expect = 7e-11
Identities = 34/104 (32%), Positives = 58/104 (55%)
Query: 84 NLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGH 143
+LSGT+ +G ++L + + +N++SG IPS L + L+L N+ SG +P +
Sbjct: 396 SLSGTIPPKLGWYSDLWVLDMSDNHLSGRIPSYLCLHSNMIILNLGTNNLSGNIPTGITT 455
Query: 144 LRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPR 187
++L LRL N+LVG P +L ++ ++L N G +PR
Sbjct: 456 CKTLVQLRLARNNLVGRFPSNLCKQVNVTAIELGQNRFRGSIPR 499
Score = 65.5 bits (158), Expect = 9e-11
Identities = 45/152 (29%), Positives = 73/152 (47%), Gaps = 4/152 (2%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L+GT+ IGNL+ + N ++G IP LG + L+ L L N +G +P L L
Sbjct: 301 LNGTIPREIGNLSYAIEIDFSENALTGEIPLELGNIEGLELLYLFENQLTGTIPVELSTL 360
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILA--KSFSILGNPLVC 202
++L L L+ N+L G P + L L L N+LSG +P L +L
Sbjct: 361 KNLSKLDLSINALTGPIPLGFQYLRGLFMLQLFQNSLSGTIPPKLGWYSDLWVLDMSDNH 420
Query: 203 ATGKEPN--CHGITLMPMSMNLNNTEDALPSG 232
+G+ P+ C ++ +++ NN +P+G
Sbjct: 421 LSGRIPSYLCLHSNMIILNLGTNNLSGNIPTG 452
Score = 56.6 bits (135), Expect = 4e-08
Identities = 37/111 (33%), Positives = 52/111 (46%)
Query: 85 LSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGHL 144
L+G + L L + L N++SG IP LG L LD+S+N SG +P L
Sbjct: 373 LTGPIPLGFQYLRGLFMLQLFQNSLSGTIPPKLGWYSDLWVLDMSDNHLSGRIPSYLCLH 432
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKSFSI 195
++ L L N+L G P + L L L+ NNL G P L K ++
Sbjct: 433 SNMIILNLGTNNLSGNIPTGITTCKTLVQLRLARNNLVGRFPSNLCKQVNV 483
>At2g33170 putative receptor-like protein kinase
Length = 1124
Score = 290 bits (743), Expect = 1e-78
Identities = 190/517 (36%), Positives = 276/517 (52%), Gaps = 41/517 (7%)
Query: 86 SGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQ-TLDLSNNSFSGEVPPSLGHL 144
SG + TIGNLT+L + + N SG IP LG L LQ ++LS N FSGE+PP +G+L
Sbjct: 602 SGNIPFTIGNLTHLTELQMGGNLFSGSIPPQLGLLSSLQIAMNLSYNDFSGEIPPEIGNL 661
Query: 145 RSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKS----FSILGNPL 200
L YL LNNN L GE P + N++ L + SYNNL+G +P S LGN
Sbjct: 662 HLLMYLSLNNNHLSGEIPTTFENLSSLLGCNFSYNNLTGQLPHTQIFQNMTLTSFLGNKG 721
Query: 201 VCATGKEPNCHGITLMPMSMNLNNTEDALPSGKPKTHKMAIAFGLSLGCLCLILLGFGAF 260
+C G +C P + + +L +G + ++ I +G + L+L+
Sbjct: 722 LCG-GHLRSCD-----PSHSSWPHIS-SLKAGSARRGRIIIIVSSVIGGISLLLIAIVVH 774
Query: 261 LWWR-------HKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNILGKGG 313
+ H+++ FF D +Y +RF +++ AT F + I+G+G
Sbjct: 775 FLRNPVEPTAPYVHDKEPFFQESD-----IYFVPKERFTVKDILEATKGFHDSYIVGRGA 829
Query: 314 FGNVYKGVLSDGTLVAVKRLK------DGNAIGGEIQFQTEVEMISLAVHRNLLKLYGFC 367
G VYK V+ G +AVK+L+ + N+ + F+ E+ + HRN+++LY FC
Sbjct: 830 CGTVYKAVMPSGKTIAVKKLESNREGNNNNSNNTDNSFRAEILTLGKIRHRNIVRLYSFC 889
Query: 368 --MTPTERLLVYPYMSNGSVALRLKG--KPVLDWGTRKHIALGAARGLLYLHEQCDPKII 423
LL+Y YMS GS+ L G +DW TR IALGAA GL YLH C P+II
Sbjct: 890 YHQGSNSNLLLYEYMSRGSLGELLHGGKSHSMDWPTRFAIALGAAEGLAYLHHDCKPRII 949
Query: 424 HRDVKAANILLDDYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKT 483
HRD+K+ NIL+D+ EA VGDFGLAK++D S +AV G+ G+IAPEY T + +EK
Sbjct: 950 HRDIKSNNILIDENFEAHVGDFGLAKVIDMPLSKSVSAVAGSYGYIAPEYAYTMKVTEKC 1009
Query: 484 DVFGFGILLLELITGQRALEFGKAANQKGAMLDWVKK-IHLEKKLELLVDKDLKSNYDQI 542
D++ FG++LLEL+TG+ ++ Q G + W + I ++D L D +
Sbjct: 1010 DIYSFGVVLLELLTGKAPVQ---PLEQGGDLATWTRNHIRDHSLTSEILDPYLTKVEDDV 1066
Query: 543 ELEEMV---QVALLCTQYLPGHRPKMSEVVRMLEGDG 576
L M+ ++A+LCT+ P RP M EVV ML G
Sbjct: 1067 ILNHMITVTKIAVLCTKSSPSDRPTMREVVLMLIESG 1103
Score = 111 bits (277), Expect = 1e-24
Identities = 70/192 (36%), Positives = 96/192 (49%), Gaps = 16/192 (8%)
Query: 7 VSVLCFAIFLVWSSASALLSPKGVNFEVQALMGIKEL-LHDPRGVLDNWDGDAVDPCSWA 65
V VL LVW+S S +N + Q L+ +K D L NW+G PC+W
Sbjct: 17 VGVLFLLTLLVWTSES-------LNSDGQFLLELKNRGFQDSLNRLHNWNGIDETPCNWI 69
Query: 66 MVTCSSDN--------LVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSAL 117
V CSS +V SL S NLSG +S +IG L NL + L N ++G IP +
Sbjct: 70 GVNCSSQGSSSSSNSLVVTSLDLSSMNLSGIVSPSIGGLVNLVYLNLAYNALTGDIPREI 129
Query: 118 GKLPKLQTLDLSNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLS 177
G KL+ + L+NN F G +P + L L+ + NN L G PE + ++ L L
Sbjct: 130 GNCSKLEVMFLNNNQFGGSIPVEINKLSQLRSFNICNNKLSGPLPEEIGDLYNLEELVAY 189
Query: 178 YNNLSGPVPRIL 189
NNL+GP+PR L
Sbjct: 190 TNNLTGPLPRSL 201
Score = 85.9 bits (211), Expect = 6e-17
Identities = 43/110 (39%), Positives = 64/110 (58%)
Query: 77 SLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGE 136
+L +L G + + IGN+ +L+ + L N ++G IP LGKL K+ +D S N SGE
Sbjct: 281 TLALYGNSLVGPIPSEIGNMKSLKKLYLYQNQLNGTIPKELGKLSKVMEIDFSENLLSGE 340
Query: 137 VPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
+P L + L+ L L N L G P L+ + L+ LDLS N+L+GP+P
Sbjct: 341 IPVELSKISELRLLYLFQNKLTGIIPNELSKLRNLAKLDLSINSLTGPIP 390
Score = 84.3 bits (207), Expect = 2e-16
Identities = 85/281 (30%), Positives = 125/281 (44%), Gaps = 34/281 (12%)
Query: 78 LGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEV 137
LG +SG L IG L LQ V+L N SG IP +G L L+TL L NS G +
Sbjct: 234 LGLAQNFISGELPKEIGMLVKLQEVILWQNKFSGFIPKDIGNLTSLETLALYGNSLVGPI 293
Query: 138 PPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAK-----S 192
P +G+++SL+ L L N L G P+ L ++++ +D S N LSG +P L+K
Sbjct: 294 PSEIGNMKSLKKLYLYQNQLNGTIPKELGKLSKVMEIDFSENLLSGEIPVELSKISELRL 353
Query: 193 FSILGNPLVCATGKEPN--CHGITLMPMSMNLNNTEDALPSGKPKTHKM--AIAFGLSLG 248
+ N L TG PN L + +++N+ +P G M F SL
Sbjct: 354 LYLFQNKL---TGIIPNELSKLRNLAKLDLSINSLTGPIPPGFQNLTSMRQLQLFHNSLS 410
Query: 249 CLCLILLGFGAFLWWRHKHNQQAFFDVKDRHHEEVYLGNLKRFPFRELQIATHNFSNKNI 308
+ LG + LW V D E G + F ++ + N + I
Sbjct: 411 GVIPQGLGLYSPLW------------VVD-FSENQLSGKIPPFICQQSNLILLNLGSNRI 457
Query: 309 LGKGGFGNVYKGVLSDGTLVAVKRLKDGNAIGGEIQFQTEV 349
FGN+ GVL +L+ ++ + GN + G QF TE+
Sbjct: 458 -----FGNIPPGVLRCKSLLQLRVV--GNRLTG--QFPTEL 489
Score = 84.3 bits (207), Expect = 2e-16
Identities = 47/124 (37%), Positives = 66/124 (52%)
Query: 73 NLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNS 132
N + + + SG + IG NL+ + L N ISG +P +G L KLQ + L N
Sbjct: 205 NKLTTFRAGQNDFSGNIPTEIGKCLNLKLLGLAQNFISGELPKEIGMLVKLQEVILWQNK 264
Query: 133 FSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILAKS 192
FSG +P +G+L SL+ L L NSLVG P + NM L L L N L+G +P+ L K
Sbjct: 265 FSGFIPKDIGNLTSLETLALYGNSLVGPIPSEIGNMKSLKKLYLYQNQLNGTIPKELGKL 324
Query: 193 FSIL 196
++
Sbjct: 325 SKVM 328
Score = 80.1 bits (196), Expect = 3e-15
Identities = 43/125 (34%), Positives = 69/125 (54%), Gaps = 5/125 (4%)
Query: 82 SQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSL 141
+ LSG L IG+L NL+ ++ NN++GP+P +LG L KL T N FSG +P +
Sbjct: 166 NNKLSGPLPEEIGDLYNLEELVAYTNNLTGPLPRSLGNLNKLTTFRAGQNDFSGNIPTEI 225
Query: 142 GHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRILA-----KSFSIL 196
G +L+ L L N + GE P+ + + +L + L N SG +P+ + ++ ++
Sbjct: 226 GKCLNLKLLGLAQNFISGELPKEIGMLVKLQEVILWQNKFSGFIPKDIGNLTSLETLALY 285
Query: 197 GNPLV 201
GN LV
Sbjct: 286 GNSLV 290
Score = 74.7 bits (182), Expect = 1e-13
Identities = 39/112 (34%), Positives = 56/112 (49%)
Query: 75 VISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFS 134
++ L L+G + L NL + L N SGP+P +G KLQ L L+ N FS
Sbjct: 471 LLQLRVVGNRLTGQFPTELCKLVNLSAIELDQNRFSGPLPPEIGTCQKLQRLHLAANQFS 530
Query: 135 GEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
+P + L +L +++NSL G P +AN L LDLS N+ G +P
Sbjct: 531 SNLPNEISKLSNLVTFNVSSNSLTGPIPSEIANCKMLQRLDLSRNSFIGSLP 582
Score = 69.3 bits (168), Expect = 6e-12
Identities = 36/122 (29%), Positives = 62/122 (50%), Gaps = 1/122 (0%)
Query: 69 CSSDNLVISLGTPSQNLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDL 128
C NL++ L S + G + + +L + + N ++G P+ L KL L ++L
Sbjct: 442 CQQSNLIL-LNLGSNRIFGNIPPGVLRCKSLLQLRVVGNRLTGQFPTELCKLVNLSAIEL 500
Query: 129 SNNSFSGEVPPSLGHLRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVPRI 188
N FSG +PP +G + LQ L L N P ++ ++ L ++S N+L+GP+P
Sbjct: 501 DQNRFSGPLPPEIGTCQKLQRLHLAANQFSSNLPNEISKLSNLVTFNVSSNSLTGPIPSE 560
Query: 189 LA 190
+A
Sbjct: 561 IA 562
Score = 65.1 bits (157), Expect = 1e-10
Identities = 35/103 (33%), Positives = 53/103 (50%)
Query: 84 NLSGTLSATIGNLTNLQTVLLQNNNISGPIPSALGKLPKLQTLDLSNNSFSGEVPPSLGH 143
+LSG + +G + L V N +SG IP + + L L+L +N G +PP +
Sbjct: 408 SLSGVIPQGLGLYSPLWVVDFSENQLSGKIPPFICQQSNLILLNLGSNRIFGNIPPGVLR 467
Query: 144 LRSLQYLRLNNNSLVGECPESLANMTQLSFLDLSYNNLSGPVP 186
+SL LR+ N L G+ P L + LS ++L N SGP+P
Sbjct: 468 CKSLLQLRVVGNRLTGQFPTELCKLVNLSAIELDQNRFSGPLP 510
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,265,454
Number of Sequences: 26719
Number of extensions: 633746
Number of successful extensions: 10886
Number of sequences better than 10.0: 1189
Number of HSP's better than 10.0 without gapping: 996
Number of HSP's successfully gapped in prelim test: 193
Number of HSP's that attempted gapping in prelim test: 1602
Number of HSP's gapped (non-prelim): 3730
length of query: 623
length of database: 11,318,596
effective HSP length: 105
effective length of query: 518
effective length of database: 8,513,101
effective search space: 4409786318
effective search space used: 4409786318
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0215a.1


